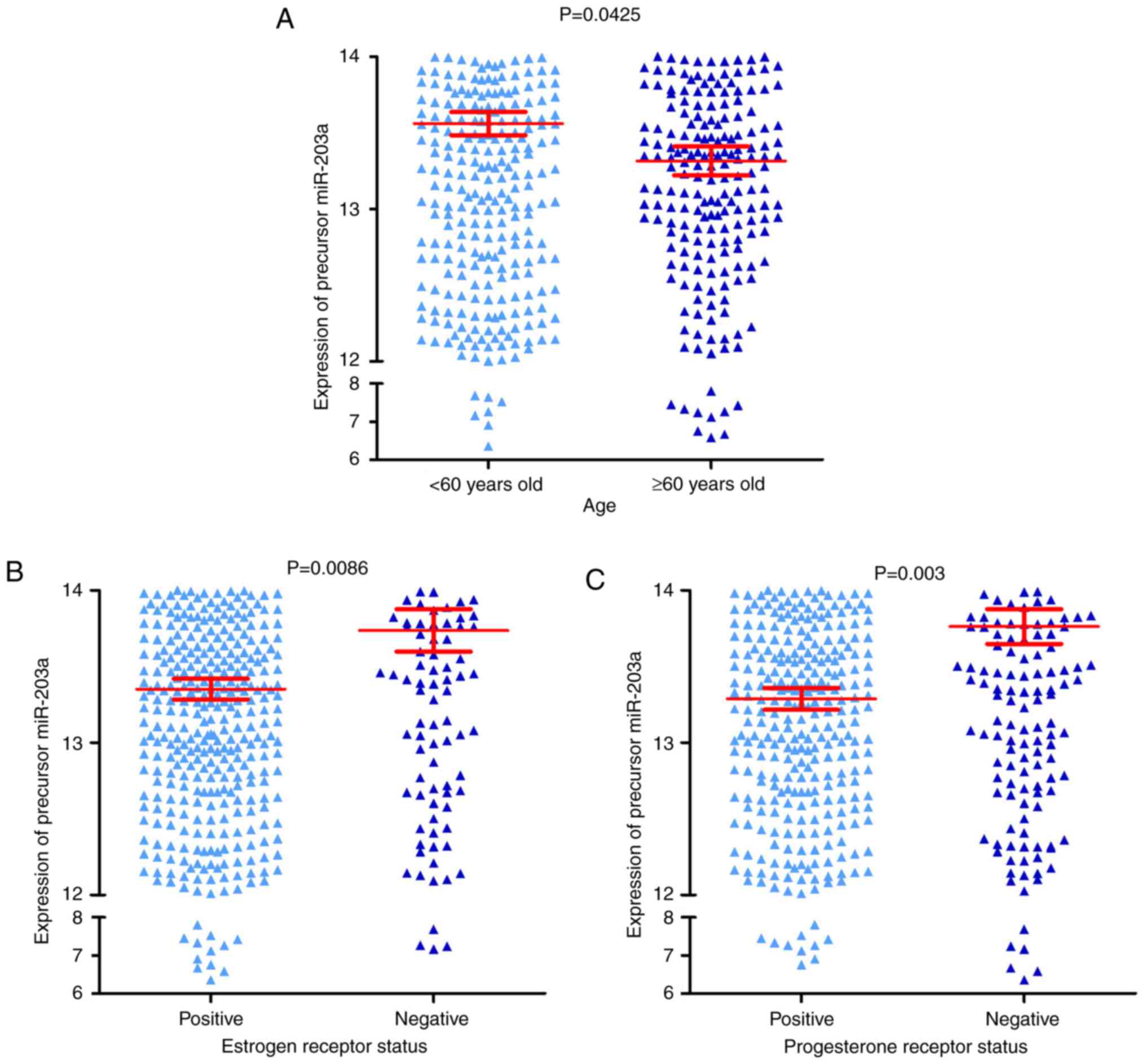
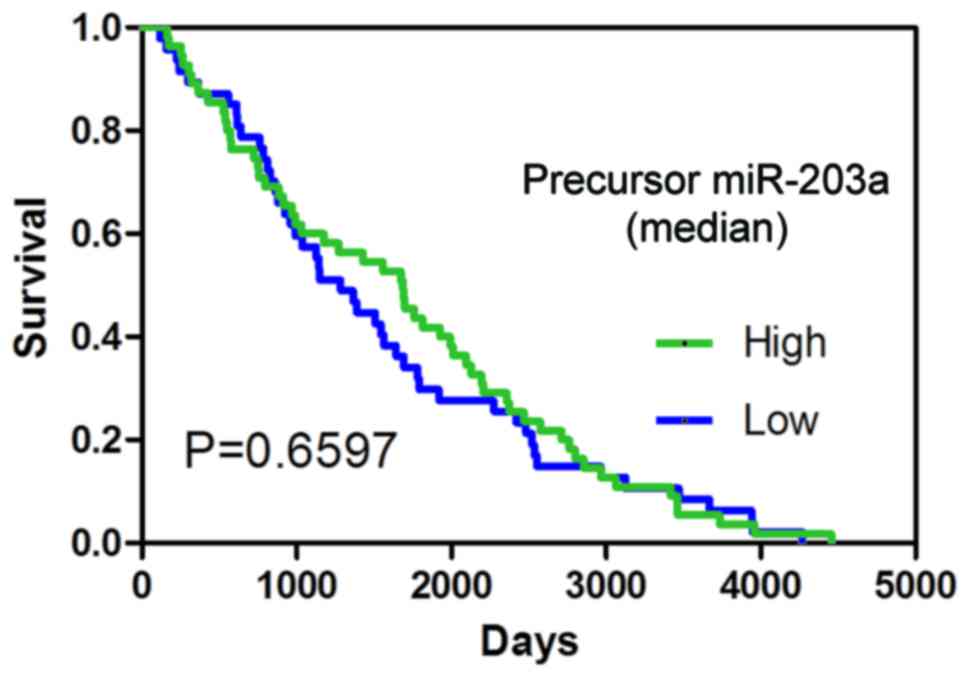
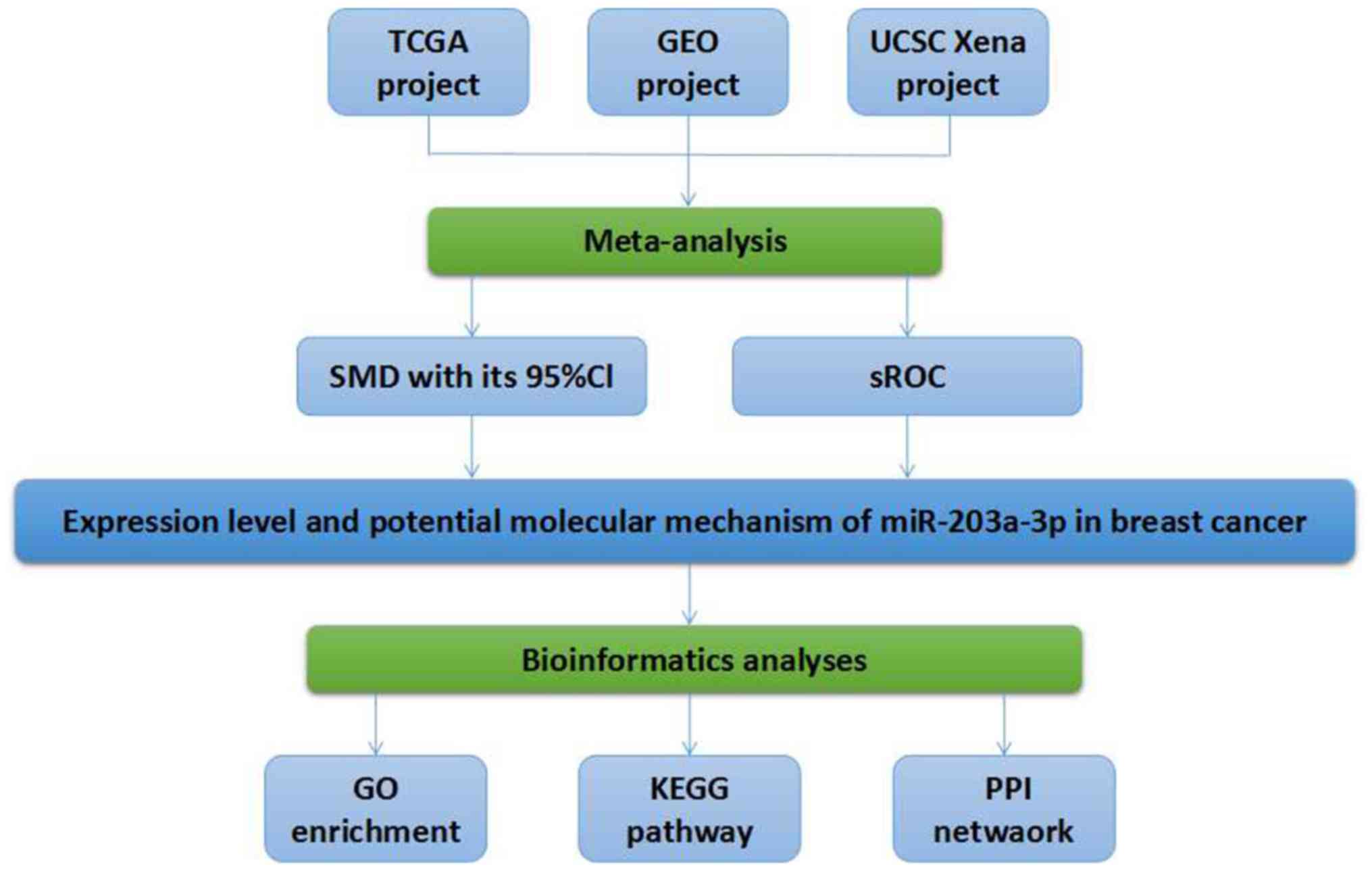
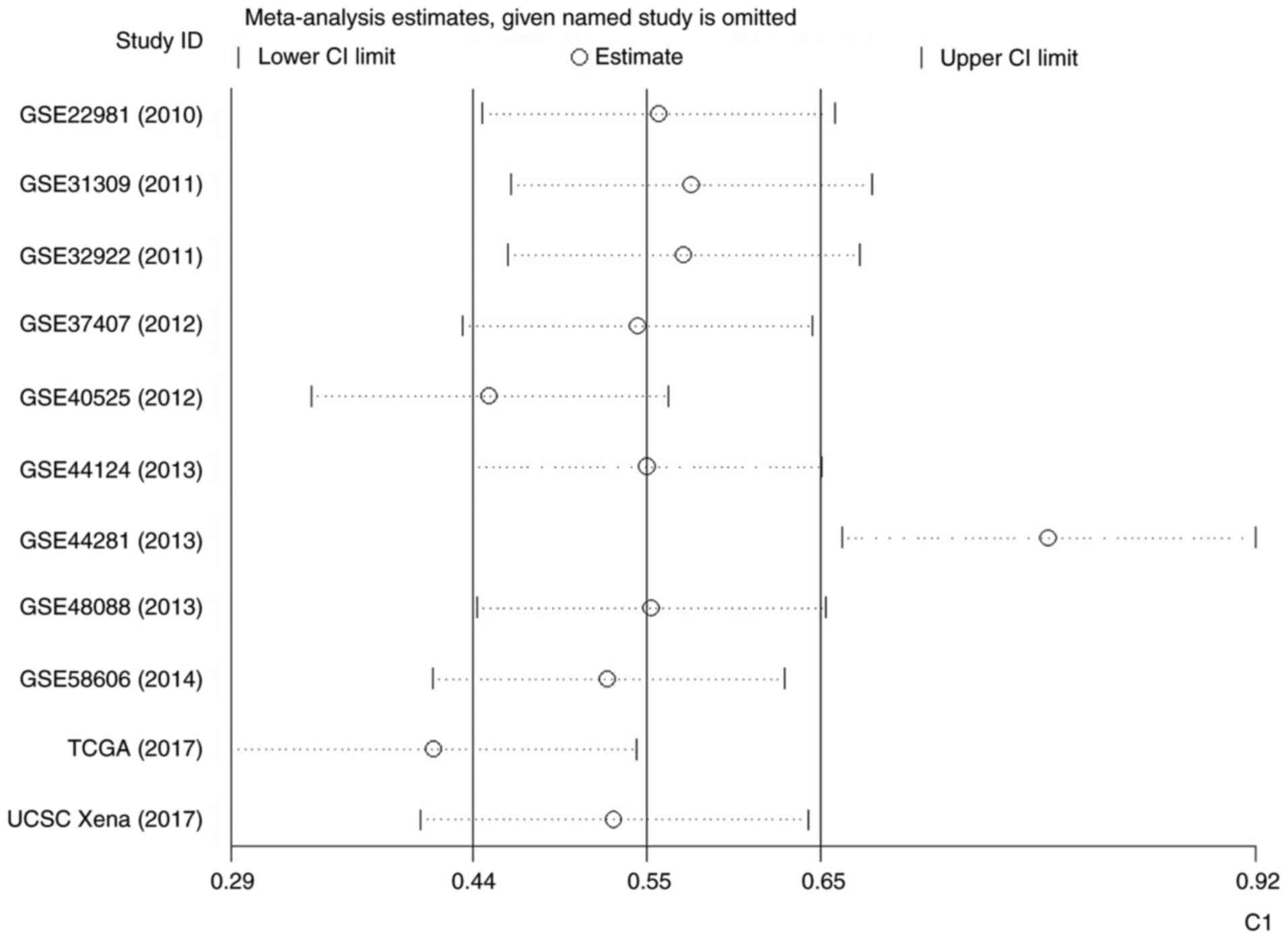
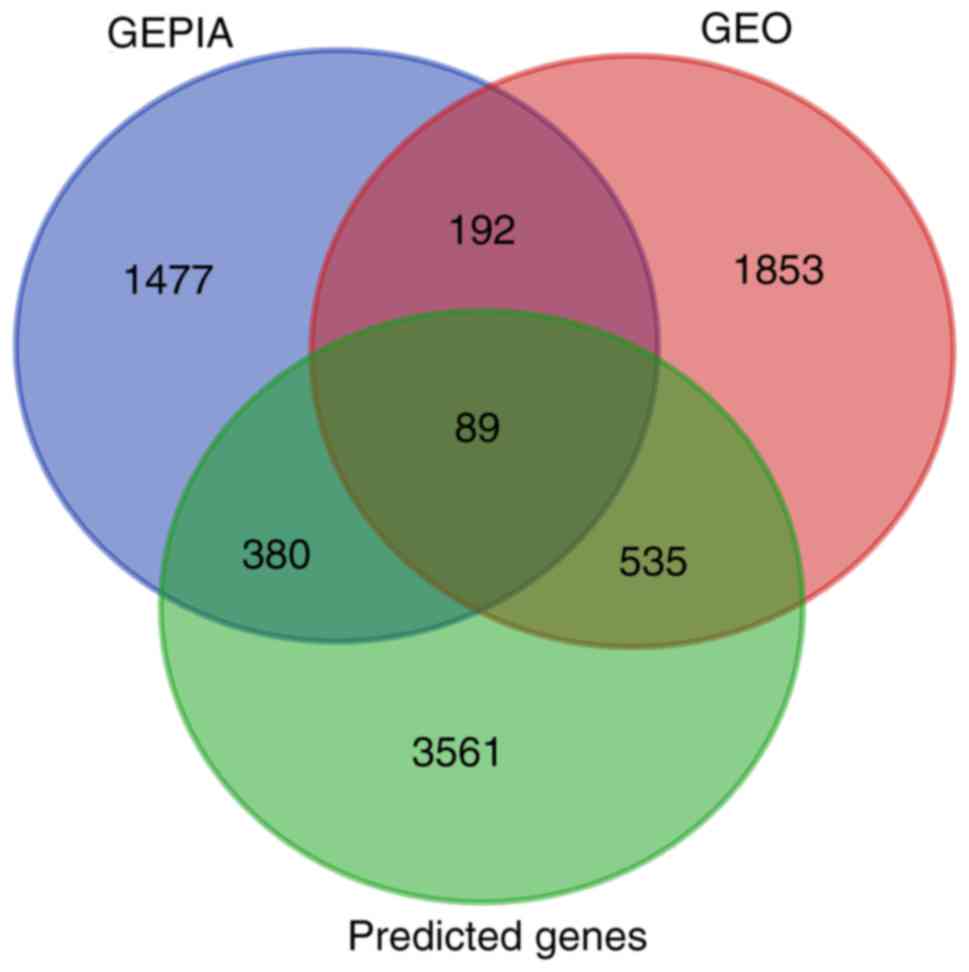
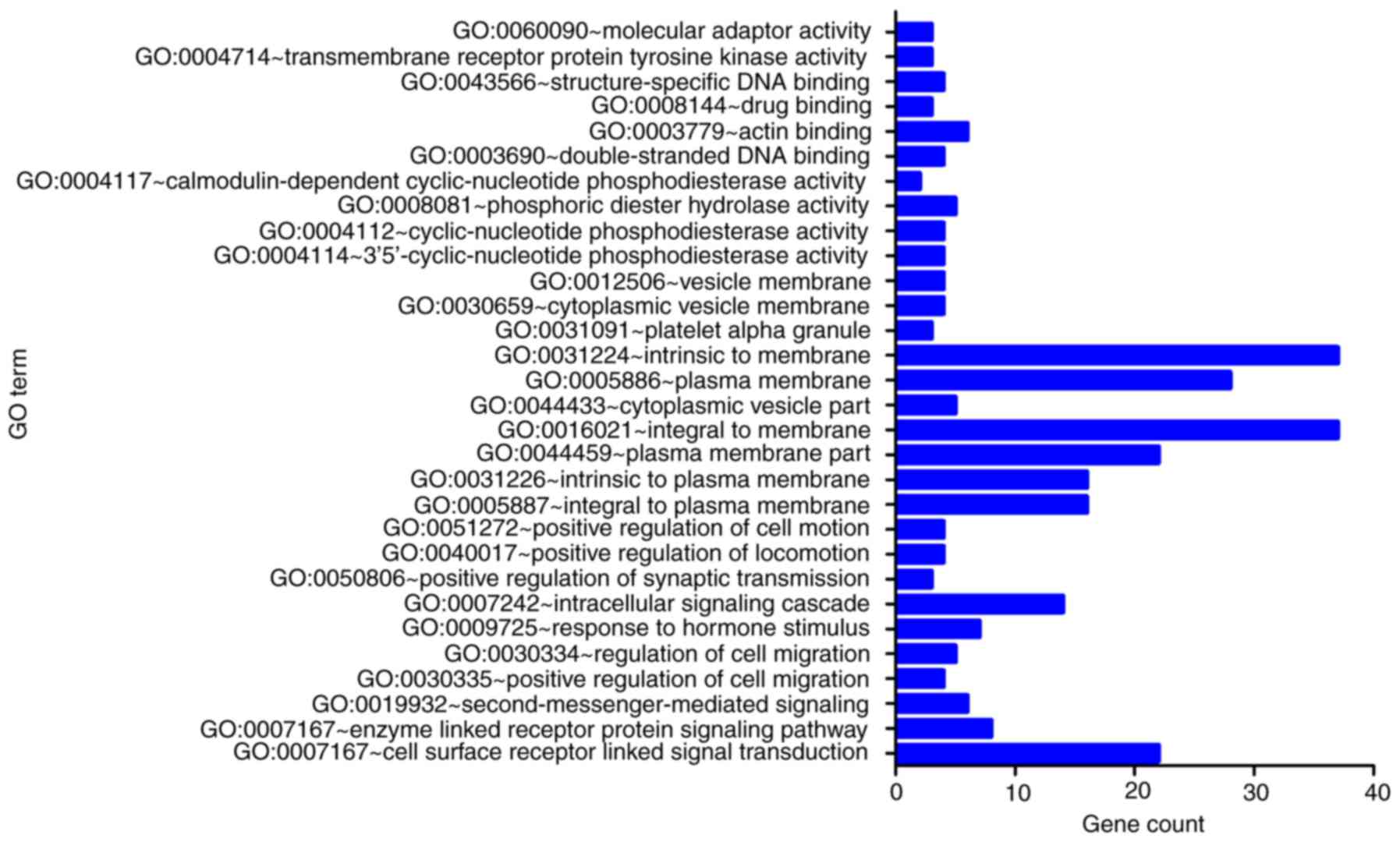
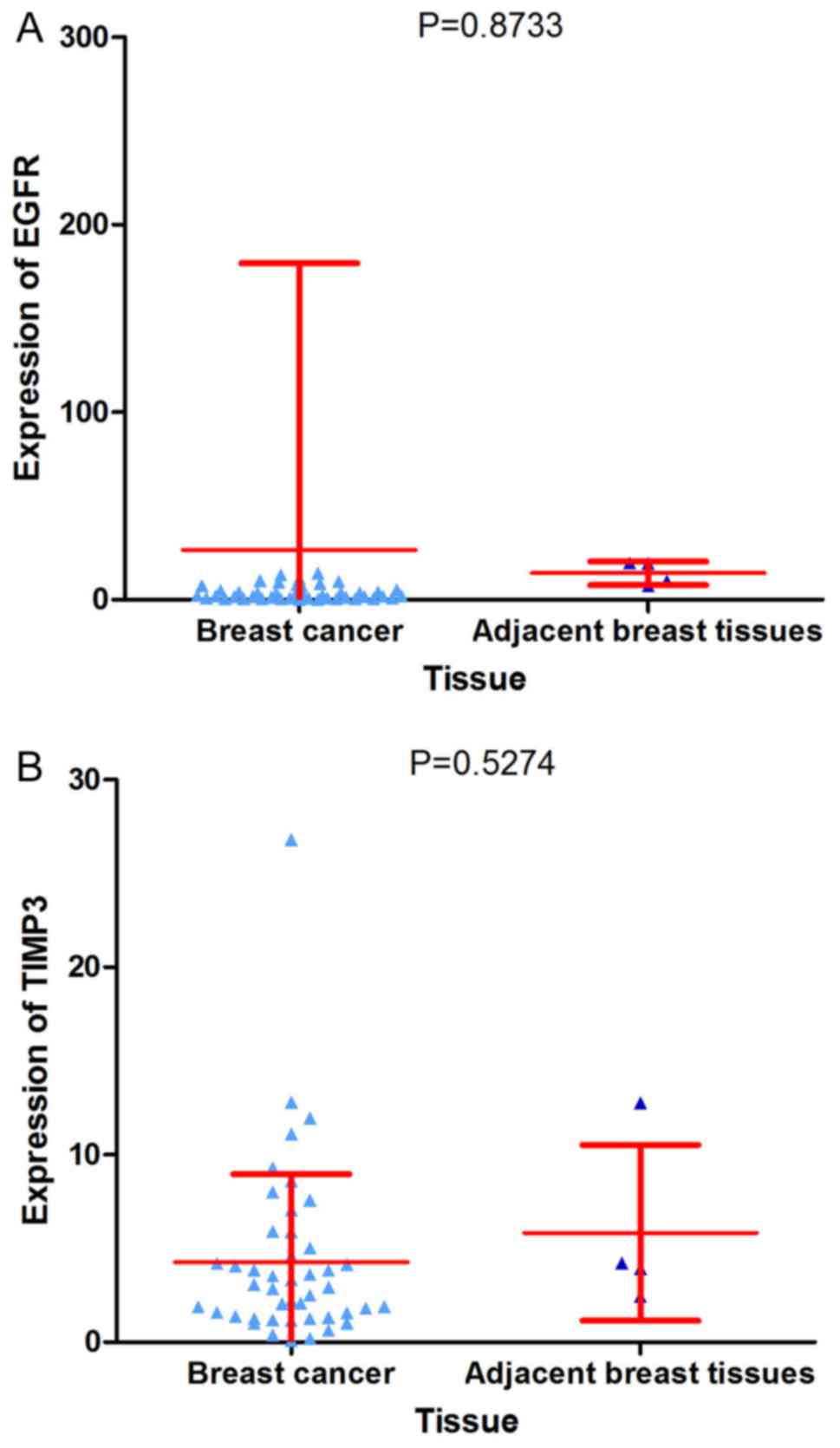
|
1
|
Jalali C, Ghaderi B, Amini S, Abdi M and
Roshani D: Association of XRCC1 Trp194 allele with risk of breast
cancer, and Ki67 protein status in breast tumor tissues. Saudi Med
J. 37:624–630. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liang Z and Xi Y: MicroRNAs mediate
therapeutic and preventive effects of natural agents in breast
cancer. Chin J Nat Med. 14:881–887. 2016.PubMed/NCBI
|
|
3
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shou D, Wen L, Song Z, Yin J, Sun Q and
Gong W: Suppressive role of myeloid-derived suppressor cells
(MDSCs) in the microenvironment of breast cancer and targeted
immunotherapies. Oncotarget. 7:64505–64511. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xing L, He Q, Wang YY, Li HY and Ren GS:
Advances in the surgical treatment of breast cancer. Chin Clin
Oncol. 5:342016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hennequin C, Barillot I, Azria D,
Belkacémi Y, Bollet M, Chauvet B, Cowen D, Cutuli B, Fourquet A,
Hannoun-Lévi JM, et al: Radiotherapy of breast cancer. Cancer
Radiother. 20 Suppl:S139–S146. 2016.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ejlertsen B: Adjuvant chemotherapy in
early breast cancer. Dan Med J. 63:B52222016.PubMed/NCBI
|
|
8
|
Zhang X, Ren D, Guo L, Wang L, Wu S, Lin
C, Ye L, Zhu J, Li J, Song L, et al: Thymosin beta 10 is a key
regulator of tumorigenesis and metastasis and a novel serum marker
in breast cancer. Breast Cancer Res. 19:152017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Z and Kang Y: Emerging therapeutic
targets in metastatic progression: A focus on breast cancer.
Pharmacol Ther. 161:79–96. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song JL, Chen C, Yuan JP and Sun SR:
Progress in the clinical detection of heterogeneity in breast
cancer. Cancer Med. 5:3475–3488. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shen H and Li Z: miRNAs in NMDA
receptor-dependent synaptic plasticity and psychiatric disorders.
Clin Sci (Lond). 130:1137–1146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Su W, Aloi MS and Garden GA: MicroRNAs
mediating CNS inflammation: Small regulators with powerful
potential. Brain Behav Immun. 52:1–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhou W, Zou B, Liu L, Cui K, Gao J, Yuan S
and Cong N: MicroRNA-98 acts as a tumor suppressor in
hepatocellular carcinoma via targeting SALL4. Oncotarget.
7:74059–74073. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang L, Liang H, Wang Y, Gao S, Yin K, Liu
Z, Zheng X, Lv Y, Wang L, Zhang CY, et al: MiRNA-203 suppresses
tumor cell proliferation, migration and invasion by targeting Slug
in gastric cancer. Protein Cell. 7:383–387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gao Y, Feng B, Han S, Lu L, Chen Y, Chu X,
Wang R and Chen L: MicroRNA-129 in human cancers: From
tumorigenesis to clinical treatment. Cell Physiol Biochem.
39:2186–2202. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ren W, Li C, Duan W, Du S, Yang F, Zhou J
and Xing J: MicroRNA-613 represses prostate cancer cell
proliferation and invasion through targeting Frizzled7. Biochem
Biophys Res Commun. 469:633–638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hao W, Luo W, Bai M, Li J, Bai X, Guo J,
Wu J and Wang M: MicroRNA-206 inhibited the progression of
glioblastoma through BCL-2. J Mol Neurosci. 60:531–538. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liang AL, Zhang TT, Zhou N, Wu CY, Lin MH
and Liu YJ: MiRNA-10b sponge: An anti-breast cancer study in
vitro. Oncol Rep. 35:1950–1958. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Du S, Li H, Sun X, Li D, Yang Y, Tao Z, Li
Q and Liu K: MicroRNA-124 inhibits cell proliferation and migration
by regulating SNAI2 in breast cancer. Oncol Rep. 36:3259–3266.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang L, Sun H, Wang X, Hou N, Zhao L, Tong
D, He K, Yang Y, Song T, Yang J and Huang C: EGR1 mediates miR-203a
suppress the hepatocellular carcinoma cells progression by
targeting HOXD3 through EGFR signaling pathway. Oncotarget.
7:45302–45316. 2016.PubMed/NCBI
|
|
21
|
Gomes BC, Martins M, Lopes P, Morujão I,
Oliveira M, Araújo A, Rueff J and Rodrigues AS: Prognostic value of
microRNA-203a expression in breast cancer. Oncol Rep. 36:1748–1756.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu H and Zhang J: Decreased expression of
TFAP2B in endometrial cancer predicts poor prognosis: A study based
on TCGA data. Gynecol Oncol. 149:592–597. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nasif D, Campoy E, Laurito S, Branham R,
Urrutia G, Roqué M and Branham MT: Epigenetic regulation of ID4 in
breast cancer: Tumor suppressor or oncogene? Clin Epigenetics.
10:1112018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang J, Lan Q and Lin J: Identification
of key gene modules for human osteosarcoma by co-expression
analysis. World J Surg Oncol. 16:892018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang YW, Zhang W and Ma R: Bioinformatic
identification of chemoresistance-associated microRNAs in breast
cancer based on microarray data. Oncol Rep. 39:1003–1010.
2018.PubMed/NCBI
|
|
26
|
Hui HX, Hu ZW, Jiang C, Wu J, Gao Y and
Wang XW: ZNF418 overexpression protects against gastric carcinoma
and prompts a good prognosis. Onco Targets Ther. 11:2763–2770.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wu N, Yan J, Han T, Zou J and Shen W:
Integrated assessment of differentially expressed plasma microRNAs
in subtypes of nonsyndromic orofacial clefts. Medicine (Baltimore).
97:e112242018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yan L, Zhan C, Wu J and Wang S: Expression
profile analysis of head and neck squamous cell carcinomas using
data from the cancer genome atlas. Mol Med Rep. 13:4259–4265. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hou L, Lin Z, Ni Y, Wu Y, Chen D, Song L,
Huang X, Hu H and Yang D: Microarray expression profiling and gene
ontology analysis of long non-coding RNAs in spontaneously
hypertensive rats and their potential roles in the pathogenesis of
hypertension. Mol Med Rep. 13:295–300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li GM, Zhang CL, Rui RP, Sun B and Guo W:
Bioinformatics analysis of common differential genes of coronary
artery disease and ischemic cardiomyopathy. Eur Rev Med Pharmacol
Sci. 22:3553–3569. 2018.PubMed/NCBI
|
|
31
|
Zhou W, Ma CX, Xing YZ and Yan ZY:
Identification of candidate target genes of pituitary adenomas
based on the DNA microarray. Mol Med Rep. 13:2182–2186. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu F, Gao F, Liu Y, Wang Z, Zhuang X, Qu
Z, Ma H, Liu Y, Fu C, Zhang Q and Duan X: Bioinformatics analysis
of molecular mechanisms involved in intervertebral disc
degeneration induced by TNF-α and IL-1β. Mol Med Rep. 13:2925–2931.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huo W, Du M, Pan X, Zhu X, Gao Y and Li Z:
miR-203a-3p.1 targets IL-24 to modulate hepatocellular carcinoma
cell growth and metastasis. FEBS Open Bio. 7:1085–1091. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kara M, Yumrutas O, Ozcan O, Celik OI,
Bozgeyik E, Bozgeyik I and Tasdemir S: Differential expressions of
cancer-associated genes and their regulatory miRNAs in colorectal
carcinoma. Gene. 567:81–86. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu W, Dong Z, Liang J, Guo X, Guo Y, Shen
S, Kuang G and Guo W: Downregulation of potential tumor suppressor
mir-203a by promoter methylation contributes to the invasiveness of
gastric cardia adenocarcinoma. Cancer Invest. 34:506–516. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Riemann A, Reime S and Thews O:
Hypoxia-related tumor acidosis affects MicroRNA expression pattern
in prostate and breast tumor cells. Adv Exp Med Biol. 977:119–124.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin QH, Zhang KD, Duan HX, Liu MX, Wei WL
and Cao Y: ERGIC3, which is regulated by miR-203a, is a potential
biomarker for non-small cell lung cancer. Cancer Sci.
106:1463–1473. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Y, Dong Z, Liang J, Guo Y, Guo X, Shen
S, Kuang G and Guo W: Methylation-mediated repression of potential
tumor suppressor miR-203a and miR-203b contributes to esophageal
squamous cell carcinoma development. Tumour Biol. 37:5621–5632.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Costa-Silva DR, Barros-Oliveira MD, Borges
RS, Tavares CB, Borges US, Alves-Ribeiro FA, Silva VC and Silva BB:
Insulin-like growth factor 1 gene polymorphism and breast cancer
risk. An Acad Bras Cienc. 88:2349–2356. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
De Santi M, Annibalini G, Barbieri E,
Villarini A, Vallorani L, Contarelli S, Berrino F, Stocchi V and
Brandi G: Human IGF1 pro-forms induce breast cancer cell
proliferation via the IGF1 receptor. Cell Oncol (Dordr).
39:149–159. 2016. View Article : Google Scholar : PubMed/NCBI
|