|
1
|
IARC: World Cancer Report 2014, . Stewart
BW and Wild CP: World Health Organization; Geneva: 2015
|
|
2
|
Orditura M, Galizia G, Sforza V,
Gambardella V, Fabozzi A, Laterza MM, Andreozzi F, Ventriglia J,
Savastano B, Mabilia A, et al: Treatment of gastric cancer. World J
Gastroenterol. 20:1635–1649. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in china,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
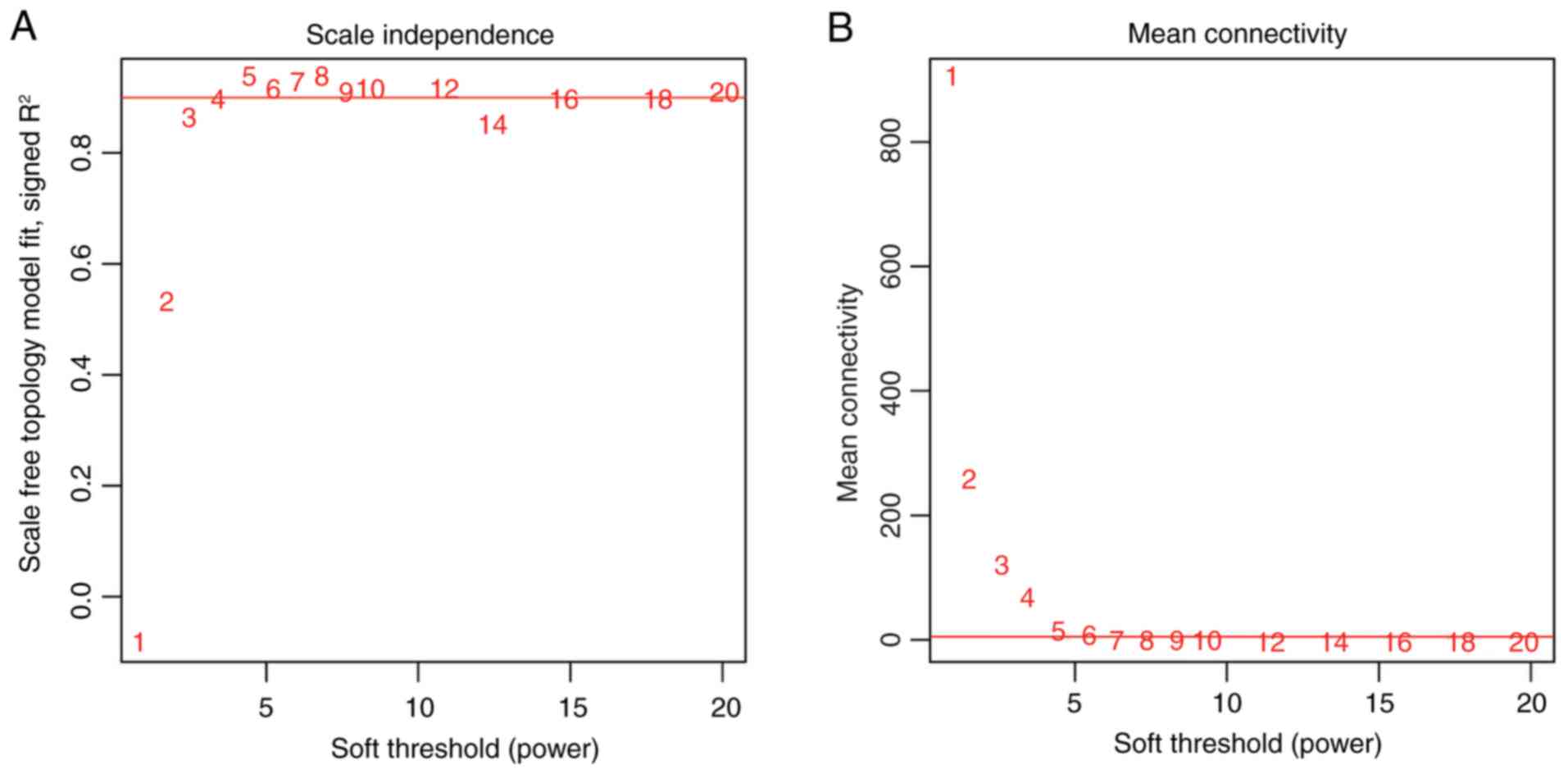
|
4
|
Sun Z, Wang ZN, Zhu Z, Xu YY, Xu Y, Huang
BJ, Zhu GL and Xu HM: Evaluation of the seventh edition of american
joint committee on cancer TNM staging system for gastric cancer:
Results from a chinese monoinstitutional study. Ann Surg Oncol.
19:1918–1927. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mikkelsen TS, Ku M, Jaffe DB, Issac B,
Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP,
et al: Genome-wide maps of chromatin state in pluripotent and
lineage-committed cells. Nature. 448:553–560. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liz J and Esteller M: lncRNAs and
microRNAs with a role in cancer development. Biochim Biophys Acta.
1859:169–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Evans JR, Feng FY and Chinnaiyan AM: The
bright side of dark matter: LncRNAs in cancer. J Clin Invest.
126:2775–2782. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou X, Yin C, Dang Y, Ye F and Zhang G:
Identification of the long non-coding RNA H19 in plasma as a novel
biomarker for diagnosis of gastric cancer. Sci Rep. 5:115162015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou X, Ye F, Yin C, Zhuang Y, Yue G and
Zhang G: The interaction between MiR-141 and lncRNA-H19 in
regulating cell proliferation and migration in gastric cancer. Cell
Physiol Biochem. 36:1440–1452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen
H, Zhou L, Yuan Q, Zhou C and Yang M: A functional lncRNA HOTAIR
genetic variant contributes to gastric cancer susceptibility. Mol
Carcinog. 55:90–96. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang EB, Kong R, Yin DD, You LH, Sun M,
Han L, Xu TP, Xia R, Yang JS, De W and Chen Jf: Long noncoding RNA
ANRIL indicates a poor prognosis of gastric cancer and promotes
tumor growth by epigenetically silencing of miR-99a/miR-449a.
Oncotarget. 5:2276–2292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Miao Y, Sui J, Xu SY, Liang GY, Pu YP and
Yin LH: Comprehensive analysis of a novel four-lncRNA signature as
a prognostic biomarker for human gastric cancer. Oncotarget.
8:75007–75024. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Danford T, Rolfe A and Gifford D: GSE: A
comprehensive database system for the representation, retrieval,
and analysis of microarray data. Pac Symp Biocomput. 539–550.
2008.PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou M, Guo M, He D, Wang X, Cui Y, Yang
H, Hao D and Sun J: A potential signature of eight long non-coding
RNAs predicts survival in patients with non-small cell lung cancer.
J Transl Med. 13:2312015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou M, Xu W, Yue X, Zhao H, Wang Z, Shi
H, Cheng L and Sun J: Relapse-related long non-coding RNA signature
to improve prognosis prediction of lung adenocarcinoma. Oncotarget.
7:29720–29738. 2016.PubMed/NCBI
|
|
19
|
Larkin MA, Blackshields G, Brown NP,
Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm
A, Lopez R, et al: Clustal W and Clustal X version 2.0.
Bioinformatics. 23:2947–2948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhai X, Xue Q, Liu Q, Guo Y and Chen Z:
Colon cancer recurrence-associated genes revealed by WGCNA
co-expression network analysis. Mol Med Rep. 16:6499–6505. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qi C, Hong L, Cheng Z and Yin Q:
Identification of metastasis-associated genes in colorectal cancer
using metaDE and survival analysis. Oncol Lett. 11:568–574. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Kang DD, Shen K, Song C, Lu S,
Chang LC, Liao SG, Huo Z, Tang S, Ding Y, et al: An R package suite
for microarray meta-analysis in quality control, differentially
expressed gene analysis and pathway enrichment detection.
Bioinformatics. 28:2534–2536. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goeman JJ: L1 penalized estimation in the
Cox proportional hazards model. Biom J. 52:70–84. 2010.PubMed/NCBI
|
|
25
|
Tibshirani R: The lasso method for
variable selection in the Cox model. Stat Med. 16:385–395. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cristescu R, Lee J, Nebozhyn M, Kim KM,
Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K, et al: Molecular
analysis of gastric cancer identifies subtypes associated with
distinct clinical outcomes. Nat Med. 21:449–456. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Parrish RS and Spencer HJ III: Effect of
normalization on significance testing for oligonucleotide
microarrays. J Biopharm Stat. 14:575–589. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
da Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li T, Mo X, Fu L, Xiao B and Guo J:
Molecular mechanisms of long noncoding RNAs on gastric cancer.
Oncotarget. 7:8601–8612. 2016.PubMed/NCBI
|
|
31
|
Sun M, Nie FQ, Wang ZX and De W:
Involvement of lncRNA dysregulation in gastric cancer. Histol
Histopathol. 31:33–39. 2016.PubMed/NCBI
|
|
32
|
Lee HS, Lee HK, Kim HS, Yang HK, Kim YI
and Kim WH: MUC1, MUC2, MUC5AC, and MUC6 expressions in gastric
carcinomas. Cancer. 92:1427–1434. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brown CJ, Ballabio A, Rupert JL,
Lafreniere RG, Grompe M, Tonlorenzi R and Willard HF: A gene from
the region of the human X inactivation centre is expressed
exclusively from the inactive X chromosome. Nature. 349:38–44.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL,
Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al: Long non-coding
RNA XIST regulates gastric cancer progression by acting as a
molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin
Cancer Res. 35:1422016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Endo H, Shiroki T, Nakagawa T, Yokoyama M,
Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, et
al: Enhanced expression of long non-coding RNA HOTAIR is associated
with the development of gastric cancer. PLoS One. 8:e770702013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z
and Zhang LW: Identification of key long non-coding RNAs as
competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma.
Eur Rev Med Pharmacol Sci. 20:2285–2295. 2016.PubMed/NCBI
|
|
37
|
Diaz-Lagares A, Crujeiras AB, Lopez-Serra
P, Soler M, Setien F, Goyal A, Sandoval J, Hashimoto Y,
Martinez-Cardús A, Gomez A, et al: Epigenetic inactivation of the
p53-induced long noncoding RNA TP53 target 1 in human cancer. Proc
Natl Acad Sci USA. 113:E7535–E7544. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chen X, Gao Y, Li D, Cao Y and Hao B:
LncRNA-TP53TG1 participated in the stress response under glucose
deprivation in glioma. J Cell Biochem. 118:4897–4904. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang H, Niu L, Jiang S, Zhai J, Wang P,
Kong F and Jin X: Comprehensive analysis of aberrantly expressed
profiles of lncRNAs and miRNAs with associated ceRNA network in
muscle-invasive bladder cancer. Oncotarget. 7:86174–86185. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li J, Li P, Zhao W, Yang R, Chen S, Bai Y,
Dun S, Chen X, Du Y, Wang Y, et al: Expression of long non-coding
RNA DLX6-AS1 in lung adenocarcinoma. Cancer Cell Int. 15:482015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lubovac-Pilav Z, Borràs DM, Ponce E and
Louie MC: Using expression profiling to understand the effects of
chronic cadmium exposure on MCF-7 breast cancer cells. PLoS One.
8:e846462013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Candido J and Hagemann T: Cancer-related
inflammation. J Clin Immunol. 33 Suppl 1:S79–S84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Elinav E, Nowarski R, Thaiss CA, Hu B, Jin
C and Flavell RA: Inflammation-induced cancer: Crosstalk between
tumours, immune cells and microorganisms. Nat Rev Cancer.
13:759–771. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Evan GI and Vousden KH: Proliferation,
cell cycle and apoptosis in cancer. Nature. 411:342–348. 2001.
View Article : Google Scholar : PubMed/NCBI
|