Introduction
Psoriasis, as characterized by a well-demarcated
erythematous plaque with silver scales, is a chronic,
immune-mediated disorder that mainly affects the skin and joints
(1). The worldwide prevalence
rates of psoriasis range from 0.9–8.5% in adults and 0–2.1% in
children (2). Although this
condition rarely poses a threat to life, the irritation, pain and
especially the aberrant appearance make these patients susceptible
to psychological problems, such as anxiety and depression (3,4).
With recent advances in the understanding of psoriasis, an
increasing number of therapies have emerged, however a high
recurrence rate persists. Therefore, it is important to better
understand the underlying molecular pathogenesis of psoriasis in
order to identify more effective therapeutic approaches for the
control of psoriasis development and progression.
Gene expression microarrays have been widely applied
in psoriatic research and represent an important new tool for use
in the identification of disease-related molecules associated with
psoriasis. Recently, comprehensive analysis of microarray data from
multiple centers has become a popular research area. Ainali et
al investigated gene expression patterns in lesional and
non-lesional psoriatic tissue samples from 2 GEO data sets to
establish a molecular sub-groups within the clinical phenotype of
plaque psoriasis (5). Mei and Mei
screened differentially expressed genes based on 4 psoriatic data
sets followed by characterization of gene functions and mutual
interactions (6). Sevimoglu and
Arga analyzed and integrated data from 12 studies to identify the
potential candidates for disease biomarkers and therapeutic targets
(7).
However, analysis of the unpaired data obtained from
lesional and non-lesional samples may lead to a potential bias
caused by disease heterogeneity. In order to eliminate or reduce
such bias, only paired lesional and the corresponding non-lesional
skin samples were selected and analyzed in this study. Information
was compiled from 5 original microarray data sets, GSE14905
(8), GSE30999 (9,10),
GSE34248 (11), GSE41662 (11) and GSE53552 (12), from the Gene Expression Omnibus
(GEO) database. A total of 175 pairs of lesional and non-lesional
skin samples from plaque psoriatic patients were selected. With use
of bioinformatic methods, integrated differentially expressed genes
(DEGs) were identified, followed by the Gene ontology (GO) and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment.
Protein-protein interaction (PPI) analysis and hug gene calculation
were subsequently performed. Finally, an additional GEO data set,
GSE75343 (13), which contained a
study of gene expression levels in scalp psoriatic patients, was
used as a means to validate whether the hub genes obtained from the
aforementioned databases exhibited a similar expression profile as
that in scalp psoriatic lesions.
Through integration of the bioinformatic analyses of
the gene expression from these 175 pairs of psoriatic skin samples,
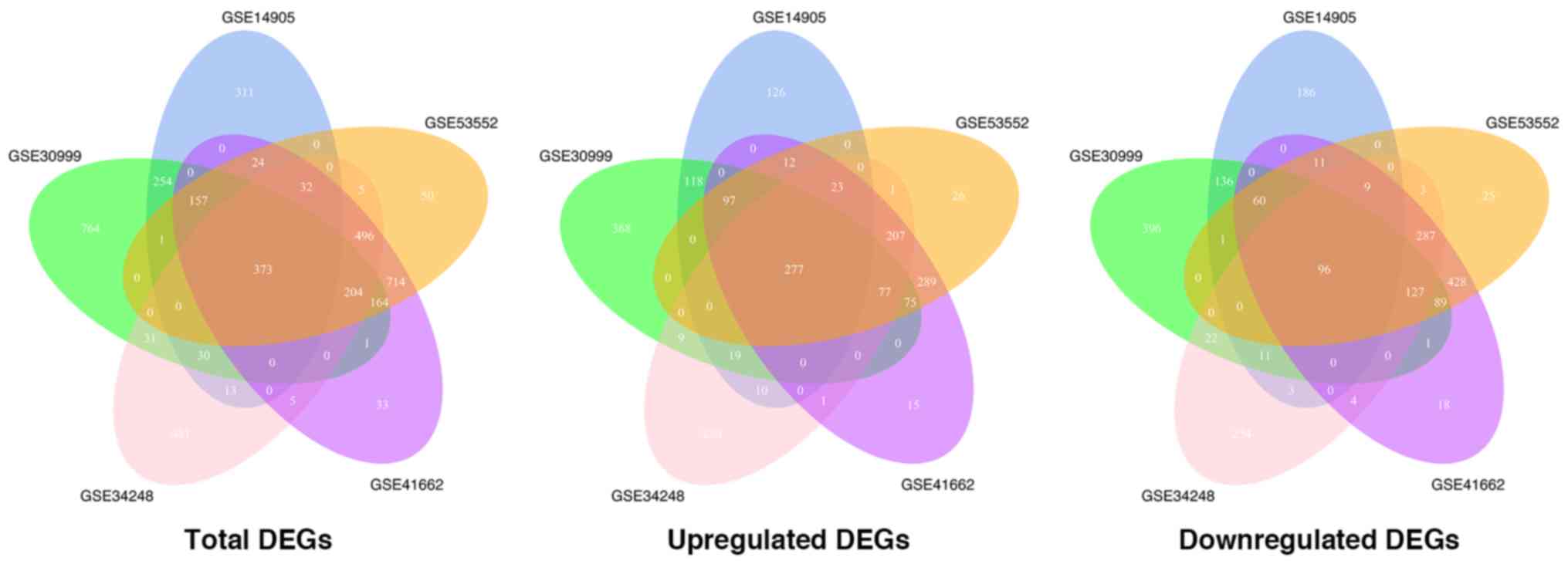
377 genes were identified as DEGs, with 277 of these genes being
upregulated and 96 genes downregulated. We revealed that these
genes covered a wide range of biological functions in epidermal
development, keratinization, immune responses, metabolic pathways,
cell cycle and extracellular spaces. These results provide a
comprehensive understanding of the molecular pathogenesis of the
disease, which may guide subsequent studies on psoriasis
research.
Materials and methods
Microarray data sets and data
calibration
Using the keyword ‘psoriasis’, data sets using the
descriptors ‘paired biopsy from both lesional and non-lesional
skin’ and ‘pre-treatment status’ were screened. The raw files of 5
enrolled microarray data sets, including GSE14905 (8), GSE30999 (9,10),
GSE34248 (11), GSE41662 (11) and GSE53552 (12) (Table
I), were downloaded from the NCBI GEO database (https://www.ncbi.nlm.nih.gov/geo/). In each data
set, only pre-treatment psoriatic skin samples and their matched
adjacent normal samples were selected, which resulted in 175 pairs
of skin samples from psoriatic patients for subsequent analysis.
The raw files were processed with R software 3.5.1 (https://www.r-project.org) to convert the gene probe
IDs to gene symbol codes. Finally, calibrations of gene expression
levels according to the quartile method were performed for
subsequent analysis.
 | Table I.Information for psoriatic GEO
data. |
Table I.
Information for psoriatic GEO
data.
| GEO series | Platform | Sample | Type | Pair no. | (Refs.) |
|---|
| GSE14905 | GPL570 | Paired LS and
NLS | Plaque
psoriasis | 28 | Yao et al
(8) |
| GSE30999 | GPL570 | Paired LS and
NLS | Moderate to severe
plaque psoriasis | 85 | Suárez-Fariñas
et al (9) |
| GSE34248 | GPL570 | Paired LS and
NLS | Mild to moderate
plaque psoriasis | 14 | Bigler et al
(11) |
| GSE41662 | GPL570 | Paired LS and
NLS | Moderate to severe
plaque psoriasis | 24 | Bigler et al
(11) |
| GSE53552 | GPL570 | Paired LS and
NLS | Moderate to severe
plaque psoriasis | 24 | Russell et
al (12) |
| GSE75343 | GPL570 | Paired scalp LS and
scalp NLS | Moderate to severe
plaque psoriasis with scalp involvement | 13 | Ruano et al
(13) |
DEGs analysis and integration
A differential expression analysis on each GEO
series, as based on paired-sample t-tests between psoriatic skin
and adjacent normal skin samples, were performed with use of R
software. A gene was defined as a differentially expressed gene
between the psoriatic and normal sample when the P-value was
<0.05 (P<0.05) and the gene expression fold change (FC) value
was >2 or <0.5 (|log2FC|≥1), which were illustrated as
Volcano plots. An overlap of total, upregulated or downregulated
DEGs, plotted as Venn charts, from all 5 data sets were listed for
subsequent function analysis.
GO term and KEGG pathway analysis of
DEGs
The DAVID knowledgebase (https://david.ncifcrf.gov/), an online gene functional
annotation tool, was used to analyze the function and pathway
enrichment of candidate genes obtained (14). With this technique, the Fisher
exact test P-value was calculated as a result of enrichment degree.
The top 10 enrichment GO term or KEGG pathway annotations for both
up- or downregulated DEGs obtained in our study were listed.
PPI network and hub gene analyses
The STRING platform, an online tool used for the
structural and functional analysis of protein interactions
(15), was used to identify
interactions among proteins of interest. The corresponding results
were analyzed and structured with the use of the Cytoscape software
3.6.1 (https://cytoscape.org). The hub genes,
which were considered to be involved in playing pivotal regulatory
roles in the PPI network, were subsequently calculated based on the
overlapping results obtained by MCC (Maximal Clique Centrality) and
DMNC (Density of Maximum Neighborhood Component) topological
analysis methods, respectively, with use of the cytoHubba app built
in the software (16).
GEO2R analysis of gene expression
levels
The gene expression levels of hub genes were
analyzed in GSE75343 (13), a
microarray data set comparing gene expression levels of scalp
psoriatic skin and adjacent normal skin samples (Table I). The GEO2R, an online analysis
tool built in the GEO website, was used for this analysis.
Statistical analyses were performed using paired-sample t-tests and
a P-value <0.05 was required for results to be considered
statistically significant. Scatter charts were plotted using
GraphPad Prism 7.0 (GraphPad Software, Inc., La Jolla, CA,
USA).
Results
Microarray data standardization and
DEG identification in plaque psoriasis
With use of the quartile division method, gene
expression levels of each of the 5 GEO series were standardized and
the results of pre- and post-standardization are presented in
Fig. 1A. After pre-processing of
the data, DEGs were analyzed using paired-sample t-tests within
each series using a screening criteria of P<0.05 and |log2FC|≥1
(Fig. 1B). The number of DEGs in
each series, including up- and downregulated DEGs are presented in
Table II. When DEGs in each
series were intersected with one another, 373 genes, considered as
integrated DEGs were obtained and used for subsequent analysis with
277 of these genes being upregulated and 96 downregulated (Fig. 2). The ratio of the number of
upregulated genes to that of downregulated genes was close to 1:1
in each of the GEO data sets, however, in the integrated results
this ratio was equal to approximately 3:1, indicating a possible
commonality in the upregulated genes during psoriasis development
while the downregulated genes differ in individuals.
 | Table II.DEGs in each GEO series. |
Table II.
DEGs in each GEO series.
| GEO series | No. of total
DEGs | No. of upregulated
DEGs | No. of upregulated
DEGs |
|---|
| GSE14905 | 1195 | 682 | 513 |
| GSE30999 | 1979 | 1040 | 939 |
| GSE34248 | 1670 | 854 | 816 |
| GSE41662 | 2203 | 1073 | 1130 |
| GSE53552 | 2220 | 1084 | 1136 |
GO and KEGG pathway enrichment
analysis of DEGs
GO enrichment analysis, which is comprised of 3
functional groups (biological processes, cellular components and
molecular functions), was performed using the DAVID online tool.
Within each of the functional groups, the top 10 enrichment terms
for both up- or downregulated DEGs as identified according to the
Fisher's exact test P-value are listed in Tables III and IV. The corresponding visual diagrams are
presented in Fig. 3A and B. Within
the biological process function group, upregulated DEGs were mainly
enriched in GO terms of immune responses, ectoderm development,
defense responses, keratinization and epidermal development while
downregulated DEGs were mainly enriched in the regulation of system
processes, regulation of smooth muscle contraction and muscle organ
development. Notably, with the exception of enrichment of the
cornified envelope, which is an extremely tough structure formed
beneath the cell membrane (17) in
the upregulated DEGs group, both up- and downregulated DEGs were
enriched in the extracellular space within the cellular component
enrichment analysis. Within the molecular function enrichment
group, the upregulated DEGs were mainly enriched in chemokine
activity, chemokine receptor binding and endopeptidase inhibitor
activity, while downregulated DEGs were enriched in cytoskeletal
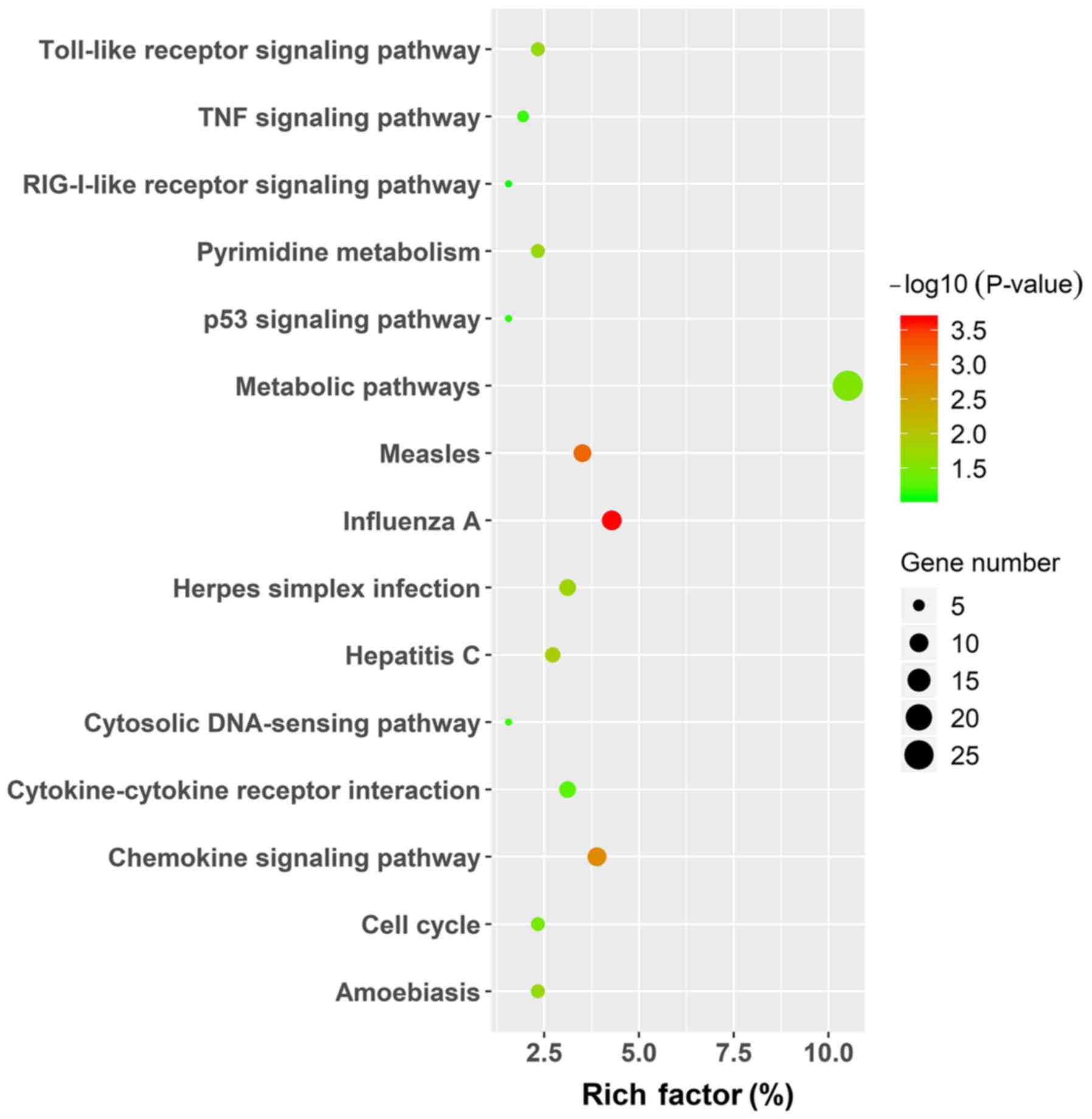
protein binding processes. The KEGG pathway enrichment was
performed using the same analysis tool and the results, in which
only the pathways for the upregulated genes are displayed by
figure, due to the limited number of enrichment pathways in the
downregulated group, are presented in Table V and Fig. 4. In upregulated DEGs, signaling
pathways were mainly enriched in metabolic pathways, measles,
influenza A and chemokine signaling pathways, while
aldosterone-regulated sodium reabsorption and PPAR signaling
pathways were enriched in downregulated DEGs.
 | Table III.GO analysis of upregulated genes
associated with psoriasis. |
Table III.
GO analysis of upregulated genes
associated with psoriasis.
| Term | Count | Rich factor
(%) | P-value | Functional
group |
|---|
| GO:0006955 immune
response | 39 | 16.04938272 | 4.38742E-13 | BP |
| GO:0007398 ectoderm
development | 17 | 6.995884774 | 2.50742E-08 | BP |
| GO:0006952 defense
response | 29 | 11.93415638 | 4.97398E-08 | BP |
| GO:0031424
keratinization | 9 | 3.703703704 | 1.40384E-07 | BP |
| GO:0008544
epidermis development | 15 | 6.172839506 | 3.81508E-07 | BP |
| GO:0009611 response
to wounding | 25 | 10.28806584 | 5.4487E-07 | BP |
| GO:0006954
inflammatory response | 18 | 7.407407407 | 4.12036E-06 | BP |
| GO:0030855
epithelial cell differentiation | 12 | 4.938271605 | 4.1551E-06 | BP |
| GO:0030216
keratinocyte differentiation | 9 | 3.703703704 | 4.20155E-06 | BP |
| GO:0009913
epidermal cell differentiation | 9 | 3.703703704 | 8.13933E-06 | BP |
| GO:0006935
chemotaxis | 12 | 4.938271605 | 1.83209E-05 | BP |
| GO:0001533
cornified envelope | 6 | 2.469135802 | 1.19845E-05 | CC |
| GO:0005576
extracellular region | 49 | 20.16460905 | 0.000143627 | CC |
| GO:0005615
extracellular space | 23 | 9.465020576 | 0.000294727 | CC |
| GO:0031262 Ndc80
complex | 3 | 1.234567901 | 0.001187266 | CC |
| GO:0044421
extracellular region part | 25 | 10.28806584 | 0.004874319 | CC |
| GO:0005792
microsome | 10 | 4.115226337 | 0.00688315 | CC |
| GO:0042598
vesicular fraction | 10 | 4.115226337 | 0.008255849 | CC |
| GO:0001772
immunological synapse | 3 | 1.234567901 | 0.008416898 | CC |
| GO:0000777
condensed chromosome kinetochore | 5 | 2.057613169 | 0.009278963 | CC |
| GO:0000793
condensed chromosome | 7 | 2.880658436 | 0.010321628 | CC |
| GO:0008009
chemokine activity | 7 | 2.880658436 | 3.62741E-05 | MF |
| GO:0004867
serine-type endopeptidase inhibitor activity | 9 | 3.703703704 | 3.72441E-05 | MF |
| GO:0042379
chemokine receptor binding | 7 | 2.880658436 | 5.23427E-05 | MF |
| GO:0004866
endopeptidase inhibitor activity | 10 | 4.115226337 | 0.000171511 | MF |
| GO:0030414
peptidase inhibitor activity | 10 | 4.115226337 | 0.00025684 | MF |
| GO:0004252
serine-type endopeptidase activity | 9 | 3.703703704 | 0.001273205 | MF |
| GO:0008236
serine-type peptidase activity | 9 | 3.703703704 | 0.003155874 | MF |
| GO:0017171 serine
hydrolase activity | 9 | 3.703703704 | 0.003376462 | MF |
| GO:0005125 cytokine
activity | 9 | 3.703703704 | 0.005452577 | MF |
| GO:0004175
endopeptidase activity | 13 | 5.349794239 | 0.005566809 | MF |
 | Table IV.GO analysis of downregulated genes
associated with psoriasis. |
Table IV.
GO analysis of downregulated genes
associated with psoriasis.
| Term | Count | Rich factor
(%) | P-value | Functional
group |
|---|
| GO:0044057
regulation of system process | 6 | 6.976744 | 0.008882 | BP |
| GO:0006940
regulation of smooth muscle contraction | 3 | 3.488372 | 0.010755 | BP |
| GO:0007517 muscle
organ development | 5 | 5.813953 | 0.011225 | BP |
| GO:0030003 cellular
cation homeostasis | 5 | 5.813953 | 0.020798 | BP |
| GO:0051241 negative
regulation of multicellular organismal process | 4 | 4.651163 | 0.030364 | BP |
| GO:0055080 cation
homeostasis | 5 | 5.813953 | 0.03044 | BP |
| GO:0042698
ovulation cycle | 3 | 3.488372 | 0.031344 | BP |
| GO:0006937
regulation of muscle contraction | 3 | 3.488372 | 0.035764 | BP |
| GO:0019432
triglyceride biosynthetic process | 2 | 2.325581 | 0.036656 | BP |
| GO:0008016
regulation of heart contraction | 3 | 3.488372 | 0.040411 | BP |
| GO:0005576
extracellular region | 25 | 29.06977 | 0.00021 | CC |
| GO:0005615
extracellular space | 12 | 13.95349 | 0.001648 | CC |
| GO:0044421
extracellular region part | 14 | 16.27907 | 0.002814 | CC |
| GO:0031012
extracellular matrix | 6 | 6.976744 | 0.047088 | CC |
| GO:0008092
cytoskeletal protein binding | 8 | 9.302326 | 0.009079 | MF |
| GO:0003779 actin
binding | 6 | 6.976744 | 0.018296 | MF |
| GO:0004857 enzyme
inhibitor activity | 5 | 5.813953 | 0.037901 | MF |
| GO:0003995 acyl-CoA
dehydrogenase activity | 2 | 2.325581 | 0.068241 | MF |
| GO:0042803 protein
homodimerization activity | 5 | 5.813953 | 0.07167 | MF |
| GO:0008201 heparin
binding | 3 | 3.488372 | 0.084388 | MF |
| GO:0030246
carbohydrate binding | 5 | 5.813953 | 0.084603 | MF |
 | Table V.KEGG analysis of DEGs associated with
psoriasis. |
Table V.
KEGG analysis of DEGs associated with
psoriasis.
| Regulation | ID | Term | Count | Rich factor
(%) | P-value | Genes |
|---|
| Upregulated | hsa05164 | Influenza A | 11 | 4.280155642 | 0.00022787 | IRF7, OAS3, CXCL8,
IL1B, RSAD2, OAS1, OAS2, MX1, STAT1, TMPRSS4, CXCL10 |
| Upregulated | hsa05162 | Measles | 9 | 3.501945525 | 0.00073285 | CCNE1, PRKCQ, IRF7,
OAS3, IL1B, OAS1, OAS2, MX1, STAT1 |
| Upregulated | hsa04062 | Chemokine signaling
pathway | 10 | 3.891050584 | 0.00162187 | CXCL1, CCL22,
CCL20, CXCL13, CXCL9, CXCL8, CXCR2, STAT1, CCL18, CXCL10 |
| Upregulated | hsa05160 | Hepatitis C | 7 | 2.723735409 | 0.01355117 | IFIT1, IRF7, OAS3,
CXCL8, OAS1, OAS2, STAT1 |
| Upregulated | hsa05168 | Herpes simplex
infection | 8 | 3.112840467 | 0.01820027 | CDK1, IFIT1, IRF7,
OAS3, IL1B, OAS1, OAS2, STAT1 |
| Upregulated | hsa00240 | Pyrimidine
metabolism | 6 | 2.33463035 | 0.01847329 | TYMP, NT5C3A, RRM2,
UPP1, PNP, CMPK2 |
| Upregulated | hsa04620 | Toll-like receptor
signaling pathway | 6 | 2.33463035 | 0.01989591 | IRF7, CXCL9, CXCL8,
IL1B, STAT1, CXCL10 |
| Upregulated | hsa05146 | Amoebiasis | 6 | 2.33463035 | 0.01989591 | ARG1, CXCL8, IL1B,
SERPINB4, SERPINB13, SERPINB3 |
| Upregulated | hsa01100 | Metabolic
pathways | 27 | 10.50583658 | 0.03117996 | XDH, GDA, KYNU,
HSD17B2, NT5C3A, GALNT6, CYP2C18, UPP1, AASS, PNP, CMPK2, ARG1,
TYMP, HPSE, ALOX12B, FUT2, SPTLC2, DHRS9, HYAL4, ST6GALNAC1, SQLE,
RRM2, AKR1B10, LIPG, GK, SMPD3, PLA2G4D |
| Upregulated | hsa04110 | Cell cycle | 6 | 2.33463035 | 0.0359891 | CCNB1, CCNE1, CDK1,
CDC45, TTK, CDC20 |
| Upregulated | hsa04060 | Cytokine-cytokine
receptor interaction | 8 | 3.112840467 | 0.05301106 | CCL20, CXCL13,
CXCL9, CXCL8, IL1B, CXCR2, IL7R, CXCL10 |
| Upregulated | hsa04623 | Cytosolic
DNA-sensing pathway | 4 | 1.556420233 | 0.06819191 | IRF7, IL1B, AIM2,
CXCL10 |
| Upregulated | hsa04668 | TNF signaling
pathway | 5 | 1.945525292 | 0.07110534 | CXCL1, NOD2, CCL20,
IL1B, CXCL10 |
| Upregulated | hsa04115 | p53 signaling
pathway | 4 | 1.556420233 | 0.07600871 | CCNB1, CCNE1, CDK1,
RRM2 |
| Upregulated | hsa04622 | RIG-I-like receptor
signaling pathway | 4 | 1.556420233 | 0.08420339 | ISG15, IRF7, CXCL8,
CXCL10 |
| Downregulated | hsa04960 |
Aldosterone-regulated sodium
reabsorption | 3 | 3.488372093 | 0.01441893 | HSD11B1, NR3C2,
ATP1A2 |
| Downregulated | hsa03320 | PPAR signaling
pathway | 3 | 3.488372093 | 0.03821865 | ACOX2, LPL,
ACADL |
PPI network construction and hub gene
selection
The online database STRING was used to obtain PPI
information on the 373 DEGs, including the 277 upregulated and 96
downregulated genes and the PPI network, with 2 notable functional
modules, was constructed with use of Cytoscape software (Fig. 5A). Hub genes were then calculated
using the cytoHubba app from the network we constructed. As a
result of these calculations, 17 genes with the highest scores were
considered as hub genes and were automatically divided into 2
groups exactly corresponding to the modules in Fig. 5A. One group consisted of TTK,
AURKA, DLGAP5, HMMR, CDC45, CENPE, SPC25, MCM10, NDC80, RRM2,
CKS2 and NUF2, which are genes involved in the cell
cycle, mitosis and proliferation. The second group consisted of
IFI6, ISG20, IFIT1, RSAD2 and IFI27, all of which
belong to IFN-α-inducible genes (Fig.
5B). Notably, these 17 hub genes all belong to the upregulated
genes of the DEGs we obtained, which further demonstrated the
importance of these upregulated genes in the molecular pathogenesis
of psoriasis.
Hub gene expression levels in scalp
psoriasis
To investigate whether scalp psoriasis displayed a
similar gene expression profile as that of skin psoriasis, 13 pairs
of scalp lesional and adjacent non-lesional samples were selected
from GSE75343 (Table I). With use
of the online analysis tool, GEO2R, expression levels of these 17
hub genes were determined. The results revealed that statistically
significant differences were obtained in the expression of IFI6,
IFI27, RSAD2, ISG20, MCM10 and SPC25 (Fig. 6), but not in the other hub genes
(data not shown). Further analysis revealed that 4 out of 6
IFN-α-inducible genes exhibited significant differences with regard
to gene expression, while in genes involved in the cell cycle,
mitosis and proliferation, only 2 of them exhibited differences in
gene expression.
Discussion
Psoriasis, one of the most common skin ailments,
afflicts millions of people worldwide. In addition to its negative
aspects on physical and mental health, the cost of psoriasis places
a huge burden on both individuals and society (17). Although dozens of medications are
available for relief of the symptoms of this disease, no cure for
psoriasis currently exists. Therefore, it is clear that the
identification of pivotal molecules that play critical roles in the
pathogenesis of this disease for potential development of
therapeutic targets represents an important area of
investigation.
Gene expression microarrays provide a comprehensive
view of genome-wide expression profiles of clinical samples and
have been widely used to analyze genes which are differentially
expressed in psoriasis. However, few studies exist which have
integrated such high-throughput gene expression microarray data of
paired lesional and non-lesional skin samples. In the present
study, gene expression profiles of 175 pairs of psoriatic skin
samples and the corresponding normal tissues from 5 GEO data sets
were integrated and analyzed with use of bioinformatic methods. Our
results demonstrated several important pathways and the pivotal
genes associated with the molecular pathogenesis of psoriasis.
Psoriasis is an immune-mediated inflammatory
cutaneous disease characterized by an overt proliferation and
differentiation of keratinocytes (1). Our GO biological process enrichment
results, especially with regard to upregulated genes, included
immune responses, keratinization, inflammatory responses and
keratinocyte differentiation, all of which are commonly accepted
components of the pathogenesis and pathological changes of
psoriasis. In addition, the enrichment in defense responses and
responses to wound healing processes indicates two important
psoriatic precipitating factors: infection (18) and trauma (Koebner phenomena)
(19), respectively, both of which
are associated with the activation of innate immunity involved in
the initial pathogenesis of psoriasis (20). Additional reported risk factors
include smoking (21), alcohol
(22) and obesity (23). In the cellular component enrichment
analysis, in addition to mitosis-associated components such as
chromosome kinetochore and the Ndc80 complex, enrichment in the
extracellular matrix of both up- or downregulated genes revealed
the significance of this component. The extracellular matrix (ECM)
is a collection of non-cellular molecular networks that regulate
diverse cellular functions, such as growth, migration and
homeostasis (24,25). The ECM is composed of interstitial
matrix and basement membrane, both of which are reported to be
involved in the development of psoriasis. Findings from a guttate
psoriasis prognosis study, indicate that psoriasis disease
progression is believed to be governed by the triggering of humoral
immune responses, which could produce extracellular antibodies to
neutralize the streptococcal lytic enzyme and prevent disruption of
the laminin layer in the basement membrane caused by the enzyme
(26). Recently, neutrophil
extracellular traps (NETs), which are web-like structures
consisting of DNA associated with histones, antimicrobial peptides
and enzymes (27,28), were reported to act as a source of
autoantigens which contribute to the occurrence of several
autoimmune diseases (29,30), including psoriasis. For example,
Lin et al reported that mast cells and neutrophils release
IL-17 through extracellular trap formation in psoriasis (31). In molecular function enrichment
analysis, it was observed that in upregulated genes, several GO
terms indicated endopeptidase inhibitor activity, which contains a
family of serine protease inhibitors (serpins). Serpins, such as
SERPINA3, SERPINB4, SERPINA1, SERPINB12, SERPINB3 and
SERPINB13 in our enrichment gene list represent a broad
family of protease inhibitors that utilize conformational changes
to inhibit target enzymes (32);
and it has been suggested that these serpins play a role in
psoriatic pathogenesis. Similar to the results obtained in our
analysis, Johnston et al detected upregulation of two
endogenous protease inhibitors, serpins A1 and A3, both of which
are present in psoriasis vulgaris and generalized pustular
psoriasis. These serpins may play a counter-regulated role to
control the activity of IL-36, whose activation requires N-terminal
peptide cleavage by neutrophil serine protease (33). Such a negative regulatory effect,
although unlikely to balance the protease expression revealed in
the study by Lin et al (31), may provide for new insights into
the development of psoriasis therapy.
The results from our KEGG pathway analysis revealed
that a high enrichment in metabolic and viral infection pathways
was present in upregulated genes. There were 26 genes enriched in
metabolic pathways, including XDH, GDA, KYNU, HSD17B2, NT5C3A,
GALNT6, CYP2C18, UPP1, AASS, PNP, CMPK2, ARG1, TYMP, HPSE, ALOX12B,
FUT2, SPTLC2, DHRS9, HYAL4, ST6GALNAC1, SQLE, RRM2, AKR1B10, LIPG,
GK, SMPD3 and PLA2G4D. Among these genes,
ALOX12B, one of the lipoxygenases, is reported to play an
important role in the regulation of epithelial proliferation,
differentiation, wound healing and inflammatory skin diseases
(34). PLA2G4D, a member of
phospholipase A2, was revealed to have a strong gene expression in
the upper spinous layer of psoriatic epidermis, while in normal
skin the expression of PLA2G4D was not detected (35). The expression or functions of the
other genes in our list have received little attention with regard
to their roles in cutaneous disorders. Therefore, these genes may
provide important new research targets for the understanding and
treatment of psoriasis. Most of the genes enriched in viral
infection KEGG pathways are IFN-α-inducible genes which belong to
one group of the hub genes in the PPI network.
The PPI network was constructed by Cytoscape
software and hub genes were then determined. With this analysis, 17
genes were identified and divided into 2 groups according to
protein-protein interactions. One group of these were
IFN-α-inducible genes, which were also enriched in KEGG viral
infection pathways, and included IFI6, IFI27, IFIT1, RSAD2
and ISG20. A role for IFN-α in psoriasis development has
been gradually revealed. For example, Garcia-Romo et al
demonstrated that in the initial phase of disease development,
cutaneous accumulated plasmacytoid pre-dendritic cells become
activated and produce IFN-α, which then drives the stimulation of
autoimmune T cells in pre-psoriatic skin (30). Such a mechanism provides an
explanation for the role of innate immunity in connecting
environmental triggers, such as viral or bacterial infection and
wound healing with disease-associated autoimmune T cells. This also
clarifies the reason for an absence of IFN-α in our analysis, since
the samples we selected were all from chronic plaque psoriasis
patients. The expression of IFN-α-inducible genes in our study was
also observed in scalp psoriatic samples, demonstrating an
important role for IFN-α in the pathogenesis of psoriasis within
different skin areas. The other group of genes including, CSK2,
CDC45, MCM10, SPC25, NDC80, NUF2, AURKA, CENPE, RRM2, DLGP5,
HMMR and TTK, were associated with the regulation of the
cell cycle, mitosis and proliferation (36,37).
Notably, there are two kinases in this group of hub genes: Aurora
kinase A, essential for chromosome segregation (38) and TTK, whose expression is at high
levels in tissues which contain large numbers of proliferating
cells (39). While the
relationship between these kinases and psoriasis development is yet
unclear, there exists a potentially important role that they may
play in the pathogenesis of psoriasis. In contrast to the results
in the IFN-α-inducible gene group, the expression of most hub genes
associated with the cell cycle and proliferation in scalp psoriatic
samples (except for MCM10 and SPC25), revealed no
significant differences from that of paired control samples. Within
the scalp area a large proportion of follicles are in anagen, which
may contribute to a set of highly expressed genes associated with
the cell cycle and proliferation. This can be contrasted with that
of skin samples from other areas where most hair follicles are in
catagen or telophase. Histologically, in the initial stages of this
disease scalp psoriasis mainly affects the interfollicular
epidermis with perifollicular inflammation (13), while later stages include
destruction of hair follicles with perifollicular fibrosis and hair
loss (40). Based on these
findings, it was hypothesized that the expression of these cell
cycle-related genes, which are assumed to be at high expression
levels in psoriatic samples, is relatively reduced in scalp
psoriatic samples where hair follicle destruction occurs as
compared to normal scalp tissues, as reflected in our results.
Although a similar bioinformatical study on
psoriasis has been performed (7),
in our present study, we limited our data sets to paired lesional
and non-lesional psoriatic skin samples and performed DEG analysis
with use of paired-sample t-tests in each data set. An overlap
method was subsequently employed to combine these DEG results as a
means to obtain an overall set of DEGs corresponding with that of
each data set. With use of these strict screening methods, we
consider that our results have a relatively high degree of
specificity for detecting pivotal disease-associated molecules,
however the resultant low sensitivity would be considered as a
limitation of this study. In our future research, if ethical
approval is obtained, RT-qPCR validation of these identified target
genes in clinical samples will be conducted. In conclusion, through
a comprehensive bioinformatic re-analysis of these original GEO
data, an overall view regarding the molecular pathogenesis of
psoriasis and the potential for identification of therapeutic
targets for this disease was provided.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81673070) and the
National Key Basic Research Program of China (grant no.
2013CB531604).
Availability of data and materials
All data generated or analyzed in the present study
are included in this published article.
Authors' contributions
YJZ collected the online microarray data and the
corresponding clinical information and drafted the manuscript. YJZ
and YZS performed the bioinformatic and statistical analysis. XHG
and RQQ contributed to the study design and performed the
proofreading and revision of the manuscript. All authors read and
approved the manuscript and agree to be accountable for all aspects
of the research in ensuring that the accuracy or integrity of any
part of the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
All authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
GEO
|
Gene Expression Omnibus
|
|
DEG
|
differentially expressed gene
|
|
GO
|
gene ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
PPI
|
protein-protein interaction
|
References
|
1
|
Greb JE, Goldminz AM, Elder JT, Lebwohl
MG, Gladman DD, Wu JJ, Mehta NN, Finlay AY and Gottlieb AB:
Psoriasis. Nat Rev Dis Primers. 2:160822016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parisi R, Symmons DP, Griffiths CE and
Ashcroft DM; Identification Management of Psoriasis, Associated
ComorbidiTy (IMPACT) project team, : Global epidemiology of
psoriasis: A systematic review of incidence and prevalence. J
Invest Dermatol. 133:377–385. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martinez-Garcia E, Arias-Santiago S,
Valenzuela-Salas I, Garrido-Colmenero C, Garcia-Mellado V and
Buendia-Eisman A: Quality of life in persons living with psoriasis
patients. J Am Acad Dermatol. 71:302–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Egeberg A, Thyssen JP, Wu JJ and Skov L:
Risk of first-time and recurrent depression in patients with
psoriasis: A population-based cohort study. Br J Dermatol.
180:116–121. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ainali C, Valeyev N, Perera G, Williams A,
Gudjonsson JE, Ouzounis CA, Nestle FO and Tsoka S: Transcriptome
classification reveals molecular subtypes in psoriasis. BMC
genomics. 13:4722012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mei R and Mei X: Screening of skin
lesion-associated genes in patients with psoriasis by
meta-integration analysis. Dermatology. 233:277–288. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sevimoglu T and Arga KY: Computational
systems biology of psoriasis: Are we ready for the age of omics and
systems biomarkers? OMICS. 19:669–687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yao Y, Richman L, Morehouse C, de los
Reyes M, Higgs BW, Boutrin A, White B, Coyle A, Krueger J, Kiener
PA and Jallal B: Type I interferon: Potential therapeutic target
for psoriasis? PLoS One. 3:e27372008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Suárez-Fariñas M, Li K, Fuentes-Duculan J,
Hayden K, Brodmerkel C and Krueger JG: Expanding the psoriasis
disease profile: Interrogation of the skin and serum of patients
with moderate-to-severe psoriasis. J Invest Dermatol.
132:2552–2564. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Correa da Rosa J, Kim J, Tian S, Tomalin
LE, Krueger JG and Suárez-Fariñas M: Shrinking the psoriasis
assessment gap: Early gene-expression profiling accurately predicts
response to long-term treatment. J Invest Dermato. 137:305–312.
2017. View Article : Google Scholar
|
|
11
|
Bigler J, Rand HA, Kerkof K, Timour M and
Russell CB: Cross-study homogeneity of psoriasis gene expression in
skin across a large expression range. PLoS One. 8:e522422013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Russell CB, Rand H, Bigler J, Kerkof K,
Timour M, Bautista E, Krueger JG, Salinger DH, Welcher AA and
Martin DA: Gene expression profiles normalized in psoriatic skin by
treatment with brodalumab, a human anti-IL-17 receptor monoclonal
antibody. J Immunol. 192:3828–3836. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ruano J, Suárez-Fariñas M, Shemer A, Oliva
M, Guttman-Yassky E and Krueger JG: Molecular and cellular
profiling of scalp psoriasis reveals differences and similarities
compared to skin psoriasis. PLoS One. 11:e01484502016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sherman BT, Huang da W, Tan Q, Guo Y, Bour
S, Liu D, Stephens R, Baseler MW, Lane HC and Lempicki RA: DAVID
Knowledgebase: A gene-centered database integrating heterogeneous
gene annotation resources to facilitate high-throughput gene
functional analysis. BMC Bioinformatics. 8:4262007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45:D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ishida-Yamamoto A and Iizuka H: Structural
organization of cornified cell envelopes and alterations in
inherited skin disorders. Exp Dermatol. 7:1–10. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Telfer NR, Chalmers RJ, Whale K and Colman
G: The role of streptococcal infection in the initiation of guttate
psoriasis. Arch Dermatol. 128:39–42. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Morhenn VB: The relationship of wound
healing with psoriasis and multiple sclerosis. Adv Wound Care (New
Rochelle). 7:185–188. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sweeney CM, Tobin AM and Kirby B: Innate
immunity in the pathogenesis of psoriasis. Arch Dermatol Res.
303:691–705. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Armstrong AW, Harskamp CT, Dhillon JS and
Armstrong EJ: Psoriasis and smoking: A systematic review and
meta-analysis. Br J Dermatol. 170:304–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qureshi AA, Dominguez PL, Choi HK, Han J
and Curhan G: Alcohol intake and risk of incident psoriasis in US
women: A prospective study. Arch Dermatol. 146:1364–1369. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jensen P and Skov L: Psoriasis and
obesity. Dermatology. 232:633–639. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Frantz C, Stewart KM and Weaver VM: The
extracellular matrix at a glance. J Cell Sci. 123:4195–4200. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Theocharis AD, Skandalis SS, Gialeli C and
Karamanos NK: Extracellular matrix structure. Adv Drug Deliv Rev.
97:4–27. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McFadden J, Fry L, Powles AV and Kimber I:
Concepts in psoriasis: Psoriasis and the extracellular matrix. Br J
Dermatol. 167:980–986. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Brinkmann V, Reichard U, Goosmann C,
Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y and Zychlinsky A:
Neutrophil extracellular traps kill bacteria. Science.
303:1532–1535. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brinkmann V and Zychlinsky A: Neutrophil
extracellular traps: Is immunity the second function of chromatin?
J Cell Biol. 198:773–783. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kessenbrock K, Krumbholz M, Schönermarck
U, Back W, Gross WL, Werb Z, Gröne HJ, Brinkmann V and Jenne DE:
Netting neutrophils in autoimmune small-vessel vasculitis. Nat Med.
15:623–625. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Garcia-Romo GS, Caielli S, Vega B,
Connolly J, Allantaz F, Xu Z, Punaro M, Baisch J, Guiducci C,
Coffman RL, et al: Netting neutrophils are major inducers of type I
IFN production in pediatric systemic lupus erythematosus. Sci
Transl Med. 3:73ra202011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin AM, Rubin CJ, Khandpur R, Wang JY,
Riblett M, Yalavarthi S, Villanueva EC, Shah P, Kaplan MJ and Bruce
AT: Mast cells and neutrophils release IL-17 through extracellular
trap formation in psoriasis. J Immunol. 187:490–500. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Law RH, Zhang Q, McGowan S, Buckle AM,
Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI
and Whisstock JC: An overview of the serpin superfamily. Genome
Biol. 7:2162006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Johnston A, Xing X, Wolterink L, Barnes
DH, Yin Z, Reingold L, Kahlenberg JM, Harms PW and Gudjonsson JE:
IL-1 and IL-36 are dominant cytokines in generalized pustular
psoriasis. J Allergy Clin Immunol. 140:109–120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Krieg P and Fürstenberger G: The role of
lipoxygenases in epidermis. Biochim Biophys Acta. 1841:390–400.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chiba H, Michibata H, Wakimoto K, Seishima
M, Kawasaki S, Okubo K, Mitsui H, Torii H and Imai Y: Cloning of a
gene for a novel epithelium-specific cytosolic phospholipase A2,
cPLA2delta, induced in psoriatic skin. J Biol Chem.
279:12890–12897. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kudalkar EM, Scarborough EA, Umbreit NT,
Zelter A, Gestaut DR, Riffle M, Johnson RS, MacCoss MJ, Asbury CL
and Davis TN: Regulation of outer kinetochore Ndc80 complex-based
microtubule attachments by the central kinetochore Mis12/MIND
complex. Proc Natl Acad Sci USA. 112:E5583–E5589. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Santaguida S and Musacchio A: The life and
miracles of kinetochores. EMBO J. 28:2511–2531. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
DeLuca KF, Meppelink A, Broad AJ, Mick JE,
Peersen OB, Pektas S, Lens SMA and DeLuca JG: Aurora A kinase
phosphorylates Hec1 to regulate metaphase kinetochore-microtubule
dynamics. J Cell Bio. 217:163–177. 2018. View Article : Google Scholar
|
|
39
|
Mills GB, Schmandt R, McGill M, Amendola
A, Hill M, Jacobs K, May C, Rodricks AM, Campbell S and Hogg D:
Expression of TTK, a novel human protein kinase, is associated with
cell proliferation. J Biol Chem. 267:16000–16006. 1992.PubMed/NCBI
|
|
40
|
George SM, Taylor MR and Farrant PB:
Psoriatic alopecia. Clin Exp Dermatol. 40:717–721. 2015. View Article : Google Scholar : PubMed/NCBI
|