Introduction
Pancreatic cancer, a highly lethal malignancy, is
the sixth leading cause of cancer-associated mortalities due to
malignant tumours in China (1).
Pancreatic cancer is difficult to diagnose early since the clinical
symptoms are not obvious; therefore, patients with pancreatic
cancer are frequently diagnosed at advanced stages and are
unsuitable for surgical resection (2). Despite significant developments in
diagnosis and therapy, the treatment outcomes of patients with
pancreatic cancer remain poor with high recurrence and an
unsatisfactory five-year survival rate (3). Numerous genetic and epigenetic
alterations that cause inactivation of tumour suppressor genes and
activation of oncogenes, contribute to pancreatic cancer
pathogenesis and development (4,5).
However, the causes of pancreatic cancer remain largely unknown.
Therefore, further elucidation of the underlying mechanisms of
pancreatic cancer initiation and progression is urgently required
to identify effective therapeutic strategies for the treatment of
patients with this disease.
MicroRNAs (miRNAs) are a class of highly conserved
and non-coding RNA molecules that are involved in the regulation of
gene expression (6). miRNAs
negatively modulate gene expression by recognising and directly
binding to the 3′-untranslated regions (3′-UTRs) of their target
genes, inhibiting translation and/or inducing mRNAs degradation
(7). miRNAs are aberrantly
expressed in numerous human malignancy types, including pancreatic
(8), gastric (9), thyroid (10) and lung (11) cancer. By modulating the expression
of their targets, miRNAs affect numerous biological processes and
thus are involved in the regulation of tumourigenesis and tumour
progression (12,13). Specifically, miRNAs that are
downregulated in human cancer may serve tumour suppressive roles
(14). By contrast, oncogenic
miRNAs are generally upregulated and serve oncogenic roles during
carcinogenesis and cancer progression (15). Therefore, understanding the
expression patterns, biological roles and the mechanisms associated
with miRNA regulation is crucial for anticancer therapy.
miR-584-5p (miR-584) serves tumour suppressive roles
in multiple types of human malignancy (16–18).
However, the expression pattern and detailed roles of miR-584 in
pancreatic cancer and its underlying mechanisms require further
examination and investigation. In the present study, it was
demonstrated that miR-584 is downregulated in pancreatic cancer. In
addition, miR-584 inhibited cell proliferation and invasion in
pancreatic cancer by directly targeting cyclin D1 (CCND1).
Materials and methods
Tissue collection
A total of 26 primary pancreatic cancer tissues and
their matched normal adjacent specimens were collected from
patients (17 males, 9 females; age, 42–67 years) The First
Affiliated Hospital of Jiamusi University (Jiamusi, China) between
June 2014 and February 2016. None of these patients had received
chemotherapy, radiotherapy or other treatments prior to surgical
resection. Patients that had been treated with chemotherapy,
radiotherapy or other treatments prior to surgery were excluded
from the study. All tissue specimens were quickly snap-frozen in
liquid nitrogen subsequent to surgical resection and were stored at
−80°C. The present study was approved by The Ethics Committee of
the First Affiliated Hospital of Jiamusi University and was
performed in accordance with the Declaration of Helsinki and the
guidelines of the Ethics Committee of The First Affiliated Hospital
of Jiamusi University. Informed written consent was obtained from
all patients enrolled in the present study.
Cell culture
A normal human pancreatic cell line HPDE6c7 was
purchased from The American Type Culture Collection (Manassas, VA,
USA). A total of three human pancreatic cancer cell lines, Panc-1,
Sw1990 and Bxpc-3, were purchased from Shanghai Institute of
Biochemistry and Cell Biology (Shanghai, China). All these cell
lines were cultured at 37°C under 5% CO2, and were
maintained in Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% v/v fetal bovine serum (FBS), 100 U/ml
penicillin and 100 U/ml streptomycin (all from Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
Transfection
miR-584 mimics, negative control miRNA mimics
(miR-NC), small interfering RNA (siRNA) targeting CCND1 (si-CCND1)
and negative control siRNA (si-NC) were chemically synthesised by
Guangzhou RiboBio Co., Ltd. (Guangzhou, China). The miR-584 mimics
sequence was 5′-UUAUGGUUUGCCUGGGACUGAG-3′ and the miR-NC sequence
was 5′-UUCUCCGAACGUGUCACGUTT-3′. The si-CCND1 sequence was
5′-CUACCUGGACCGUUUCUUGUU-3′ and the si-NC sequence was
5′-UUCUCCGAACGUGUCACGUUU-3′. The cytomegalovirus promoter
(pCMV)-CCND1 and empty pCMV plasmids were ordered from Amspring
(Changsha, China). Cells were seeded (7×105 cells/well)
into six-well plates and cultured to 60–70% confluence. Prior to
transfection, the culture medium was replaced with fresh FBS-free
DMEM. Cells were transfected with miR-584 mimics (100 pmol), miR-NC
(100 pmol), si-CCND1 (100 pmol), si-NC (100 pmol), pCMV-CCDN1 (4
µg) or pCMV (4 µg) using Lipofectamine® 2000 Reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Transfected cells were incubated at 37°C
with 5% CO2 for 8 h. Subsequently, the culture medium
was replaced with 10% v/v FBS-containing DMEM. The efficiency of
miR-584 mimics and pCMV-CCND1 transfection was determined using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blot analysis, respectively.
RT-qPCR
Total RNA of the tissues or cultured cells was
isolated using TRIzol® (Thermo Fisher Scientific, Inc.)
reagent, according to the manufacturer's protocol. For the analysis
of miR-584 expression, synthesis of miR-584 cDNA was conducted
using a TaqMan™ miRNA RT kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.) with the following parameters: 16°C for 30
min, 42°C for 30 min and 85°C for 5 min. Subsequently, qPCR was
conducted using the Applied Biosystems 7500 Sequence Detection
System (Thermo Fisher Scientific, Inc.) using a TaqMan miRNA PCR
kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) and qPCR
was performed as follows: 50°C for 2 min, 95°C for 10 min; followed
by 40 cycles of denaturation at 95°C for 15 sec; and
annealing/extension at 60°C for 60 sec. For the detection of CCND1
mRNA expression, total RNA was subjected to RT for the synthesis of
cDNA using a PrimeScript RT Reagent kit (Takara Biotechnology Co.,
Ltd., Dalian, China) as follows: 37°C for 15 min and 85°C for 5
sec. A SYBR Premix Ex Taq kit (Takara Biotechnology Co.,
Ltd.) was used to conduct the PCR amplification. qPCR was performed
with thermocycling conditions as follows: initial denaturation for
5 min at 95°C, followed by 40 cycles of 95°C for 30 sec and 65°C
for 45 sec. U6 small nuclear RNA and GAPDH served as the endogenous
controls for miR-584 and CCND1 mRNA, respectively. Data were
analysed using the 2−ΔΔCq method (19). The primers were designed as
follows: miR-584, forward 5′-TGCAATGTGTGTGTTAGCCA-3′, reverse
5′-ATCATTGCTCCTTGGATGGT-3′; U6, forward 5′-CTCGCTTCGGCAGCACA-3′,
reverse 5′-AACGCTTCACGAATTTGCGT-3′; CCND1, forward
5′-GACGGCCGAGAAGCTGTGCA-3′, reverse 5′-GCCACCATGGAGGGCGGATT-3′; and
GAPDH, forward 5′-CGGAGTCAACGGATTTGGTCGTAT-3′, reverse
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′.
Cell Counting Kit-8 (CCK-8) assay
Transfected cells were collected at 24 h
post-transfection and were inoculated into 96-well plates at a
density of 3,000 cells/well. Following culturing for 0, 24, 48 and
72 h, CCK-8 assays were performed to determine cell proliferation.
In total, 10 µl CCK-8 reagent (Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany) was added into each well at each time point.
The cells were further incubated at 37°C under 5% CO2
for 2 h, and the absorbance of each well was detected at 450 nm
wavelength using a microplate reader (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Matrigel invasion assay
The invasive ability of transfected cells was
evaluated using 24-well Transwell chambers that were precoated with
Matrigel (Becton, Dickinson and Company, Franklin Lakes, NJ, USA).
Transfected cells were incubated for 48 h and subsequently
harvested and washed twice with FBS-free DMEM. A total of
1×105 transfected cells, suspended in FBS-free DMEM,
were added to the upper Transwell chambers. In total, 500 µl DMEM
containing 20% FBS was added to the lower chambers as a
chemoattractant. Following 24 h of incubation at 37°C with 5%
CO2, cotton swabs were used to remove the non-invasive
cells remaining on the upper surface of the membranes. The invasive
cells were fixed with 100% methanol at room temperature for 20 min
and stained with 0.1% crystal violet at room temperature for 20
min. The number of invasive cells was counted in five randomly
chosen fields under an inverted light (Olympus Corporation, Tokyo,
Japan; magnification, ×200).
Bioinformatics prediction
TargetScan (http://www.targetscan.org) and Pictar (http://www.pictar.org/) were used to predict the
potential targets of miR-584.
Luciferase reporter gene assay
The 3′-UTRs of CCND1 containing the wild-type (WT)
or mutant (Mut) target sequences of miR-584 were amplified by
Shanghai GenePharma Co., Ltd. (Shanghai, China), inserted
downstream of the pMIR-REPORT™ Luciferase Plasmid
(Thermo Fisher Scientific, Inc.) and defined as WT-CCND1-3′-UTR and
Mut-CCND1-3′-UTR, respectively. For reporter assays,
WT-CCND1-3′-UTR or Mut-CCND1-3′-UTR, together with miR-584 mimics
or miR-NC were transfected into Panc-1 and Sw1990 cells using
Lipofectamine® 2000 reagent. Luciferase activity was
determined 48 h following transfection using the
Dual-Luciferase® Reporter Assay System (Promega
Corporation, Madison, WI, USA) according to the manufacturer's
protocol. Firefly luciferase activity was normalised to
Renilla luciferase activity.
Western blot analysis
Tissue specimens or cells were lysed in cold
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.) to isolate the total protein, which was subsequently
quantified using the Bicinchoninic Acid Protein Assay kit (Pierce;
Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocol. Subsequently, equal amounts of protein (30 µg) were
separated by 10% SDS-PAGE and transferred onto polyvinylidene
difluoride membranes, followed by blocking with 5% skimmed milk
that was dissolved in Tris-buffered saline containing 0.1% Tween-20
(TBST) for 2 h at room temperature. The membranes were incubated
overnight at 4°C with rabbit anti-human CCND1 monoclonal antibody
(1:1,000; cat. no. ab16663; Abcam, Cambridge, UK) or rabbit
anti-human GAPDH monoclonal antibody (1:1,000; cat. no. ab128915;
Abcam). The membranes were washed three times with TBST and
subsequently probed with goat anti-rabbit horseradish
peroxidase-conjugated immunoglobulin G secondary antibody (1:5,000;
cat. no. ab205718; Abcam) at room temperature for 2 h.
Chemiluminescence detection was performed using an enhanced
chemiluminescence kit (Thermo Fisher Scientific, Inc.). GAPDH
served as the loading control. Quantity One software version 4.62
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) was used to analyse
the densitometry.
Statistical analysis
Data are presented as the mean ± standard deviation
from at least three independent experiments and analysed using SPSS
software (version 19.0; IBM Corp., Armonk, NY, USA). Student's
t-test and one-way analysis of variance (ANOVA) were used to
compare the differences between groups. Student-Newman-Keuls test
was used as the post hoc test following ANOVA. P<0.05 was
considered to indicate a statistically significant difference.
Results
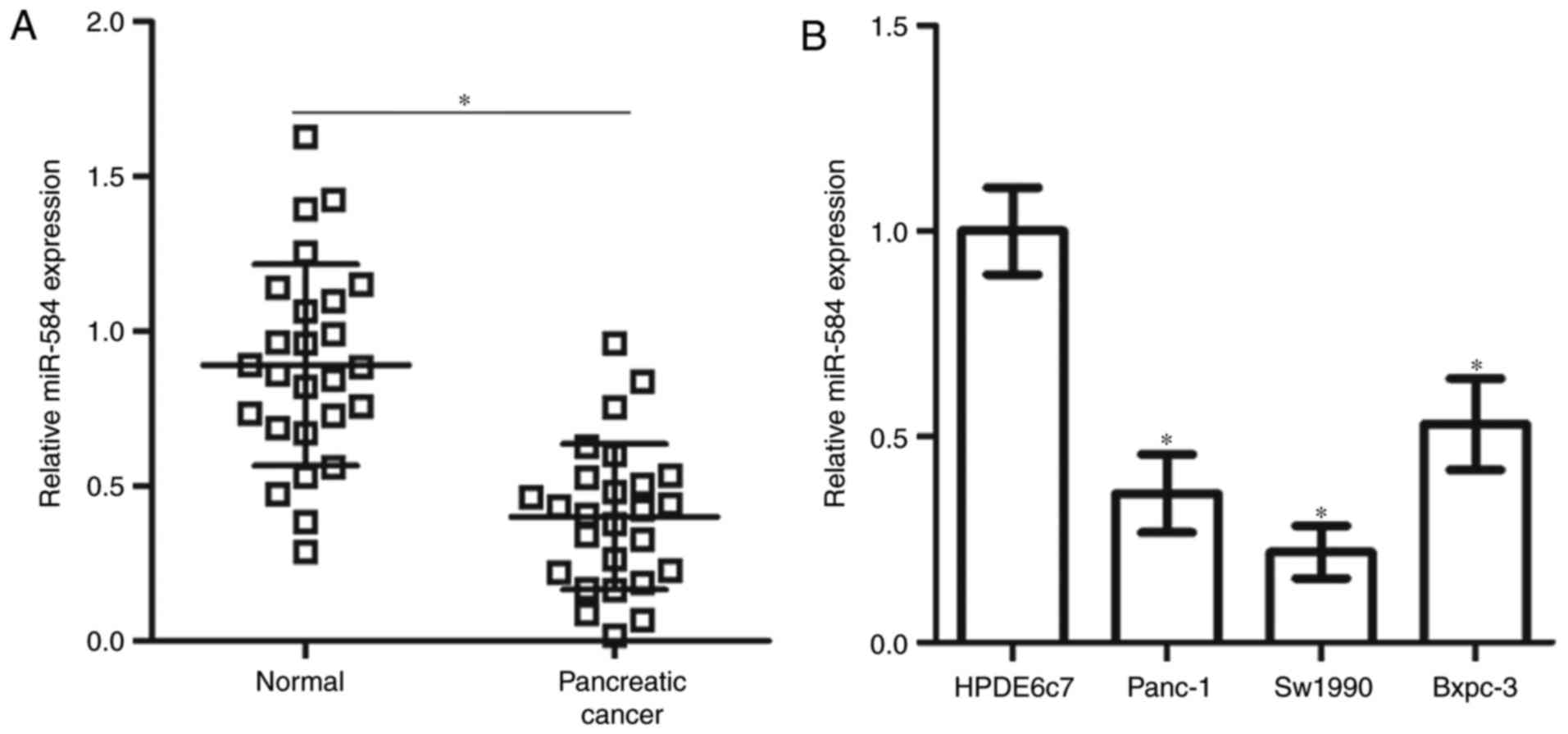
miR-584 expression is decreased in
pancreatic cancer tissues and cell lines
To measure the miR-584 expression level in
pancreatic cancer, RT-qPCR was performed in 26 primary pancreatic
cancer tissues and their matched normal adjacent specimens. miR-584
expression was significantly downregulated in pancreatic cancer
tissues compared with the normal adjacent specimens (Fig. 1A). The expression level of miR-584
was additionally determined in three human pancreatic cancer cell
lines (Panc-1, Sw1990 and Bxpc-3). As depicted in Fig. 1B, the miR-584 expression level was
significantly decreased in all three pancreatic cancer cell lines
compared with the normal human pancreatic cell line HPDE6c7.
Therefore, these findings suggested that miR-584 may be associated
with the development of pancreatic cancer.
miR-584 overexpression suppresses the
proliferation and invasion of pancreatic cancer cells
The significant decrease in miR-584 expression level
in pancreatic cancer suggested that miR-584 may serve tumour
suppressive roles in the progression of pancreatic cancer. To test
this hypothesis, Panc-1 and Sw1990 cell lines that expressed
relatively lower miR-584 expression were selected for the following
functional experiments. miR-584 mimics or miR-NC was transfected
into Panc-1 and Sw1990 cells, and RT-qPCR analysis was used to
evaluate the transfection efficiency. miR-584 expression was
significantly upregulated in Panc-1 and Sw1990 cells transfected
with miR-584 mimics compared with cells transfected with miR-NC
(Fig. 2A). CCK-8 and Matrigel
invasion assays were performed to examine the effects of miR-584
overexpression on cell proliferation and invasion in pancreatic
cancer, respectively. miR-584 overexpression resulted in a
significant decrease in the proliferation at 48 and 72 h (Fig. 2B) and invasion (Fig. 2C) of Panc-1 and Sw1990 cells. The
present results suggested that miR-584 may serve as a tumour
suppressor in pancreatic cancer.
CCND1 is a direct target gene of
miR-584 in pancreatic cancer cells
The mechanism underlying the action of miR-584 in
pancreatic cancer was further examined. Bioinformatics analysis was
conducted to predict the potential targets of miR-584. CCND1, a
predicted target of miR-584 (Fig.
3A), serves crucial roles in pancreatic cancer progression
(20–25) and was thus selected for further
investigation. To test whether the 3′-UTR of CCND1 is directly
targeted by miR-584, luciferase reporter plasmids were constructed
and co-transfected into Panc-1 and Sw1990 cells together with
miR-584 mimics or miR-NC. The overexpression of miR-584
significantly decreased the luciferase activity of WT-CCND1-3′-UTR;
however, had no effect on Mut-CCND1-3′-UTR in Panc-1 and Sw1990
cells (Fig. 3B). To test whether
miR-584 may affect endogenous CCND1 expression, RT-qPCR and western
blot analysis were performed to measure CCND1 mRNA and protein
expression levels, respectively, in Panc-1 and Sw1990 cells that
were transfected with miR-584 mimics or miR-NC. The overexpression
of miR-584 significantly decreased CCND1 mRNA (Fig. 3C) and protein (Fig. 3D) expression levels in Panc-1 and
Sw1990 cells. The present results demonstrated that CCND1 was
directly targeted by miR-584 in pancreatic cancer cells.
CCND1 knockdown mimics the suppressive
effect of miR-584 in pancreatic cancer cells
To elucidate the roles of CCND1 in pancreatic
cancer, a knockdown of endogenous CCND1 expression was performed in
Panc-1 and Sw1990 cells via transfection with si-CCND1. Western
blot analysis demonstrated that the CCND1 protein was significantly
downregulated in Panc-1 and Sw1990 cells following transfection
with si-CCND1 (Fig. 4A).
Functional experiments indicated that the inhibition of CCND1
decreased the proliferative (Fig. 4B
and C) and invasive ability (Fig.
4D) of Panc-1 and Sw1990 cells, mimicking miR-584
overexpression. The present results suggested that miR-584 directly
targeted CCND1 in pancreatic cancer cells.
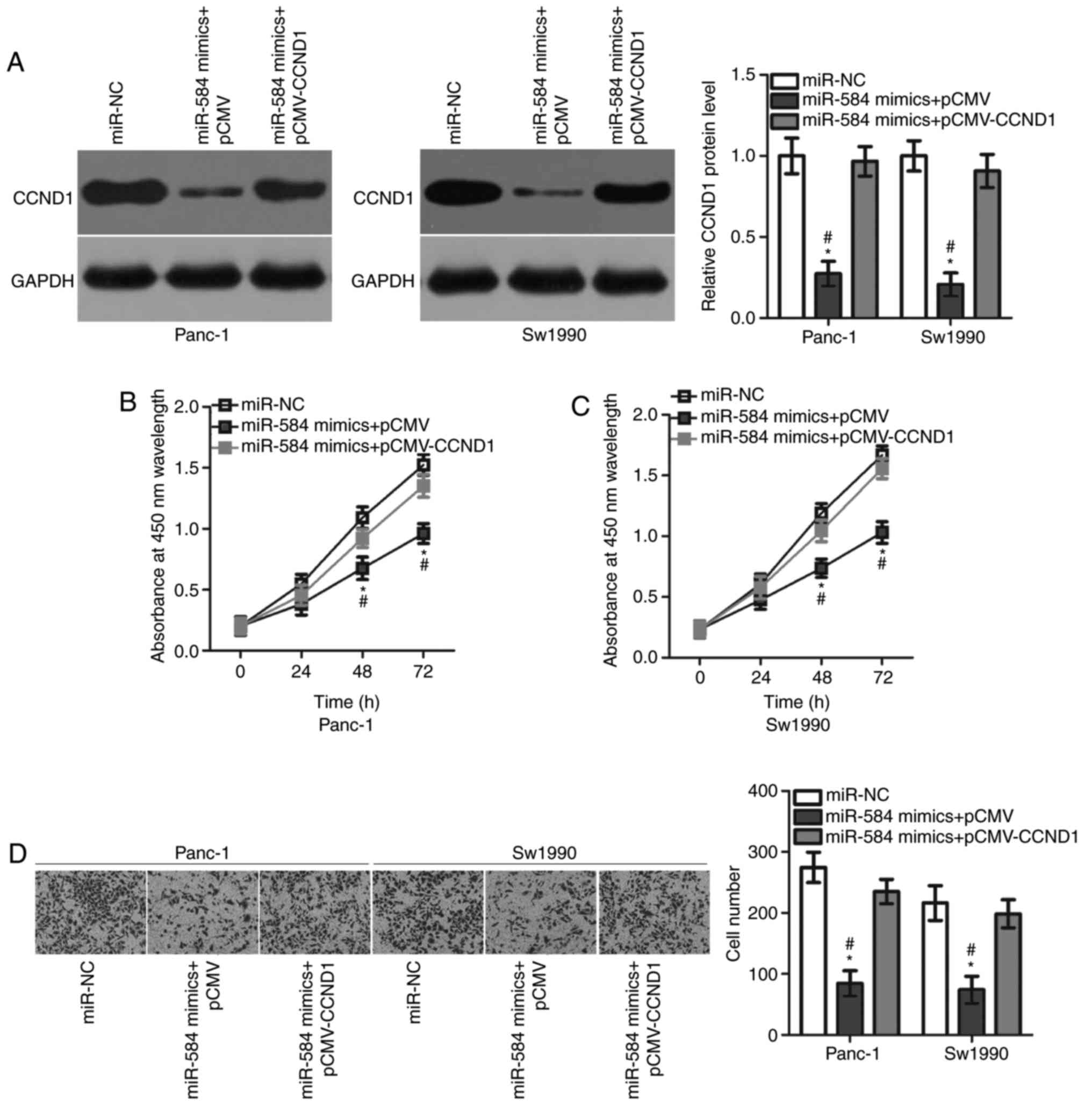
CCND1 overexpression attenuates the
suppressive roles of miR-584 in pancreatic cancer cells
Rescue experiments were performed to test the role
of CCND1 in mediating the action of miR-584 in pancreatic cancer
cells. Panc-1 and Sw1990 cells were transfected with miR-584
mimics, in combination with pCMV-CCND1 or empty pCMV plasmids.
Western blot analysis demonstrated that the CCND1 overexpressing
plasmid pCMV-CCND1 was able to effectively restore the CCND1
protein expression level in Panc-1 and Sw1990 cells (Fig. 5A). In addition, CCK-8 and Matrigel
invasion assays demonstrated that restoring CCND1 expression
inhibited the suppressive effects of miR-584 overexpression on the
proliferation (Fig. 5B and C) and
invasion (Fig. 5D) of Panc-1 and
Sw1990 cells. The present results suggested that miR-584 served
tumour suppressing roles in pancreatic cancer cells by repressing
CCND1 expression.
Discussion
Multiple previous studies have demonstrated that the
dysregulation of miRNAs is implicated in the occurrence and
development of pancreatic cancer (26–28).
Nevertheless, the detailed roles of miRNAs in pancreatic cancer and
their underlying mechanisms of action remain largely unknown.
Hence, the further characterization of deregulated miRNAs in
pancreatic cancer may provide insight into the oncogenesis and
progression of pancreatic cancer, and guide the identification of
novel therapeutic targets for treating patients with this disease.
In the present study, the expression, biological roles and
underlying mechanisms of miR-584 in pancreatic cancer were
examined. The expression of miR-584 in pancreatic cancer was
investigated and it was identified that the miR-584 expression
level was significantly decreased in pancreatic cancer tissues and
cell lines. Notably, ectopic miR-584 expression decreased the
proliferation and invasion of pancreatic cancer cells and CCND1 was
identified as a direct target gene of miR-584 in pancreatic cancer
cells. It was observed that inhibition of CCND1 and miR-584
overexpression led to similar effects on pancreatic cancer cells.
Importantly, restoring CCND1 expression inhibited the suppressive
roles of miR-584 overexpression in pancreatic cancer cells. The
present results demonstrated that miR-584 inhibited pancreatic
cancer progression by directly targeting CCND1, suggesting that
this miRNA may represent a novel potential therapeutic target for
the treatment of patients with pancreatic cancer.
miR-584 has been identified to be differently
expressed in numerous types of human malignancy. miR-584 expression
was downregulated in non-small cell lung cancer, and its
downregulation was strongly associated with tumour size, tumour
node, metastasis stage and distant metastasis (16). miR-584 expression was decreased in
neuroblastoma tissues and cell lines, and was an independent
prognostic biomarker for predicting the prognosis of patients with
neuroblastoma (17). In gastric
cancer, low expression of miR-584 was detected and its expression
was negatively correlated with tumour size (18). Aberrant downregulation of miR-584
was additionally reported in glioma (29) and clear cell renal cell carcinoma
(30). Collectively, numerous
findings suggested that miR-584 may be a valuable biomarker for the
diagnosis and prediction of the therapeutic outcomes of patients
with these specific cancer types.
miR-584 serves tumour suppressive roles in multiple
types of human cancer. Restoration of miR-584 expression suppressed
the proliferative and invasive capabilities of non-small cell lung
cancer cells (16). Xiang et
al (17) observed that miR-584
overexpression prohibited the proliferation, metastasis and
angiogenesis of neuroblastoma cells in vitro and in
vivo. Li et al (18)
demonstrated that the upregulation of miR-584 expression inhibited
gastric cancer cell proliferation and induced apoptosis in
vitro. Wang et al (29)
demonstrated that miR-584 restoration supressed cell growth and
motility, and induced apoptosis of glioma. Ueno et al
(30) demonstrated that miR-584
overexpression attenuated cell migration and invasion in clear cell
renal cell carcinoma. These findings suggested that miR-584 may
serve crucial roles in the tumourigenesis and tumour development of
these human cancer types. Therefore, miR-584 restoration may be an
attractive therapeutic strategy for treating patients with these
specific human cancer types.
Identifying the direct targets of miR-584 may
elucidate the mechanisms underlying its biological roles in human
cancer. Numerous target genes of miR-584 have been previously
identified, including metadherin (16) in non-small cell lung cancer, matrix
metalloproteinase-14 (17) in
neuroblastoma, WW domain containing E3 ubiquitin protein ligase 1
(18) in gastric cancer, pituitary
tumour-transforming gene 1 (29)
in glioma and Rho-associated protein kinase 1 (30) in clear cell renal cell carcinoma.
The CCND1 gene, located on chromosome 11q13, was demonstrated to be
a direct target gene of miR-584 in pancreatic cancer. It is highly
expressed in numerous types of human cancer, including colorectal
cancer (31), breast cancer
(32), lung cancer (33), hepatocellular carcinoma (34) and glioma (35). CCND1 expression was upregulated in
pancreatic cancer tissues and cell lines and the high expression of
CCND1 was correlated with differentiation and prognosis of patients
with pancreatic cancer (20,21).
Additionally, the dysregulation of CCND1 is implicated in the
development of pancreatic cancer by affecting cell growth and
metastasis in vitro and in vivo (22–25).
The present results suggested that the miR-584/CCND1 pathway has
potential therapeutic applications in treating patients with this
malignancy.
In conclusion, miR-584 was downregulated in
pancreatic cancer tissues and cell lines. miR-584 may have tumour
suppressive roles in pancreatic cancer partly by directly targeting
CCND1, suggesting that this miRNA may represent a promising
therapeutic target for treating patients with this aggressive
disease. However, the association between miR-584 and the prognosis
of patients with pancreatic cancer was not analysed, representing a
limitation of the present study, that requires investigation in
future studies.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Heilongjiang
Natural Science Foundation (China; grant no. H2017072) and Key
Topics of Jiamusi University (Jiamusi, China; grant no.
Sz2013-006).
Availability of data and materials
The datasets used and/or analysed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YQ, GC and MH designed the study. GC, MH, XQ and KW
performed the functional assays. KW analysed the data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Research
Ethics Committee of The First Affiliated Hospital of Jiamusi
University (Jiamusi, China) and was performed in accordance with
the Declaration of Helsinki and the guidelines of the Ethics
Committee of The First Affiliated Hospital of Jiamusi University.
Written informed consent was obtained from all patients for the use
of their clinical tissues.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Zhang S, Zhao P, Li G, Wu
L and He J: Report of incidence and mortality in China cancer
registries, 2009. Chin J Cancer Res. 25:10–21. 2013.PubMed/NCBI
|
|
2
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cameron JL, Riall TS, Coleman J and
Belcher KA: One thousand consecutive pancreaticoduodenectomies. Ann
Surg. 244:10–15. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Niedergethmann M, Alves F, Neff JK,
Heidrich B, Aramin N, Li L, Pilarsky C, Grützmann R, Allgayer H,
Post S and Gretz N: Gene expression profiling of liver metastases
and tumour invasion in pancreatic cancer using an orthotopic SCID
mouse model. Br J Cancer. 97:1432–1440. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng H, Wang Y, Su J, Liang H, Zhang CY,
Chen X and Yao W: MicroRNA-148a suppresses the proliferation and
migration of pancreatic cancer cells by down-regulating ErbB3.
Pancreas. 45:1263–1271. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Galasso M, Sandhu SK and Volinia S:
MicroRNA expression signatures in solid malignancies. Cancer J.
18:238–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wald P, Liu XS, Pettit C, Dillhoff M,
Manilchuk A, Schmidt C, Wuthrick E, Chen W and Williams TM:
Prognostic value of microRNA expression levels in pancreatic
adenocarcinoma: A review of the literature. Oncotarget.
8:73345–73361. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sekar D, Krishnan R, Thirugnanasambantham
K, Rajasekaran B, Islam VI and Sekar P: Significance of microRNA 21
in gastric cancer. Clin Res Hepatol Gastroenterol. 40:538–545.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Celano M, Rosignolo F, Maggisano V, Pecce
V, Iannone M, Russo D and Bulotta S: MicroRNAs as biomarkers in
thyroid carcinoma. Int J Genomics. 2017:64965702017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fadejeva I, Olschewski H and Hrzenjak A:
MicroRNAs as regulators of cisplatin-resistance in non-small cell
lung carcinomas. Oncotarget. 8:115754–115773. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Z, Tan Y, Yu W, Zheng S, Zhang S, Sun
L and Ding K: Small role with big impact: MiRNAs as communicators
in the cross-talk between cancer-associated fibroblasts and cancer
cells. Int J Biol Sci. 13:339–348. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim YK, Yu J, Han TS, Park SY, Namkoong B,
Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK and Kim VN: Functional links
between clustered microRNAs: Suppression of cell-cycle inhibitors
by microRNA clusters in gastric cancer. Nucleic Acids Res.
37:1672–1681. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fu F, Wan X, Wang D, Kong Z, Zhang Y,
Huang W, Wang C, Wu H and Li Y: MicroRNA-19a acts as a prognostic
marker and promotes prostate cancer progression via inhibiting
VPS37A expression. Oncotarget. 9:1931–1943. 2017.PubMed/NCBI
|
|
16
|
Zhang Y, Wang Y and Wang J: MicroRNA-584
inhibits cell proliferation and invasion in non-small cell lung
cancer by directly targeting MTDH. Exp Ther Med. 15:2203–2211.
2018.PubMed/NCBI
|
|
17
|
Xiang X, Mei H, Qu H, Zhao X, Li D, Song
H, Jiao W, Pu J, Huang K, Zheng L and Tong Q: miRNA-584-5p exerts
tumor suppressive functions in human neuroblastoma through
repressing transcription of matrix metalloproteinase 14. Biochim
Biophys Acta. 1852:1743–1754. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Q, Li Z, Wei S, Wang W, Chen Z, Zhang
L, Chen L, Li B, Sun G, Xu J, et al: Overexpression of miR-584-5p
inhibits proliferation and induces apoptosis by targeting WW
domain-containing E3 ubiquitin protein ligase 1 in gastric cancer.
J Exp Clin Cancer Res. 36:592017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gansauge S, Gansauge F, Ramadani M, Stobbe
H, Rau B, Harada N and Beger HG: Overexpression of cyclin D1 in
human pancreatic carcinoma is associated with poor prognosis.
Cancer Res. 57:1634–1637. 1997.PubMed/NCBI
|
|
21
|
Li YJ and Ji XR: Relationship between the
expression of beta-cat, cyclin D1 and c-myc and the occurance and
biological behavior of pancreatic cancer. Zhonghua Bing Li Xue Za
Zhi. 32:238–241. 2003.(In Chinese). PubMed/NCBI
|
|
22
|
Xu Y, Liu T and Gao J: Effects of
antisense cyclin D1 expressing vector on the cell growth and
apoptosis of pancreatic carcinoma. Zhonghua Bing Li Xue Za Zhi.
27:348–351. 1998.(In Chinese). PubMed/NCBI
|
|
23
|
Yan L, Wang Y, Wang ZZ, Rong YT, Chen LL,
Li Q, Liu T, Chen YH, Li YD, Huang ZH and Peng J: Cell motility and
spreading promoted by CEACAM6 through cyclin D1/CDK4 in human
pancreatic carcinoma. Oncol Rep. 35:418–426. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee Y, Ko D, Min HJ, Kim SB, Ahn HM, Lee Y
and Kim S: TMPRSS4 induces invasion and proliferation of prostate
cancer cells through induction of Slug and cyclin D1. Oncotarget.
7:50315–50332. 2016.PubMed/NCBI
|
|
25
|
Zhang Y, Su Y, Zhao Y, Lv G and Luo Y:
MicroRNA720 inhibits pancreatic cancer cell proliferation and
invasion by directly targeting cyclin D1. Mol Med Rep.
16:9256–9262. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao W, Jin H, Zhang L, Chen X and Qian H:
Identification of miR-601 as a novel regulator in the development
of pancreatic cancer. Biochem Biophys Res Commun. 483:638–644.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Le Large TY, Meijer LL, Prado MM, Kazemier
G, Frampton AE and Giovannetti E: Circulating microRNAs as
diagnostic biomarkers for pancreatic cancer. Expert Rev Mol Diagn.
15:1525–1529. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takikawa T, Masamune A, Yoshida N, Hamada
S, Kogure T and Shimosegawa T: Exosomes derived from pancreatic
stellate cells: MicroRNA signature and effects on pancreatic cancer
cells. Pancreas. 46:19–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang XP, Deng XL and Li LY: MicroRNA-584
functions as a tumor suppressor and targets PTTG1IP in glioma. Int
J Clin Exp Pathol. 7:8573–8582. 2014.PubMed/NCBI
|
|
30
|
Ueno K, Hirata H, Shahryari V, Chen Y,
Zaman MS, Singh K, Tabatabai ZL, Hinoda Y and Dahiya R: Tumour
suppressor microRNA-584 directly targets oncogene Rock-1 and
decreases invasion ability in human clear cell renal cell
carcinoma. Br J Cancer. 104:308–315. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Y, Wei J, Xu C, Zhao Z and You T:
Prognostic significance of cyclin D1 expression in colorectal
cancer: A meta-analysis of observational studies. PLoS One.
9:e945082014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li X, Huo X, Li W, Yang Q, Wang Y and Kang
X: Genetic association between cyclin D1 polymorphism and breast
cancer susceptibility. Tumour Biol. 35:11959–11965. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Betticher DC, Heighway J, Hasleton PS,
Altermatt HJ, Ryder WD, Cerny T and Thatcher N: Prognostic
significance of CCND1 (cyclin D1) overexpression in primary
resected non-small-cell lung cancer. Br J Cancer. 73:294–300. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lu JW, Lin YM, Chang JG, Yeh KT, Chen RM,
Tsai JJ, Su WW and Hu RM: Clinical implications of deregulated CDK4
and Cyclin D1 expression in patients with human hepatocellular
carcinoma. Med Oncol. 30:3792013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xu Z, Zeng X, Tian D, Xu H, Cai Q, Wang J
and Chen Q: MicroRNA-383 inhibits anchorage-independent growth and
induces cell cycle arrest of glioma cells by targeting CCND1.
Biochem Biophys Res Commun. 453:833–838. 2014. View Article : Google Scholar : PubMed/NCBI
|