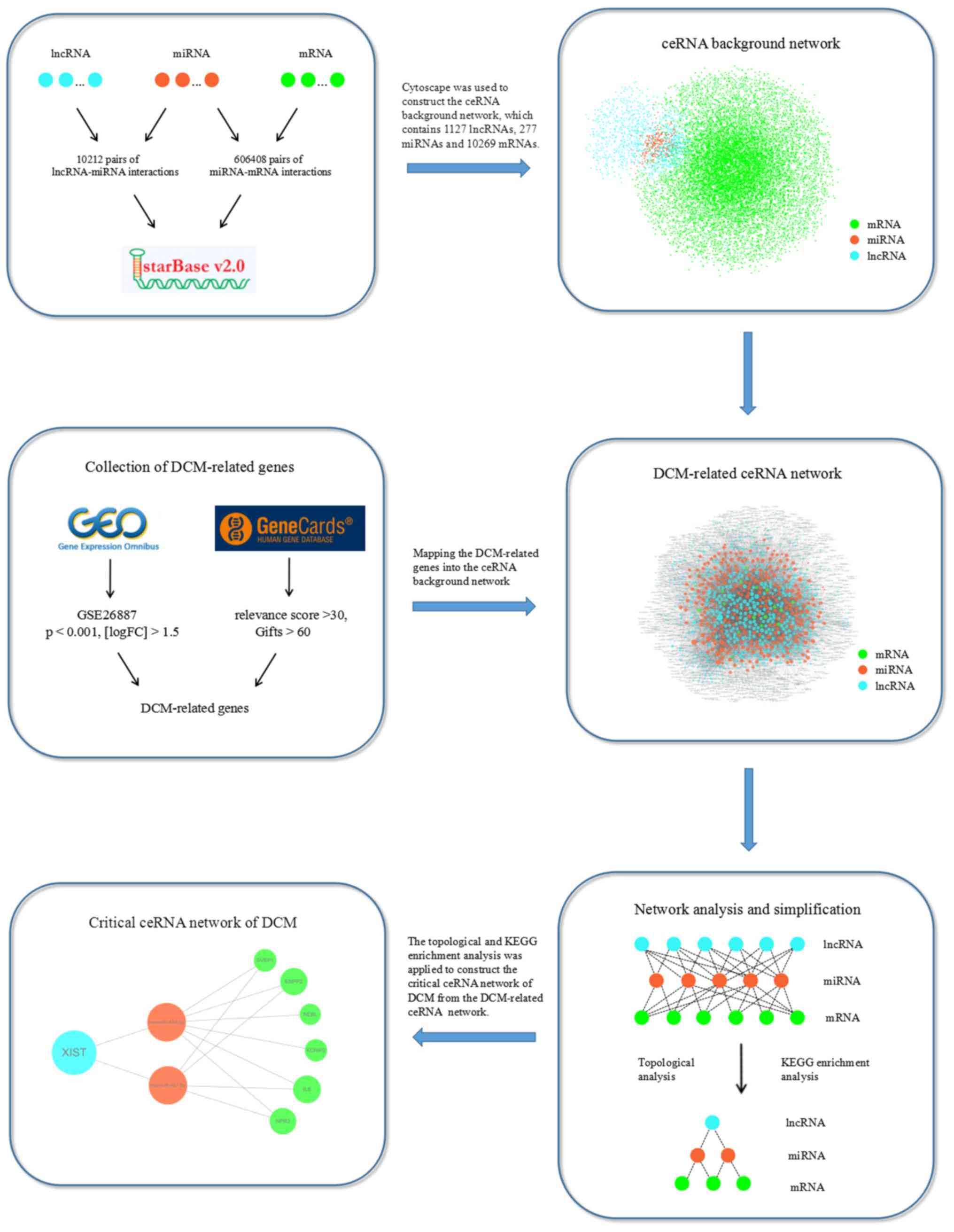
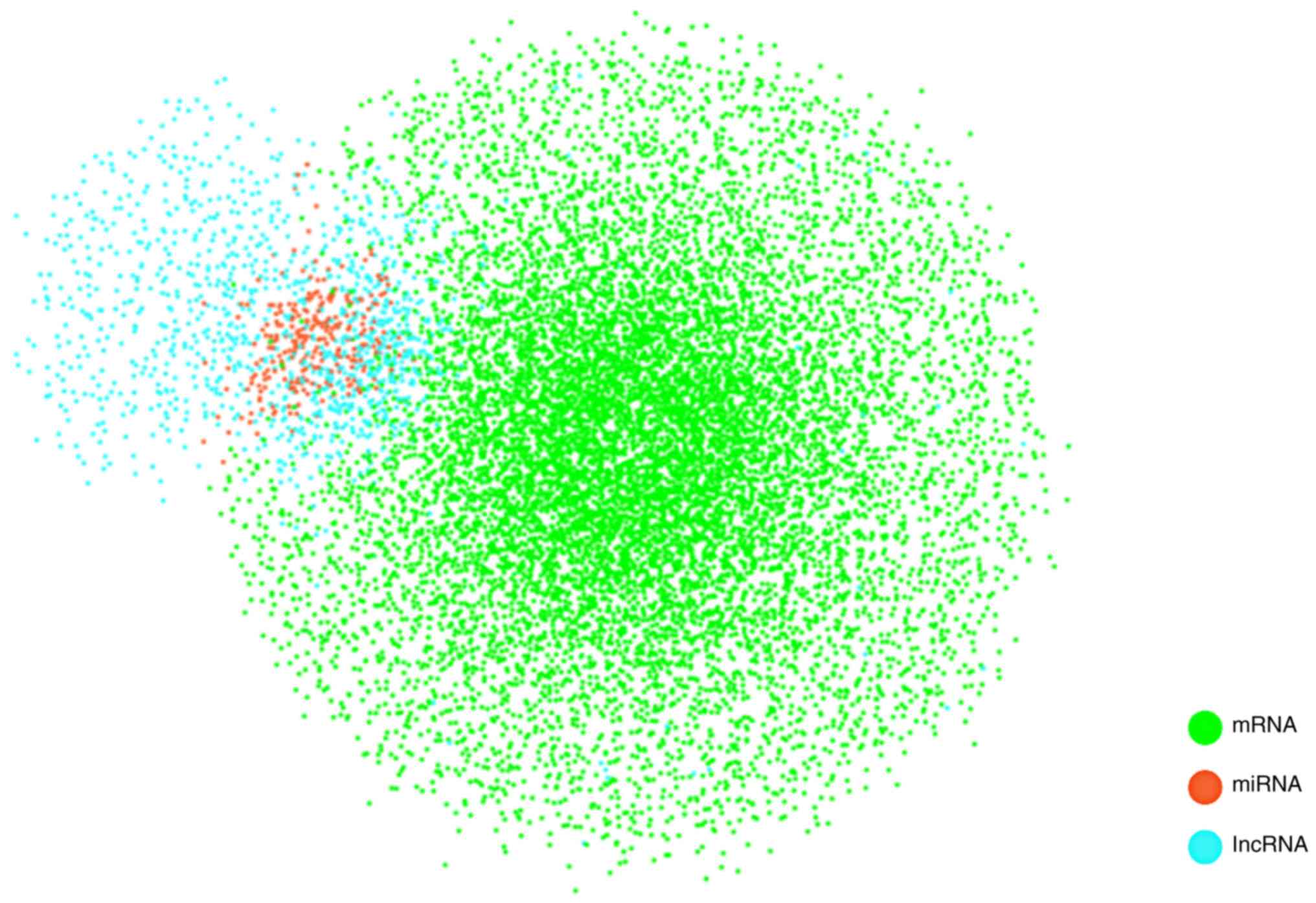
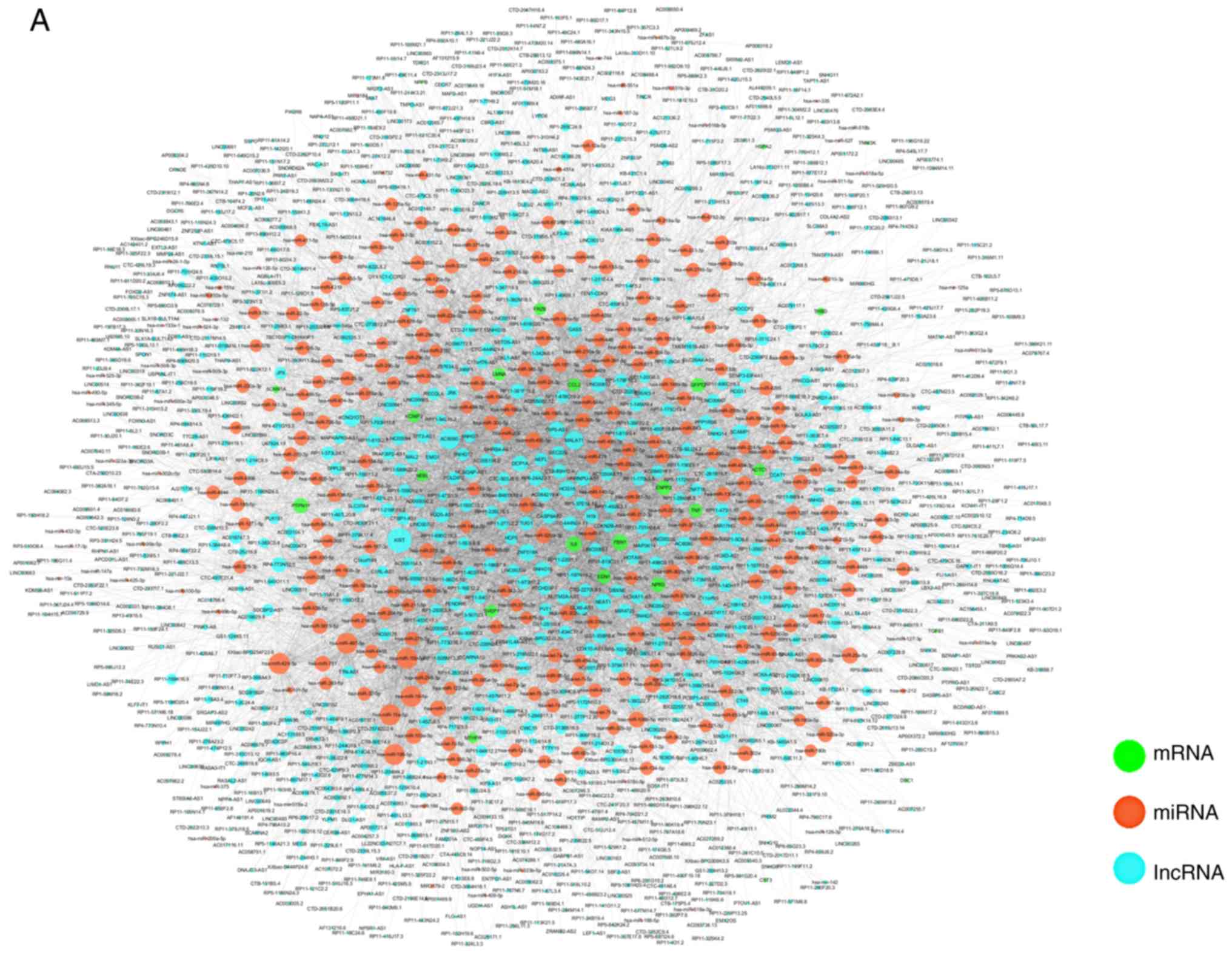
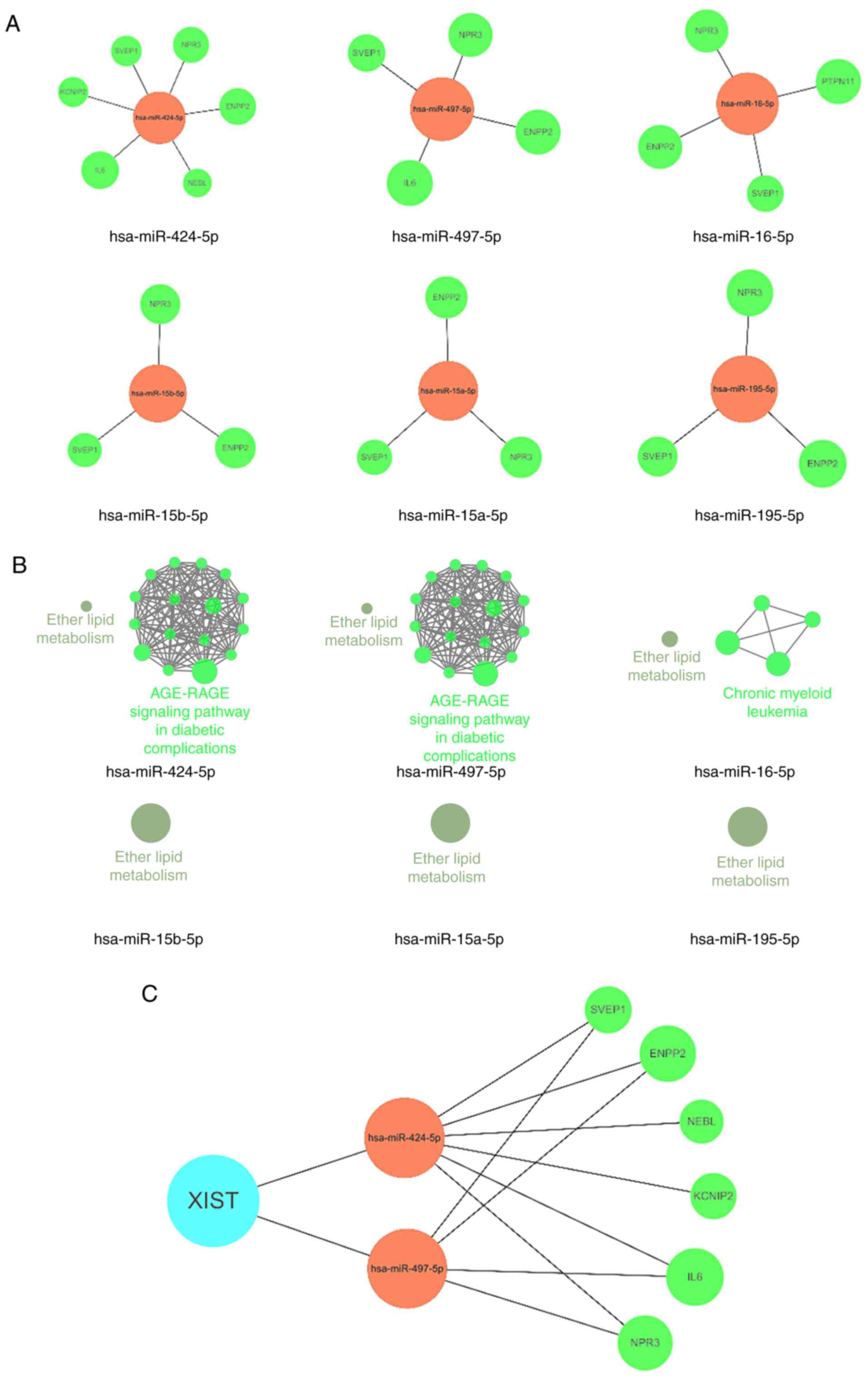
|
1
|
Jia G, Whaley-Connell A and Sowers JR:
Diabetic cardiomyopathy: A hyperglycaemia-and
insulin-resistance-induced heart disease. Diabetologia. 61:21–28.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Seferović PM and Paulus WJ: Clinical
diabetic cardiomyopathy: A two-faced disease with restrictive and
dilated phenotypes. Eur Heart J. 36:1718–1727. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lam CS: Diabetic cardiomyopathy: An
expression of stage B heart failure with preserved ejection
fraction. Diab Vasc Dis Res. 12:234–238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Aneja A, Tang WH, Bansilal S, Garcia MJ
and Farkouh ME: Diabetic cardiomyopathy: Insights into
pathogenesis, diagnostic challenges, and therapeutic options. Am J
Med. 121:748–757. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Taft RJ, Pang KC, Mercer TR, Dinger M and
Mattick JS: Non-coding RNAs: Regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iwakawa H and Tomari Y: The functions of
MicroRNAs: MRNA decay and translational repression. Trends Cell
Biol. 25:651–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jandura A and Krause HM: The New RNA
World: Growing Evidence for long noncoding RNA functionality.
Trends Genet. 33:665–676. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li W, Notani D and Rosenfeld MG: Enhancers
as non-coding RNA transcription units: Recent insights and future
perspectives. Nat Rev Genet. 17:207–223. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thum T and Condorelli G: Long Noncoding
RNAs and MicroRNAs in cardiovascular pathophysiology. Circ Res.
116:751–762. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA Hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smillie CL, Sirey T and Ponting CP:
Complexities of post-transcriptional regulation and the modeling of
ceRNA crosstalk. Crit Rev Biochem Mol Biol. 53:231–245. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dong Y, Liu C, Zhao Y, Ponnusamy M, Li P
and Wang K: Role of noncoding RNAs in regulation of cardiac cell
death and cardiovascular diseases. Cell Mol Life Sci. 75:291–300.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang M, Gu H, Chen J and Zhou X:
Involvement of long noncoding RNA MALAT1 in the pathogenesis of
diabetic cardiomyopathy. Int J Cardiol. 202:753–755. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou X, Zhang W, Jin M, Chen J, Xu W and
Kong X: lncRNA MIAT functions as a competing endogenous RNA to
upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy.
Cell Death Dis. 8:e29292017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Greco S, Fasanaro P, Castelvecchio S,
D'Alessandra Y, Arcelli D, Di Donato M, Malavazos A, Capogrossi MC,
Menicanti L and Martelli F: MicroRNA dysregulation in diabetic
ischemic heart failure patients. Diabetes. 61:1633–1641. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ismail M, Suresh H, Nazrath N, Anisatu R,
Marian P, Bruno V, Fabrizio S and Conrad B: GeoDiver: Differential
gene expression analysis & gene-set analysis for GEO Datasets.
Queen Mary J Intellectual Property. Apr 15–2017.doi:
https://doi.org/10.1101/127753.
|
|
18
|
Harel A, Inger A, Stelzer G,
Strichman-Almashanu L, Dalah I, Safran M and Lancet D: GIFtS:
Annotation landscape analysis with GeneCards. BMC Bioinformatics.
10:3482009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bindea G, Mlecnik B, Hackl H, Charoentong
P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z and
Galon J: ClueGO: A Cytoscape plug-in to decipher functionally
grouped gene ontology and pathway annotation networks.
Bioinformatics. 25:1091–1093. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Su G, Morris JH, Demchak B and Bader GD:
Biological network exploration with Cytoscape 3. Curr Protoc
Bioinformatics. 47:1–24. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Han JD, Bertin N, Hao T, Goldberg DS,
Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP and
Vidal M: Evidence for dynamically organized modularity in the yeast
protein-protein interaction network. Nature. 430:88–93. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cline MS, Smoot M, Cerami E, Kuchinsky A,
Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross
B, et al: Integration of biological networks and gene expression
data using Cytoscape. Nat Protoc. 2:2366–2382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou B, Wang B and Zhe H: Degree-layer
theory of network topology. Phys Soc. 18–Sep;2014.
|
|
25
|
Fromm B, Billipp T, Peck LE, Johansen M,
Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E and
Peterson KJ: A Uniform system for the annotation of vertebrate
microRNA genes and the evolution of the human microRNAome. Annu Rev
Genet. 49:213–242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miranda KC, Huynh T, Tay Y, Ang YS, Tam
WL, Thomson AM, Lim B and Rigoutsos I: A pattern-based method for
the identification of MicroRNA binding sites and their
corresponding heteroduplexes. Cell. 126:1203–1217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nature
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Betel D, Koppal A, Agius P, Sander C and
Leslie C: Comprehensive modeling of microRNA targets predicts
functional non-conserved and non-canonical sites. Genome Biol.
11:R902010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carrel L and Willard HF: X-inactivation
profile reveals extensive variability in X-linked gene expression
in females. Nature. 434:400–404. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Marques FZ, Vizi D, Khammy O, Mariani JA
and Kaye DM: The transcardiac gradient of cardio-microRNAs in the
failing heart. Eur J Heart Fail. 18:1000–1008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ghosh G, Subramanian IV, Adhikari N, Zhang
X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y and
Ramakrishnan S: Hypoxia-induced microRNA-424 expression in human
endothelial cells regulates HIF-α isoforms and promotes
angiogenesis. J Clin Invest. 120:4141–4154. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Flavin RJ, Smyth PC, Laios A, O'Toole SA,
Barrett C, Finn SP, Russell S, Ring M, Denning KM, Li J, et al:
Potentially important microRNA cluster on chromosome 17p13.1 in
primary peritoneal carcinoma. Mod Pathol. 22:197–205. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Finnerty JR, Wang WX, Hébert SS, Wilfred
BR, Mao G and Nelson PT: The miR-15/107 group of microRNA genes:
Evolutionary biology, cellular functions, and roles in human
diseases. J Mol Biol. 402:491–509. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen X, Shi C, Wang C, Liu W, Chu Y, Xiang
Z, Hu K, Dong P and Han X: The role of miR-497-5p in myofibroblast
differentiation of LR-MSCs and pulmonary fibrogenesis. Sci Rep.
7:409582017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jafarzadeh M, Soltani BM, Dokanehiifard S,
Kay M, Aghdami N and Hosseinkhani S: Experimental evidences for
hsa-miR-497-5p as a negative regulator of SMAD3 gene expression.
Gene. 586:216–221. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gendrel AV and Heard E: Fifty years of
X-inactivation research. Development. 138:5049–5055. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Payer B and Lee JT: X chromosome dosage
compensation: How mammals keep the balance. Annu Rev Genet.
42:733–772. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wei W, Liu Y, Lu Y, Yang B and Tang L:
LncRNA XIST promotes pancreatic cancer proliferation through
miR-133a/EGFR. J Cell Biochem. 118:3349–3358. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Song H, He P, Shao T, Li Y, Li J and Zhang
Y: Long non-coding RNA XIST functions as an oncogene in human
colorectal cancer by targeting miR-132-3p. J BUON. 22:696–703.
2017.PubMed/NCBI
|
|
40
|
Zhou T, Qin G, Yang L, Xiang D and Li S:
LncRNA XIST regulates myocardial infarction by targeting
miR-130a-3p. J Cell Physiol. 234:8659–8667. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Uribarri J, Cai W, Ramdas M, Goodman S,
Pyzik R, Chen X, Zhu L, Striker GE and Vlassara H: Restriction of
advanced glycation end products improves insulin resistance in
human type 2 diabetes: Potential role of AGER1 and SIRT1. Diabetes
Care. 34:1610–1616. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Prasad K: Low levels of serum soluble
receptors for advanced glycation end products, biomarkers for
disease state: Myth or Reality. Int J Angiol. 23:11–16. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tam XH, Shiu SW, Leng L, Bucala R,
Betteridge DJ and Tan KC: Enhanced expression of receptor for
advanced glycation end-products is associated with low circulating
soluble isoforms of the receptor in Type 2 diabetes. Clin Sci
(Lond). 120:81–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gao X, Zhang H, Schmidt AM and Zhang C:
AGE/RAGE produces endothelial dysfunction in coronary arterioles in
type 2 diabetic mice. Am J Physiol Heart Circ Physiol.
295:H491–H498. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Villegas-Rodríguez ME, Uribarri J,
Solorio-Meza SE, Fajardo-Araujo ME, Cai W, Torres-Graciano S,
Rangel-Salazar R, Wrobel K and Garay-Sevilla ME: The AGE-RAGE axis
and its relationship to markers of cardiovascular disease in newly
diagnosed diabetic patients. PLoS One. 11:e01591752016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Soro-Paavonen A, Watson AM, Li J, Paavonen
K, Koitka A, Calkin AC, Barit D, Coughlan MT, Drew BG, Lancaster
GI, et al: Receptor for advanced glycation end products (RAGE)
deficiency attenuates the development of atherosclerosis in
diabetes. Diabetes. 57:2461–2469. 2008. View Article : Google Scholar : PubMed/NCBI
|