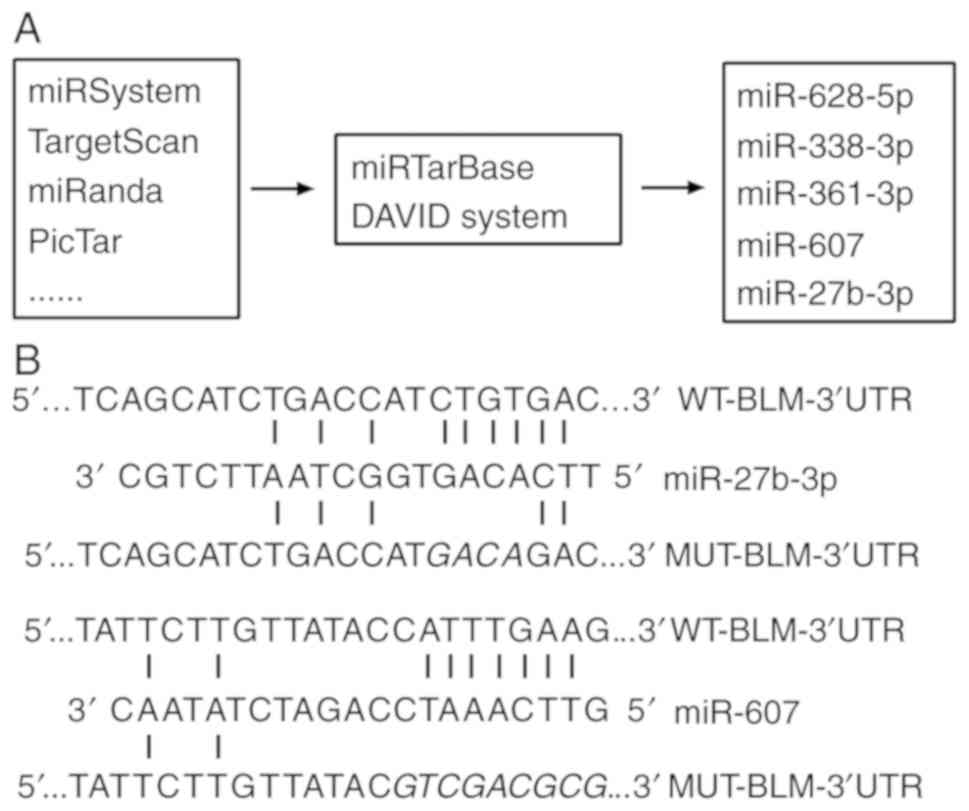
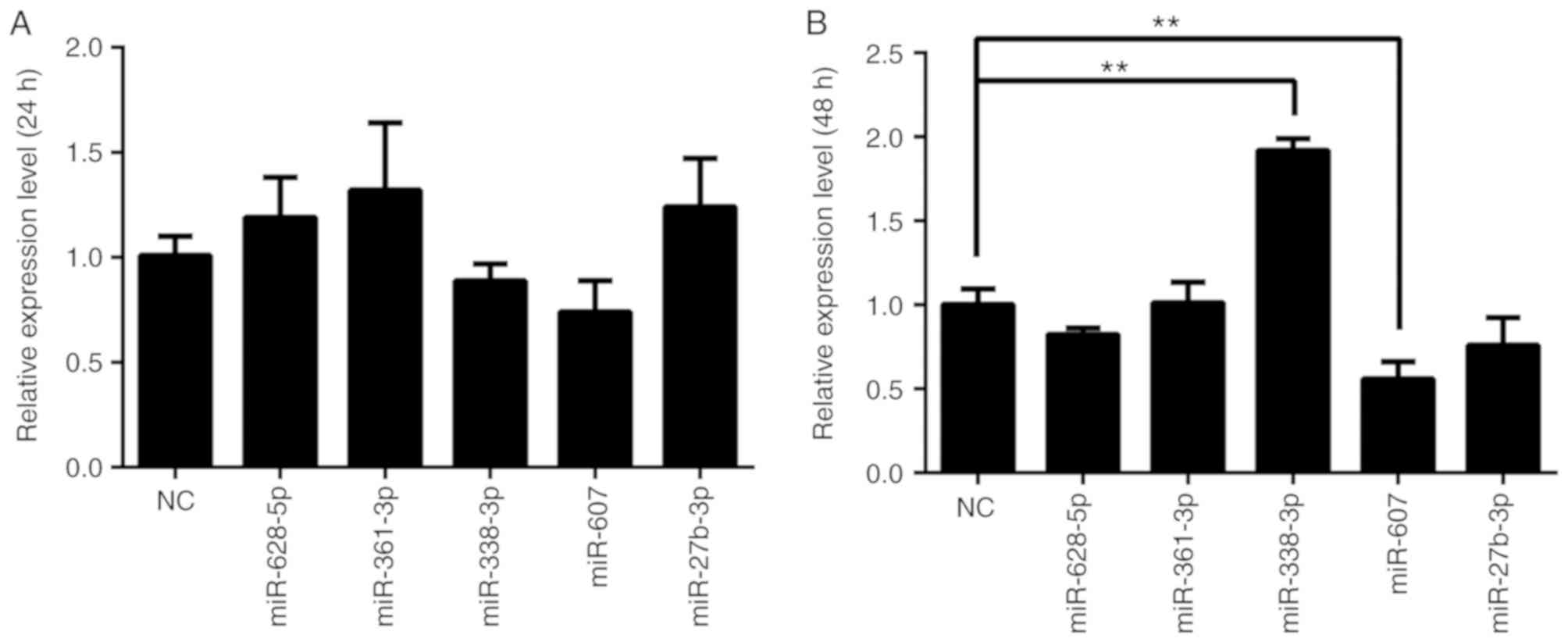
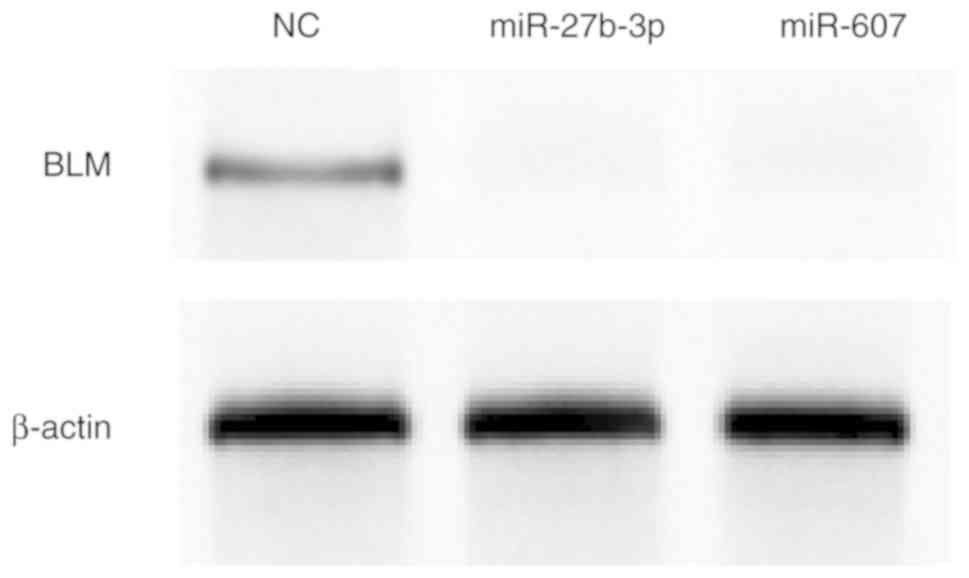
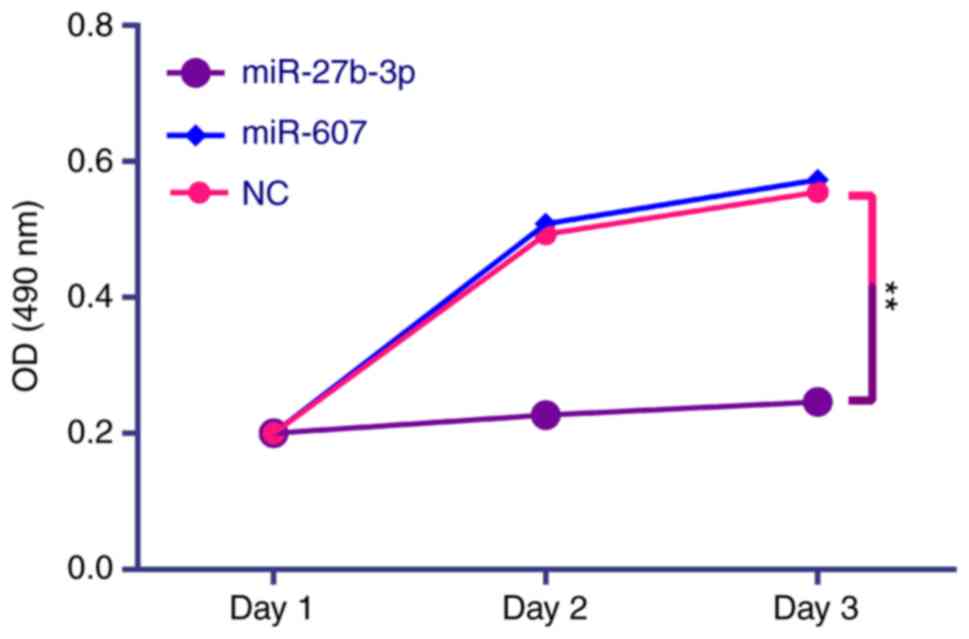
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pandita A, Manvati S, Singh SK, Vaishnavi
S and Bamezai RN: Combined effect of microRNA, nutraceuticals and
drug on pancreatic cancer cell lines. Chem Biol Interact.
233:56–64. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shi S, Han L, Deng L, Zhang Y, Shen H,
Gong T, Zhang Z and Sun X: Dual drugs (microRNA-34a and
paclitaxel)-loaded functional solid lipid nanoparticles for
synergistic cancer cell suppression. J Control Release.
194:228–237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Futami K, Ogasawara S, Goto H, Yano H and
Furuichi Y: RecQL1 DNA repair helicase: A potential tumor marker
and therapeutic target against hepatocellular carcinoma. Int J Mol
Med. 25:537–545. 2010.PubMed/NCBI
|
|
5
|
Qian X, Feng S, Xie D, Feng D, Jiang Y and
Zhang X: RecQ helicase BLM regulates prostate cancer cell
proliferation and apoptosis. Oncol Lett. 14:4206–4212. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bachrati CZ and Hickson ID: RecQ
helicases: Suppressors of tumorigenesis and premature aging.
Biochem J. 374:577–606. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sharma S, Doherty KM and Brosh RM Jr:
Mechanisms of RecQ helicases in pathways of DNA metabolism and
maintenance of genomic stability. Biochem J. 398:319–337. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wu L: Role of the BLM helicase in
replication fork management. DNA Repair (Amst). 6:936–944. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nguyen GH, Dexheimer TS, Rosenthal AS, Chu
WK, Singh DK, Mosedale G, Bachrati CZ, Schultz L, Sakurai M,
Savitsky P, et al: A small molecule inhibitor of the BLM helicase
modulates chromosome stability in human cells. Chem Biol. 20:55–62.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Laitman Y, Boker-Keinan L, Berkenstadt M,
Liphsitz I, Weissglas-Volkov D, Ries-Levavi L, Sarouk I, Pras E and
Friedman E: The risk for developing cancer in Israeli ATM, BLM, and
FANCC heterozygous mutation carriers. Cancer Genet. 209:70–74.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Voer RM, Hahn MM, Mensenkamp AR,
Hoischen A, Gilissen C, Henkes A, Spruijt L, van Zelst-Stams WA,
Kets CM, Verwiel ET, et al: Deleterious germline BLM mutations and
the risk for early-onset colorectal cancer. Sci Rep. 5:140602015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Böhm S and Bernstein KA: The role of
post-translational modifications in fine-tuning BLM helicase
function during DNA repair. DNA Repair (Amst). 22:123–132. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Suspitsin EN, Yanus GA, Sokolenko AP,
Yatsuk OS, Zaitseva OA, Bessonov AA, Ivantsov AO, Heinstein VA,
Klimashevskiy VF, Togo AV and Imyanitov EN: Development of breast
tumors in CHEK2, NBN/NBS1 and BLM mutation carriers does not
commonly involve somatic inactivation of the wild-type allele. Med
Oncol. 31:8282014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sassi A, Popielarski M, Synowiec E,
Morawiec Z and Wozniak K: BLM and RAD51 genes polymorphism and
susceptibility to breast cancer. Pathol Oncol Res. 19:451–459.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matsuyama R, Okuzaki D, Okada M and
Oneyama C: MicroRNA-27b suppresses tumor progression by regulating
ARFGEF1 and focal adhesion signaling. Cancer Sci. 107:28–35. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen C, Liu X, Ma H, Zhang W and Li H:
miR-338-3p suppresses tumor growth of ovarian epithelial carcinoma
by targeting Runx2. Int J Oncol. 46:2277–2285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen D, Si W, Shen J, Du C, Lou W, Bao C,
Zheng H, Pan J, Zhong G, Xu L, et al: miR-27b-3p inhibits
proliferation and potentially reverses multi-chemoresistance by
targeting CBLB/GRB2 in breast cancer cells. Cell Death Dis.
9:1882018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sui GQ, Fei D, Guo F, Zhen X, Luo Q, Yin S
and Wang H: MicroRNA-338-3p inhibits thyroid cancer progression
through targeting AKT3. Am J Cancer Res. 7:1177–1187.
2017.PubMed/NCBI
|
|
20
|
Hara ES, Ono M, Eguchi T, Kubota S, Pham
HT, Sonoyama W, Tajima S, Takigawa M, Calderwood SK and Kuboki T:
miRNA-720 controls stem cell phenotype, proliferation and
differentiation of human dental pulp cells. PLoS One. 8:e835452013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu S, Yi XM, Zhang ZY, Ge JP and Zhou WQ:
miR-129 predicts prognosis and inhibits cell growth in human
prostate carcinoma. Mol Med Rep. 14:5025–5032. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang G, Tian X, Li Y, Wang Z, Li X and
Zhu C: miR-27b and miR-34a enhance docetaxel sensitivity of
prostate cancer cells through inhibiting epithelial-to-mesenchymal
transition by targeting ZEB1. Biomed Pharmacother. 97:736–744.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee
DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al:
Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic
Acids Res. 33:e1792005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Braoudaki M, Lambrou GI, Giannikou K,
Milionis V, Stefanaki K, Birks DK, Prodromou N, Kolialexi A,
Kattamis A, Spiliopoulou CA, et al: Microrna expression signatures
predict patient progression and disease outcome in pediatric
embryonal central nervous system neoplasms. J Hematol Oncol.
7:962014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Favreau AJ and Sathyanarayana P:
miR-590-5p, miR-219-5p, miR-15b and miR-628-5p are commonly
regulated by IL-3, GM-CSF and G-CSF in acute myeloid leukemia. Leuk
Res. 36:334–341. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu X, Zheng Y, Han B and Dong X: Long
noncoding RNA BLACAT1 modulates ABCB1 to promote oxaliplatin
resistance of gastric cancer via sponging miR-361. Biomed
Pharmacother. 99:832–838. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang S, Liu Z, Wu L and Wang Y: MiR-361
targets Yes-associated protein (YAP) mRNA to suppress cell
proliferation in lung cancer. Biochem Biophys Res Commun.
492:468–473. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chiyomaru T, Seki N, Inoguchi S, Ishihara
T, Mataki H, Matsushita R, Goto Y, Nishikawa R, Tatarano S, Itesako
T, et al: Dual regulation of receptor tyrosine kinase genes EGFR
and c-Met by the tumor-suppressive microRNA-23b/27b cluster in
bladder cancer. Int J Oncol. 46:487–496. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ning S, Xu H, Al-Shyoukh I, Feng J and Sun
R: An application of a Hill-based response surface model for a drug
combination experiment on lung cancer. Stat Med. 33:4227–4236.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiao Q, Wang L and Xu H: Application of
kriging models for a drug combination experiment on lung cancer.
Stat Med. 38:236–246. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li C, Li L, Zhou HH, Xia C and He L:
Improving Yield of 1,3-diglyceride by whole-cell lipase fromA.
NigerGZUF36 catalyzed glycerolysis via medium optimization. J Braz
Chem Soc. 26:247–254. 2015.
|
|
32
|
Fang HB, Ross DD, Sausville E and Tan M:
Experimental design and interaction analysis of combination studies
of drugs with log-linear dose responses. Stat Med. 27:3071–3083.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Y, Shi B, Chen J, Hu L and Zhao C:
MiR-338-3p targets pyruvate kinase M2 and affects cell
proliferation and metabolism of ovarian cancer. Am J Transl Res.
8:3266–3273. 2016.PubMed/NCBI
|
|
34
|
Mezlini AM, Wang B, Deshwar A, Morris Q
and Goldenberg A: Identifying cancer specific functionally relevant
miRNAs from gene expression and miRNA-to-gene networks using
regularized regression. PLoS One. 8:e731682013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rosenthal AS, Dexheimer TS, Nguyen G,
Gileadi O, Vindigni A, Simeonov A, Jadhav A, Hickson I and Maloney
DJ: Discovery of ML216, a small molecule inhibitor of bloom (BLM)
helicase. Probe Reports from the NIH Molecular Libraries Program;
Bethesda (MD): 2010
|
|
36
|
Gupta A, Ahmad A, Singh H, Kaur S, K M N,
Ansari MM, Jayamurugan G and Khan R: Nanocarrier composed of
magnetite core coated with three polymeric shells mediates LCS-1
delivery for synthetic lethal therapy of BLM-defective colorectal
cancer cells. Biomacromolecules. 19:803–815. 2018. View Article : Google Scholar : PubMed/NCBI
|