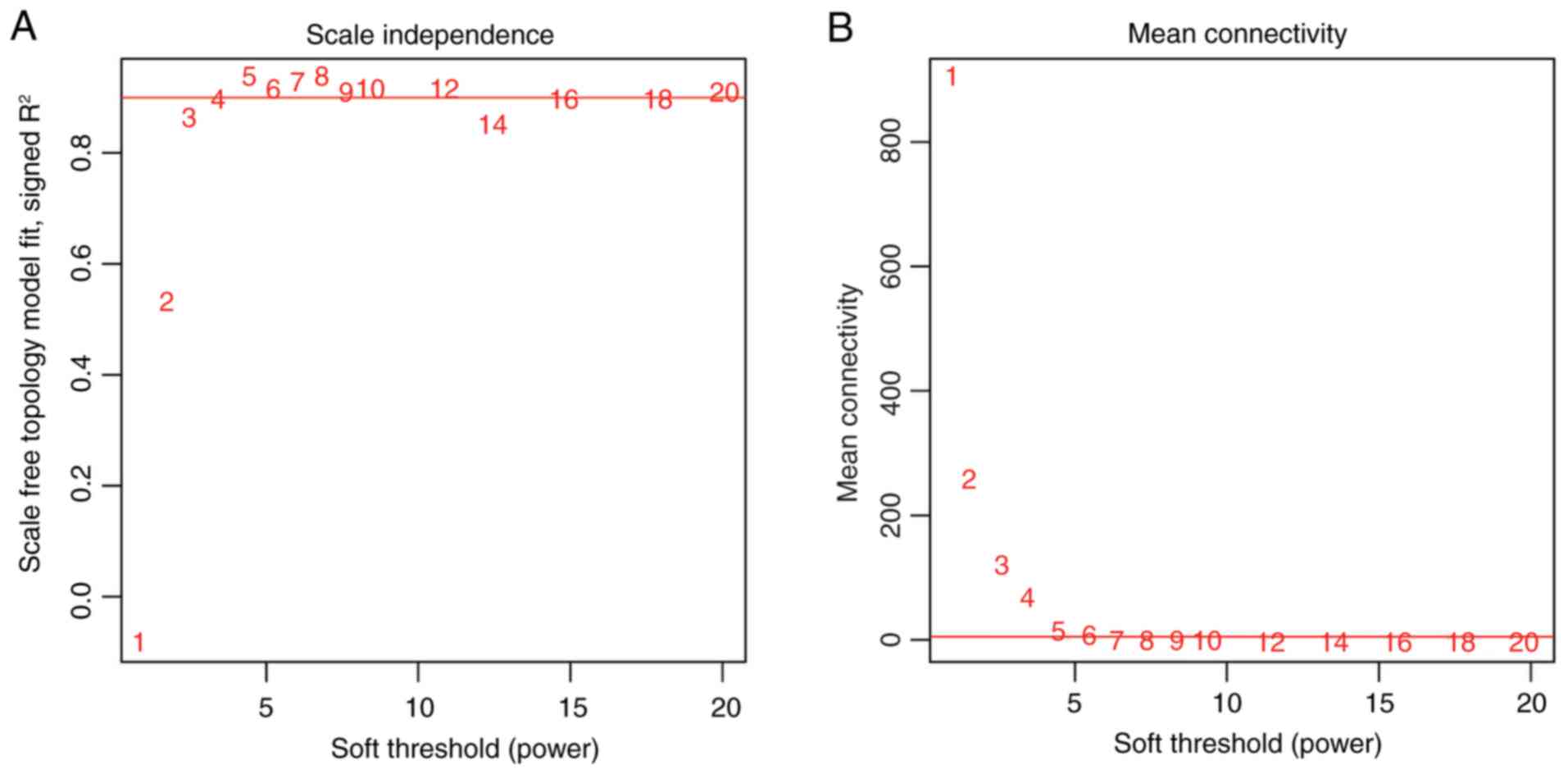
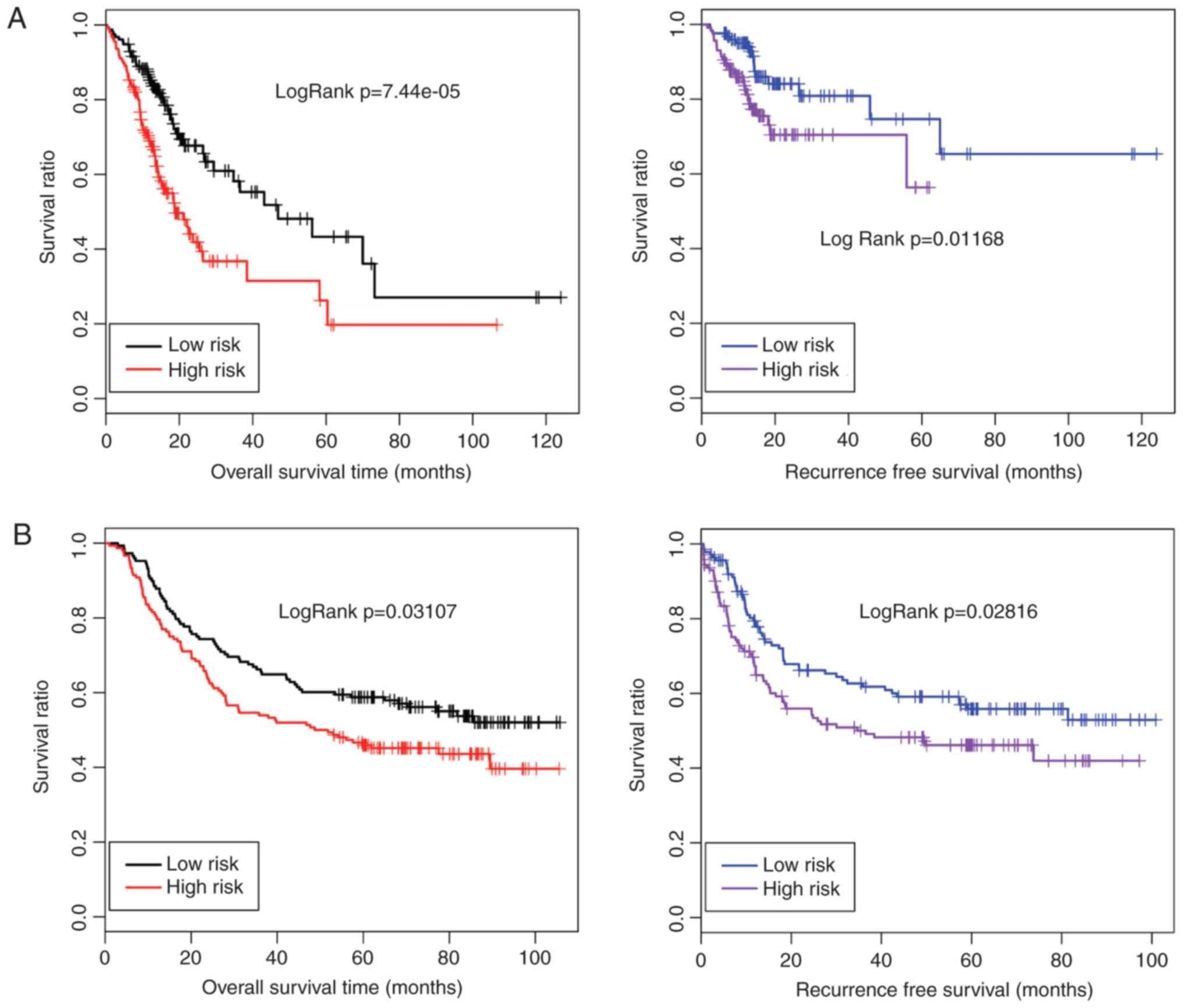
| Biology
process | Immune
response | 80 | MICB, CD8A, LY86,
HLA-DMB, HLA-DMA, C1QC, PDCD1, CD96, SH2D1A, CLEC4E, MS4A1, LTF,
FAS, FCGR3A, SPN, CIITA, LAIR1, POU2AF1, SIT1, NCF2, GZMA, NCF1,
LY96, CMKLR1, TNFRSF17, WAS, HLA-DQA1, PDCD1LG2, TRAT1, CTSW,
IGSF6, C1QB, LILRB2, IL18BP, CCR5, TNFSF13B, CCR4, LAX1, LILRB4,
HLA-DPA1, MADCAM1, GBP4, LCP1, GBP1, LCP2, HLA-DQB1, PSMB10, ITGAL,
CCR1, GPSM3, CXCL9, CX3CL1, IL7R, CCL5, CCL4, POU2F2, ZAP70,
HLA-DRB5, IL2RG, CD4, HLA-DPB1, HLA-DOA, PTPRC, IL2RA, TNFRSF13C,
CCL19, SLAMF7, CD180, AIM2, CORO1A, TNFSF10, CYBB, APOL1, CD300A,
CXCL13, CD209, IRF8, CD274, CD79B, CD79A |
5.39×10−49 |
|
| Regulation of cell
activation | 30 | KLRK1, IL7R,
HLA-DMA, CD2, ZAP70, CD4, IL2RG, FAS, HLA-DOA, LAG3, SPN, PTPRC,
SIT1, IL2RA, IKZF1, PLEK, CD3E, TNFRSF13C, CD40, PDCD1LG2, CD38,
PRKCQ, CORO1A, SIRPG, TNFSF13B, LAX1, CD274, JAK2, IRF4, SASH3 |
6.22×10−20 |
|
| Regulation of
lymphocyte activation | 28 | KLRK1, IL7R,
HLA-DMA, CD2, ZAP70, CD4, IL2RG, FAS, HLA-DOA, LAG3, SPN, PTPRC,
SIT1, IL2RA, IKZF1, CD3E, TNFRSF13C, CD40, PDCD1LG2, CD38, PRKCQ,
CORO1A, SIRPG, TNFSF13B, LAX1, CD274, IRF4, SASH3 |
1.53×10−19 |
|
| Lymphocyte
activation | 31 | ITGAL, MICB, CD8A,
IL21R, KLRK1, PTPN22, IL7R, HLA-DMA, DOCK2, CXCR5, ZAP70, MS4A1,
CD2, CD4, FAS, SPN, RHOH, PTPRC, CD3G, CD3D, IKZF1, CD3E, SLAMF7,
ITGA4, CD40, WAS, LAX1, CD79A, IRF4, BANK1, LCP1 |
1.83×10−19 |
|
| Positive regulation
of immune system process | 33 | C3AR1, MICB, CD247,
KLRK1, PTPN22, IL7R, C1QC, HLA-DMA, SH2D1A, CD2, ZAP70, CD4, IL2RG,
LAG3, SPN, PTPRC, IL2RA, IKZF1, CD3E, TNFRSF13C, CD40, PDCD1LG2,
TRAT1, CD38, PRKCQ, C1QB, CORO1A, CD37, SIRPG, TNFSF13B, LAX1,
CD79A, SASH3 |
2.32×10−19 |
|
| Leukocyte
activation | 33 | ITGAL, MICB, CD8A,
IL21R, KLRK1, PTPN22, CX3CL1, IL7R, HLA-DMA, DOCK2, CXCR5, ZAP70,
MS4A1, CD2, CD4, FAS, SPN, RHOH, PTPRC, CD3G, CD3D, IKZF1, CD3E,
SLAMF7, ITGA4, CD40, WAS, LAX1, CD79A, IRF4, BANK1, LCP1, LCP2 |
3.92×10−19 |
|
| Regulation of T
cell activation | 25 | PTPRC, SIT1, IL2RA,
IKZF1, CD3E, TNFRSF13C, KLRK1, IL7R, HLA-DMA, PDCD1LG2, PRKCQ,
CORO1A, SIRPG, TNFSF13B, LAX1, CD274, ZAP70, CD2, CD4, IL2RG, IRF4,
HLA-DOA, SPN, LAG3, SASH3 |
2.21×10−18 |
|
| Defense
response | 47 | C3AR1, ITGAL, PRF1,
AIF1, CCR1, LY86, CXCL9, ITGB2, CX3CL1, CCL5, PTPRCAP, CCL4, C1QC,
SH2D1A, AOAH, LTF, SPN, CIITA, ITK, PTPRC, IL2RA, NCF2, NCF1, LY96,
HCK, CCL19, CD40, SLAMF7, WAS, CD180, SP140, TRAT1, CD163, LSP1,
CD84, APOL3, SIGLEC1, C1QB, LILRB2, CORO1A, CYBB, APOL1, CCR5,
CCR4, CXCL13, MNDA, PLA2G7 |
3.44×10−18 |
|
| Regulation of
leukocyte activation | 28 | KLRK1, IL7R,
HLA-DMA, CD2, ZAP70, CD4, IL2RG, FAS, HLA-DOA, LAG3, SPN, PTPRC,
SIT1, IL2RA, IKZF1, CD3E, TNFRSF13C, CD40, PDCD1LG2, CD38, PRKCQ,
CORO1A, SIRPG, TNFSF13B, LAX1, CD274, IRF4, SASH3 |
3.64×10−18 |
|
| Cell
activation | 34 | ITGAL, MICB, CD8A,
IL21R, KLRK1, PTPN22, CX3CL1, IL7R, HLA-DMA, DOCK2, CXCR5, ZAP70,
MS4A1, CD2, CD4, FAS, SPN, RHOH, PTPRC, CD3G, CD3D, PLEK, IKZF1,
CD3E, SLAMF7, ITGA4, CD40, WAS, LAX1, CD79A, IRF4, BANK1, LCP1,
LCP2 |
7.10×10−18 |
|
| Positive regulation
of cell activation | 23 | PTPRC, IL2RA,
IKZF1, PLEK, CD3E, KLRK1, TNFRSF13C, CD40, IL7R, HLA-DMA, PDCD1LG2,
CD38, PRKCQ, CORO1A, SIRPG, TNFSF13B, CD2, ZAP70, JAK2, CD4, IL2RG,
SASH3, SPN |
2.55×10−16 |
|
| Positive regulation
of leukocyte activation | 21 | PTPRC, IL2RA,
IKZF1, CD3E, KLRK1, TNFRSF13C, CD40, IL7R, HLA-DMA, PDCD1LG2, CD38,
PRKCQ, CORO1A, SIRPG, TNFSF13B, CD2, ZAP70, CD4, IL2RG, SASH3,
SPN |
3.41×10−14 |
|
| Positive regulation
of lymphocyte activation | 20 | PTPRC, IL2RA,
IKZF1, CD3E, KLRK1, TNFRSF13C, CD40, IL7R, HLA-DMA, PDCD1LG2, CD38,
PRKCQ, CORO1A, SIRPG, TNFSF13B, ZAP70, CD4, IL2RG, SASH3, SPN |
1.78×10−13 |
|
| T cell
activation | 21 | ITGAL, PTPRC, MICB,
CD3G, CD3D, IKZF1, CD8A, CD3E, PTPN22, IL7R, HLA-DMA, WAS, DOCK2,
ZAP70, CD2, CD4, FAS, IRF4, LCP1, SPN, RHOH |
1.09×10−12 |
|
| Hemopoietic or
lymphoid organ development | 24 | PTPRC, CD3D, PLEK,
IKZF1, CD8A, CD3E, HCLS1, PTPN22, ITGA4, IFI16, IL7R, HLA-DMA,
DOCK2, CXCR5, CXCL13, IRF8, ZAP70, JAK2, CD4, FAS, CD79A, IRF4,
SPN, RHOH |
2.97×10−09 |
|
| Inflammatory
response | 26 | ITGAL, C3AR1, AIF1,
LY86, CCR1, CXCL9, ITGB2, CCL5, C1QC, CCL4, AOAH, CIITA, IL2RA,
LY96, CCL19, CD40, CD180, CD163, C1QB, SIGLEC1, APOL3, CYBB, CCR5,
CXCL13, CCR4, PLA2G7 |
6.83×10−09 |
|
| Immune system
development | 24 | PTPRC, CD3D, PLEK,
IKZF1, CD8A, CD3E, HCLS1, PTPN22, ITGA4, IFI16, IL7R, HLA-DMA,
DOCK2, CXCR5, CXCL13, IRF8, ZAP70, JAK2, CD4, FAS, CD79A, IRF4,
SPN, RHOH |
1.02×10−08 |
|
| Hemopoiesis | 22 | PTPRC, CD3D, PLEK,
IKZF1, CD8A, CD3E, HCLS1, PTPN22, ITGA4, IFI16, IL7R, HLA-DMA,
DOCK2, IRF8, ZAP70, JAK2, CD4, FAS, CD79A, IRF4, SPN, RHOH |
2.60×10−08 |
|
| Positive regulation
of response to stimulus | 22 | C3AR1, PTPRC, MICB,
CD3E, CD247, KLRK1, TNFRSF13C, PTPN22, CX3CL1, CCL5, HLA-DMA, C1QC,
C1QB, SH2D1A, TNFSF13B, LAX1, CCR4, ZAP70, JAK2, CD79A, SASH3,
LAG3 |
2.60×10−08 |
|
| Response to
wounding | 30 | C3AR1, ITGAL, AIF1,
LY86, CCR1, CXCL9, ITGB2, CCL5, C1QC, CCL4, AOAH, CIITA, IL2RA,
PLEK, LY96, CCL19, CD40, WAS, CD180, CD163, APOL3, PRKCQ, C1QB,
SIGLEC1, CYBB, CCR5, CCR4, CXCL13, PLA2G7, JAK2 |
4.82×10−07 |
|
| Cell surface
receptor linked signal transduction | 56 | MICB, CD8A, PTPN22,
CXCR5, CXCR6, SPN, LAG3, KLRB1, PIK3CG, CD3G, CD3D, LY96, CMKLR1,
CD3E, GPR171, CD40, IGSF6, LILRB2, DOK2, CCR5, CCR4, LAX1, LCP2,
C3AR1, ITGAL, CCR1, CD247, KLRK1, CXCL9, FPR3, ITGB2, IL7R, CCL5,
P2RY6, ITGAX, ITGB7, GPR25, ZAP70, CD2, CD4, PTPRC, IL2RA, PLEK,
DTX1, CCL19, RGS19, EVL, ITGA4, BIRC3, P2RY10, CD274, CD79B, JAK2,
JAK3, CD79A, ADAMDEC1 |
4.54×10−05 |
|
| Cell adhesion | 29 | ITGAL, CCR1,
FERMT3, ITGB2, CX3CL1, CCL5, CCL4, CD96, ITGAX, ITGB7, CD2, CD22,
CD4, CD6, SELPLG, PARVG, PTPRC, PLEK, SIGLEC10, ITGA4, SLAMF7,
EMILIN2, CD84, SIGLEC1, CORO1A, SIRPG, CD300A, CD209, MADCAM1 |
8.34×10−04 |
|
| Biological
adhesion | 29 | ITGAL, CCR1,
FERMT3, ITGB2, CX3CL1, CCL5, CCL4, CD96, ITGAX, ITGB7, CD2, CD22,
CD4, CD6, SELPLG, PARVG, PTPRC, PLEK, SIGLEC10, ITGA4, SLAMF7,
EMILIN2, CD84, SIGLEC1, CORO1A, SIRPG, CD300A, CD209, MADCAM1 |
8.58×10−04 |
| KEGG pathway | Cell adhesion
molecules (CAMs) | 26 | HLA-DQB1, ITGAL,
PTPRC, CD8A, ITGB2, CD40, ITGA4, HLA-DMB, HLA-DMA, PDCD1, HLA-DQA1,
PDCD1LG2, SIGLEC1, ITGB7, CD274, CD2, CD22, HLA-DRB5, CD4,
HLA-DPA1, MADCAM1, HLA-DPB1, HLA-DOA, CD6, SELPLG, SPN |
6.37×10−15 |
|
| Allograft
rejection | 12 | HLA-DQB1, PRF1,
HLA-DRB5, GZMB, HLA-DPA1, HLA-DPB1, FAS, CD40, HLA-DMB, HLA-DOA,
HLA-DMA, HLA-DQA1 |
8.68×10−08 |
|
| Cytokine-cytokine
receptor interaction | 24 | IL2RB, IL2RA, CCR1,
IL21R, TNFRSF13C, CXCL9, TNFRSF17, CCL19, CD40, CX3CL1, IL7R, CCL5,
CCL4, TNFSF10, TNFSF13B, CXCR5, CCR5, CCR4, CXCL13, IL10RA, CXCR6,
CSF2RB, IL2RG, FAS |
2.80×10−06 |
|
| Graft vs.host
disease | 11 | HLA-DQB1, PRF1,
HLA-DRB5, GZMB, HLA-DPA1, HLA-DPB1, FAS, HLA-DMB, HLA-DOA, HLA-DMA,
HLA-DQA1 |
4.48×10−06 |
|
| Chemokine signaling
pathway | 19 | PIK3CG, ITK, NCF1,
HCK, CCR1, CXCL9, CCL19, CX3CL1, CCL5, CCL4, WAS, DOCK2, CXCR5,
CCR5, CCR4, CXCL13, CXCR6, JAK2, JAK3 |
4.64×10−05 |
|
| Natural killer cell
mediated cytotoxicity | 15 | PIK3CG, PRF1,
ITGAL, MICB, CD247, KLRK1, GZMB, ITGB2, HCST, SH2D1A, TNFSF10,
ZAP70, FAS, FCGR3A, LCP2 |
5.69×10−04 |
|
| T cell receptor
signaling pathway | 13 | PIK3CG, ITK, PRKCQ,
PTPRC, CD3G, CD8A, CD3D, CD3E, CD247, ZAP70, CD4, PDCD1, LCP2 |
2.20×10−03 |
|
| Antigen processing
and presentation | 11 | HLA-DQB1, CIITA,
CD8A, HLA-DRB5, CD4, HLA-DPA1, HLA-DPB1, HLA-DMB, HLA-DOA, HLA-DMA,
HLA-DQA1 |
7.92×10−03 |