Introduction
Neuroblastoma (NB) is the most common extracranial
solid cancer in childhood and infancy, with a mortality rate of
15%, and an incidence between 5.9 and 10.5 per million children
under 15 years of age (1). The
majority of NB cases are metastatic, and thus are associated with a
poor prognosis and a high mortality rate (2). A total of 20–50% of high-risk cases
do not respond adequately to high-dose chemotherapy and are
progressive or refractory (3).
Novel treatments using new agents and combinations against NB are
available in phase I or II clinical trials, but the outcomes remain
poor (4). Single targeted agents
are unlikely to be sufficient for long-term treatment for high-risk
NB. New therapeutic approaches need to be developed (2).
Clinical trial results have demonstrated that
multi-target inhibitor drugs are more effective compared with
single-target drugs during cancer treatment (5). 1H-indole-2, 3-dione (isatin) is a
derivative of the anti-cancer drug indirubin, which exhibits
beneficial biological activities, including antibacterial,
antifungal and antitumor properties (6–8).
Isatin was initially considered to be an inhibitor of monoamine
oxidase-b and has been used to treat Parkinson's disease (9,10).
Recent studies have reported that isatin inhibits cell
proliferation and invasion in human neuroblastoma SH-SY5Y cells
(11,12).
mTOR has been identified as a key molecule in
tumorigenesis and cancer progression (13). Increasing evidence has identified
the mTOR pathway as a relevant target for the suppression of
tumorigenesis; thus, inhibition of mTOR may be a promising method
for targeting human malignancies (14). The aim of the present study was to
investigate the underlying molecular mechanisms of the inhibitory
effect of isatin on migration and invasion in SH-SY5Y cells, which
are associated with the mTOR pathway.
Materials and methods
Cell culture
Human neuroblastoma SH-SY5Y cells (human origin)
were purchased from Peking Union Medical College. The cell line had
been STR authenticated (ATCC no. CRL-2266). Cells were cultured in
DMEM (HyClone; GE Healthcare Life Sciences) supplemented with 10%
fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) at 37°C
in a humidified atmosphere with 5% CO2. Isatin was dissolved in
0.1% dimethyl sulfoxide and added to 6-well plates at
concentrations of 50, 100, 200 and 300 µM, which were determined
based on our previous study (11),
when the cells had reached ~70% confluence. The cells (~2×106/well)
were harvested following isatin treatment at 37°C for 48 h for
further analysis.
Cell migration and invasion assay
Cells (1×106) were seeded in 6-well culture plates
and exposed to isatin (200 µM) with or without rapamycin (10 µM) at
٣٧°C for ٤٨ h. The concentration of isatin was determined according
to previous results, which demonstrated that 200 µM isatin did not
induce necrosis of SH-SY5Y cells (11). A wound healing assay was performed
to monitor cell invasion, as previously described (11). For the Transwell invasion assay,
isatin-treated cells were seeded in Boyden chambers (EMD Millipore)
with and without Matrigel, and the assay was performed as
previously described (11). Cells
were counted in at least five fields under an inverted light
microscope (magnification, ×10/x20). All experiments were performed
in triplicate.
Microarray analysis
Microarray expression analysis was performed using
total RNA from SH-SY5Y cells that were incubated at 37°C with or
without 200 µM isatin for 48 h. Total RNA was extracted using
TRIzol® (Beyotime Institute of Biotechnology). An
Affymetrix Microarray kit (Thermo Fisher Scientific, Inc.) was used
for the gene expression analysis. Raw data intensities were
quantile-normalized, and genes exhibiting significantly higher
intensities compared with the background and a fold-change
(FC)>1.5 following isatin treatment, compared with the control,
were selected. This selection uncovered a total of 429 genes.
Expression values of the genes were rescaled to mean 0 and SD 1,
and hierarchical clustering was performed using Ward's algorithm
(15). The number of clusters was
determined using the Akaike Information Criterion (16). A Mann-Whitney test was performed to
discriminate clusters of genes with differential expression levels
between the two groups. Differentially expressed genes were
selected based on FC in average gene expression with P<0.05, as
determined by Student's t-test. Gene Ontology (GO) enrichment
(‘biological process’, ‘cellular component’ and ‘molecular
function’) (17,18) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathways (19–21)
of differentially expressed genes were analyzed using the Database
for Annotation, Visualization and Integrated Discovery
Bioinformatics Resources, version 6.7 (http://david.abcc.ncifcrf.gov). Subsequent selection
of enriched GO terms was performed based on the calculated q-values
using the threshold q<0.05.
Reverse transcription-qPCR
(RT-qPCR)
Total RNA of SH-SY5Y cells was extracted using
TRIzol and was reverse transcribed to cDNA using TransScript™
Reverse Transcriptase (Beijing Transgen Biotech Co., Ltd.) at 42°C
for 15 min and 85°C for 5 sec. Then, qPCR was performed
independently three times using SYBR Green mix (Takara Bio, Inc.).
Gene expression was analyzed using the following primers: β-actin
forward, 5′-GCGTGACATTAAGGACAAGC-3′ and reverse,
5′-CCACGTCACACTTCATGATGG-3′; mTOR forward,
5′-CTGGGACTCAAATGTGTGCAGTTC-3′ and reverse,
5′-GAACAATAGGGTGAATGATCCGGG-3′; ribosomal protein S6 kinase B1
(RPS6KB1) forward, 5′-GGTGGAGTTTGGGAGCATTA-3′ and reverse,
5′-TGTGAGGTAGGGAGGCAAAT-3′; eukaryotic translation initiation
factor 4E-binding protein 1 (EIF4EBP1) forward,
5′-GTCGGAACTCACCTGTGACC-3′ and reverse,
5′-CCGCTTATCTTCTGGGCTATT-3′; Ras homolog mTORC1-binding (RHEB)
forward, 5′-AGCTTTGGCAGAATCTTGGA-3′ and reverse,
5′-GCATGAAGACTTGCCTTGTG-3′; DNA damage-inducible transcript 4
(DDIT4) forward, 5′-AGACACGGCTTACCTGGATG-3′ and reverse,
5′-CAGTAGTTCTTTGCCCACCTG-3′. The thermocycling conditions were 94°C
for 5 min, then 40 cycles of 93°C for 30 sec, 55°C for 30 sec and
72°C for 60 sec. Data are expressed as fold change compared with
control. The 2−ΔΔCq method was employed to analyze the
relative expression of genes (22).
Protein extraction and western blot
analysis
Total protein was extracted from SH-SY5Y cells using
RIPA lysis buffer (Beyotime Institute of Biotechnology) following
isatin treatment for 48 h, and the protein concentration was
quantified using a bicinchoninic acid protein assay. Proteins
(20–40 µg/lane) were separated on SDS-polyacrylamide gels (8–12%)
and transferred onto PVDF membranes. Membranes were blocked with 5%
bovine serum albumin for 2 h at room temperature, and then
incubated with the following primary antibodies diluted in 5% BSA:
β-actin (1:3,000; cat. no. ab8227; Abcam), AMPK (1:500; cat. no.
ab131512; Abcam), phosphorylated (p)-AMPK (1:500; cat. no.
ab131357; Abcam), mTOR (1:300; cat. no. ab32028; Abcam), p-mTOR
(1:5,000; cat. no. ab109268; Abcam), microtubule associated protein
1 light chain 3α (LC3; 1:1,000; cat. no. 3868; Cell Signaling
Technology), Beclin-1 (1:1,000; cat. no. 3738; Cell Signaling
Technology) and p62 (1:1,000; cat. no. 5114; Cell Signaling
Technology) at 4°C overnight, washed with TBS with Tween-20, and
subsequently incubated with horseradish peroxidase-conjugated goat
anti-rabbit immunoglobulin G secondary antibodies (1:1,000; cat.
no. HS101-01; Beijing Transgen Biotech Co., Ltd.) for 1 h at room
temperature. Protein expression levels were detected using the
Enhanced Chemiluminescence Plus kit (Wuhan Boster Biological
Technology, Ltd.) and densitometric analysis was performed using
Quantity One analysis software (version 4.52; Bio-Rad Laboratories,
Inc.).
Statistical analysis
Each experiment was performed at least three times.
Data are expressed as the mean ± SD. Multiple comparisons were
performed by one-way ANOVA followed by a least significant
difference or Tamhane's T2 post hoc test in SPSS 22.0 (IBM Corp.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Isatin treatment affects gene
expression in SH-SY5Y cells
To investigate the underlying molecular events of
the anti-invasive activity of isatin in SH-SY5Y cells, RNA was
extracted from SH-SY5Y cells (untreated or treated with 200 µM
isatin for 48 h) for Affymetrix cDNA microarray analysis. The
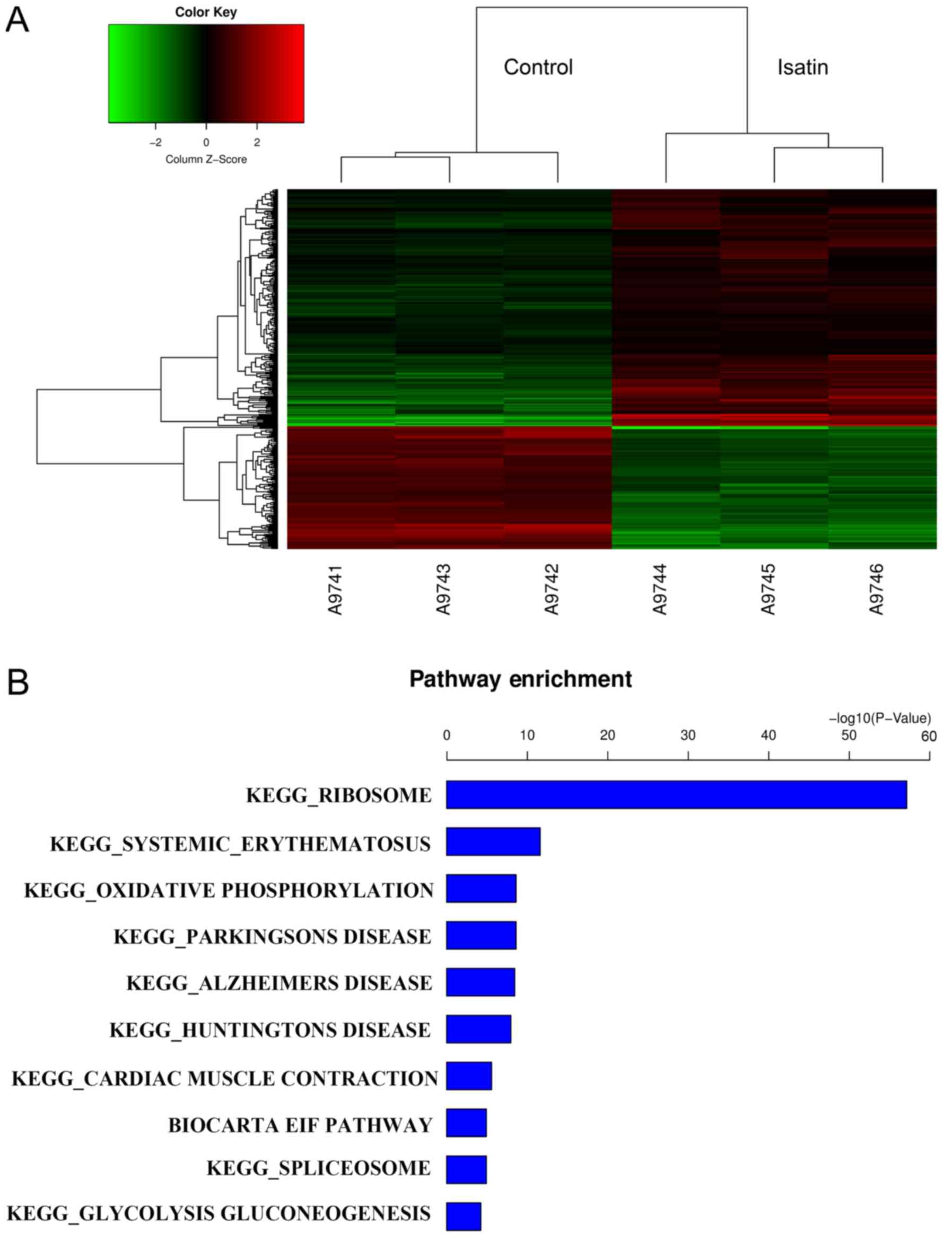
GeneChip results revealed 284 differentially upregulated and 145
downregulated genes between cells treated with isatin and controls
(FC>1.5; Fig. 1A). GO term
analysis demonstrated that the differentially expressed genes were
involved in redox activity, binding and transcription function,
cell metabolism and transport (data not shown). The results of GO
term analysis indicated that isatin is involved in cell
proliferation and the cell cycle, and functions in cell
translation, biosynthesis and metabolism. According to the results
of KEGG analysis, GeneChip predicted that isatin-modulated gene
pathways were likely to be associated with chemokine signaling
pathways, ribosome pathways and mTOR signaling pathways (Fig. 1B). The DDIT4, RHEB, EIF4EBP1 and
RPS6KB1 genes, which are associated with mTOR activation, were
selected for further analysis.
RT-qPCR verification of
mTOR-associated differentially expressed genes
The expression levels of mTOR signaling
pathway-associated genes DDIT4, RHEB, EIF4EBP1 and RPS6KB1, which
exhibited differential expression in the microarray datasets, were
verified by RT-qPCR. The results demonstrated that the differences
in the expression of the four genes were consistent with the
GeneChip data (Fig. 2A). In
addition, the mTOR mRNA expression level was also downregulated in
SH-SY5Y cells following isatin treatment compared with untreated
controls (Fig. 2B).
Rapamycin partially reverses the
effects of isatin
To validate the role of mTOR in the anti-invasive
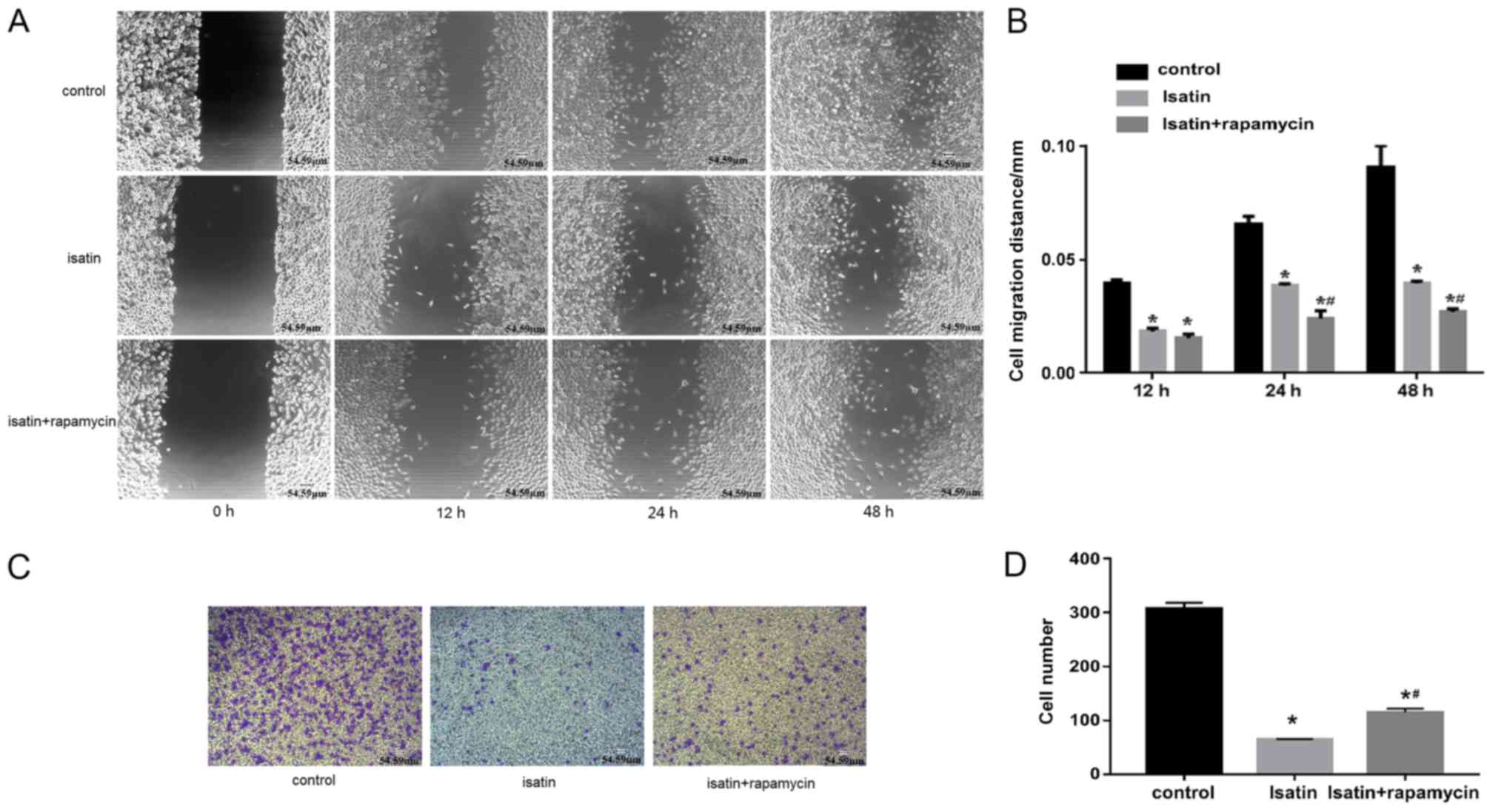
effects of isatin on SH-SY5Y cells, migration and invasion assays
were performed in the presence and absence of rapamycin. The rate
of wound healing of SH-SY5Y cells co-treated with isatin and
rapamycin was lower compared with that of isatin-treated cells
(Fig. 3A and B). In addition, the
invasion analysis showed that 200 µM isatin decreased the invasive
ability of SH-SY5Y cells compared with untreated controls, which
was partly restored by co-treatment with rapamycin (Fig. 3C and D), and the mean numbers of
invasive cells were 64 and 115, respectively.
Isatin treatment affects the
expression of proteins associated with the mTOR pathway
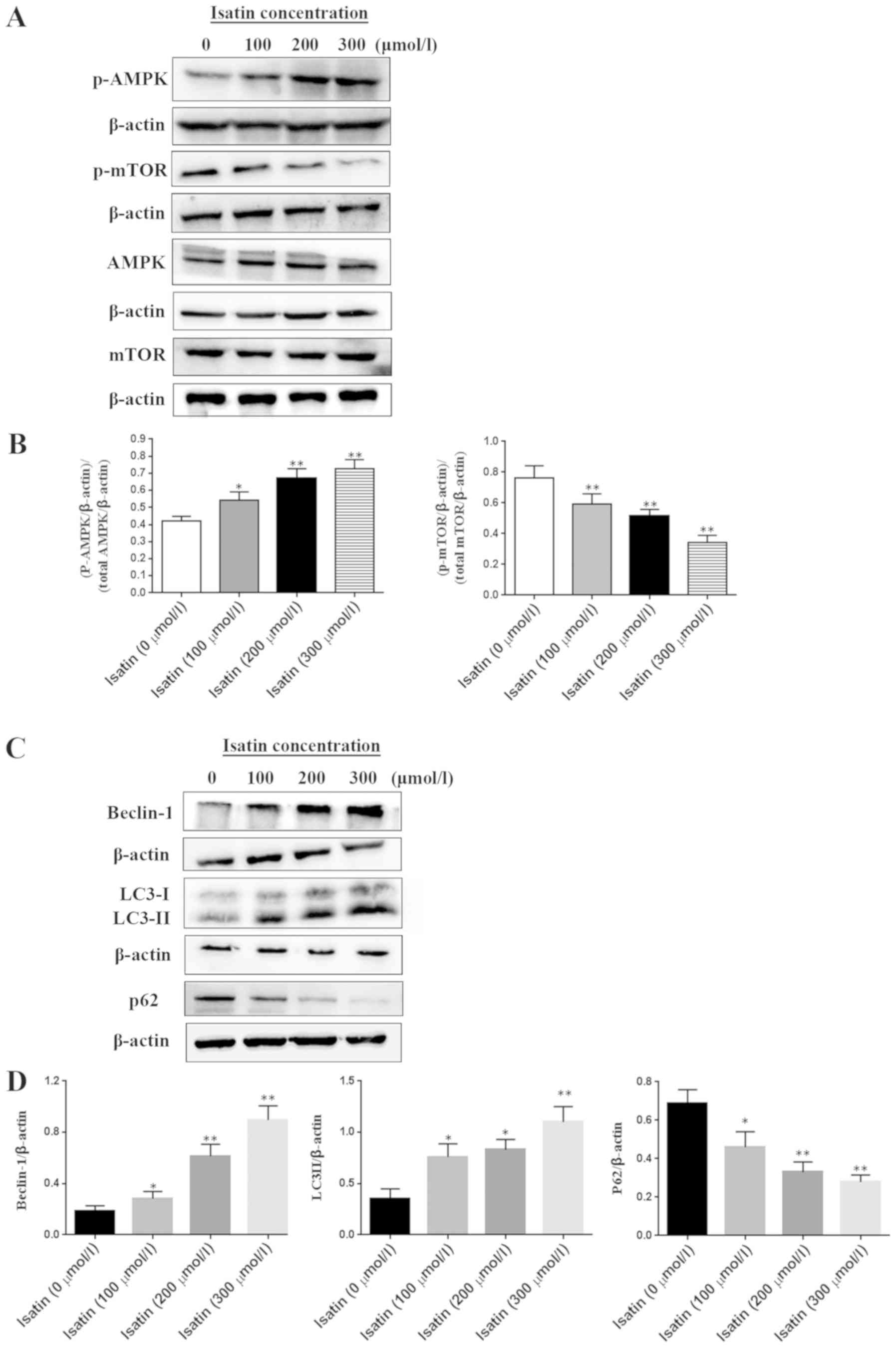
SH-SY5Y cells were treated with a range of isatin
concentrations (0–300 µM) for ٤٨ h and total or phosphorylated cell
proteins were extracted for western blotting. The mTOR
phosphorylation level was inhibited by isatin in a
concentration-dependent manner (Fig.
4A and B). By contrast, the phosphorylation of AMPK, which is
an inhibitor kinase of mTOR, was increased by isatin compared with
that in untreated cells (Fig. 4A).
LC3-II expression was upregulated and p62 was downregulated in
SH-SY5Y cells treated with 200 or 300 µM isatin compared with the
control group (P<0.01; Fig. 4C and
D). In addition, tumor suppressor Beclin-1 was also activated
following isatin treatment (Fig. 4C
and D).
Discussion
Tumor cell migration and invasion are the principal
steps in tumor metastasis (23,24).
The majority of neuroblastoma-related mortalities are due to the
infiltration of tumor cells to lymph nodes, bones and bone marrow
(25). Controlling tumor
metastasis is a promising approach for neuroblastoma treatment. In
the present study, microarray analysis was conducted to investigate
the underlying mechanism of the anti-metastatic effects of isatin
on neuroblastoma.
The mTOR gene is a key regulator of cell growth,
proliferation, differentiation and survival (26,27).
The activated mTOR signaling pathway accelerates tumor progression
and downregulates genes such as RHEB, DDIT4, EIF4EBP and RPS6KB1,
all of which are positive regulators of mTOR (28,29).
The results of the microarray analysis in the present study
demonstrated that the expression levels of these genes were among
the most significantly altered by isatin treatment, which was
further verified by RT-qPCR. These findings suggested that the mTOR
signaling pathway may be involved in tumor progression following
isatin treatment.
The results of the present study also revealed that
the anti-metastatic effect of isatin was weakened by the inhibition
of mTOR expression by rapamycin. AMPK is a serine/threonine protein
kinase that has inhibitory effects in certain types of tumor,
including prostate, pancreatic and thyroid cancer (30). Activated AMPK activates the
tuberous sclerosis complex, which leads to inhibition of mTOR
activity, subsequently inhibiting tumor angiogenesis (31). In the present study, activated
phosphorylation of AMPK was observed, which subsequently inhibited
the phosphorylation level of mTOR.
In cancer cells, autophagy is triggered in response
to cellular stress, such as nutrient or growth factor starvation
and hypoxia (32). The occurrence
of autophagy can inhibit tumor cell development in a number of
tumors, such as osteosarcoma and glioblastoma (33,34).
Further studies have demonstrated that autophagy is also closely
related to tumor invasion and metastasis (35,36).
The mTOR signaling pathway serves a crucial role in autophagy
(37,38). Among proteins related to autophagy,
LC3 is an essential marker; the level of LC3-II expression in the
membrane of autophagosomes reflects the level of autophagy
(39,40). p62 ubiquitin-like binding protein
is also a marker protein for autophagy detection; the expression
level of p62 is negatively correlated with autophagic activity
(41). The tumor suppressor
function of autophagy is induced by certain ATG-proteins, such as
Beclin-1, which exhibit anti-oncogenic functions (42). Inactivation of autophagy-related
genes, including Beclin-1, leads to increased tumorigenesis,
whereas overexpression of these genes inhibits the formation of
human breast, ovarian and prostate tumors in mouse models (43). The results of the present study
revealed high levels of autophagy-related protein expression in
isatin-treated cells, which further explained the mechanism of
action of isatin in neuroblastoma metastasis.
In conclusion, isatin is an effective inhibitor of
neuroblastoma cell invasion and migration; the mechanism of the
inhibition may be associated with the mTOR signaling pathway.
Further studies are necessary to confirm whether isatin is a
possible anti-metastasis drug for human neuroblastoma.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81472542), the Shandong
Postdoctoral Innovation Project (grant no. 201502013), the Focus on
Research and Development Plan in Shandong Province (grant no.
GG201809260275), the Qingdao Postdoctoral Application Research
Project (grant no. 2015146), the Clinical Medicine +X Project of
the Medical Department of Qingdao University and Innovation Team of
Qingdao University Medical School Youth Teacher Training Project
(grant no. 600201304).
Availability of data and materials
All data used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ, LH and WS performed statistical analyses, and
contributed towards the conception and design of the present study.
LH supervised the study. All authors including CJ, YC and XW were
involved in the acquisition and interpretation of data. LZ, WS and
LH drafted the manuscript. All authors contributed to revision of
the manuscript for important intellectual content. All authors
approved the final version of the manuscript to be submitted.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Costa RA and Seuánez HN: Investigation of
major genetic alterations in neuroblastoma. Mol Biol Rep.
45:287–295. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Swift CC, Eklund MJ, Kraveka JM and
Alazraki AL: Updates in diagnosis, management, and treatment of
neuroblastoma. Radiographics. 38:566–580. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kushner BH, Kramer K, LaQuaglia MP, Modak
S, Yataghene K and Cheung NK: Reduction from seven to five cycles
of intensive induction chemotherapy in children with high-risk
neuroblastoma. J Clin Oncol. 22:4888–4892. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ceschel S, Casotto V, Valsecchi MG, Tamaro
P, Jankovic M, Hanau G, Fossati F, Pillon M, Rondelli R, Sandri A,
et al: Survival after relapse in children with solid tumors: A
follow-up study from the Italian off-therapy registry. Pediatr
Blood Cancer. 47:560–566. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gowda R, Jones NR, Banerjee S and
Robertson GP: Use of nanotechnology to develop multi-drug
inhibitors for cancer therapy. J Nanomed Nanotechnol. 4(pii):
1842013.PubMed/NCBI
|
|
6
|
Verma M, Pandeya SN, Singh KN and Stables
JP: Anticonvulsant activity of Schiff bases of isatin derivatives.
Acta Pharm. 54:49–56. 2004.PubMed/NCBI
|
|
7
|
Gencer N, Sonmez F, Demir D, Arslan O and
Kucukislamoglu M: Synthesis, structure-activity relationships and
biological activity of new isatin derivatives as tyrosinase
inhibitors. Curr Top Med Chem. 14:1450–1462. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hamaue N: Pharmacological role of isatin,
an endogenous MAO inhibitor. Yakugaku Zasshi. 120:352–362. 2000.(In
Japanese). View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Minami M, Hamaue N, Hirafuji M, Saito H,
Hiroshige T, Ogata A, Tashiro K and Parvez SH: Isatin, an
endogenous MAO inhibitor, and a rat model of Parkinson's disease
induced by the Japanese encephalitis virus. J Neural Transm Suppl.
87–95. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Igosheva N, Lorz C, O'Conner E, Glover V
and Mehmet H: Isatin, an endogenous monoamine oxidase inhibitor,
triggers a dose- and time-dependent switch from apoptosis to
necrosis in human neuroblastoma cells. Neurochem Int. 47:216–224.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun W, Zhang L, Hou L, Ju C, Zhao S and
Wei Y: Isatin inhibits SH-SY5Y neuroblastoma cell invasion and
metastasis through MAO/HIF-1 α/CXCR4 signaling. Anti-cancer drugs.
28:645–653. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu P, Hou L, Ju C, Zhang Z, Sun W, Zhang
L, Song J, Lv Y, Liu L, Chen Z and Wang Y: Isatin inhibits the
proliferation and invasion of SH-SY5Y neuroblastoma cells. Mol Med
Rep. 13:2757–2762. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang J, Jia Z, Li Q, Wang L, Rashid A,
Zhu Z, Evans DB, Vauthey JN, Xie K and Yao JC: Elevated expression
of vascular endothelial growth factor correlates with increased
angiogenesis and decreased progression-free survival among patients
with low-grade neuroendocrine tumors. Cancer. 109:1478–1486. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shaw RJ: LKB1 and AMP-activated protein
kinase control of mTOR signalling and growth. Acta Physiol (Oxf).
196:65–80. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Murtagh F and Legendre P: Ward's
Hierarchical agglomerative clustering method: Which algorithms
implement Ward's Criterion? J Classification. 31:274–295. 2014.
View Article : Google Scholar
|
|
16
|
Vrieze SI: Model selection and
psychological theory: A discussion of the differences between the
Akaike information criterion(ALC) and the Bayesian information
criterion (BIC). Psychol Methods. 17:228–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
The Gene Ontology Consortium: The gene
ontology resource: 20 years and still Going strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Steeg PS and Theodorescu D: Metastasis: A
therapeutic target for cancer. Nat Clin Pract Oncol. 5:206–219.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Monclair T, Brodeur GM, Ambros PF, Brisse
HJ, Cecchetto G, Holmes K, Kaneko M, London WB, Matthay KK,
Nuchtern JG, et al: The International Neuroblastoma Risk Group
(INRG) staging system: An INRG Task Force report. J Clin Oncol.
27:298–303. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Morandi F, Corrias MV and Pistoia V:
Evaluation of bone marrow as a metastatic site of human
neuroblastoma. Ann N Y Acad Sci. 1335:23–31. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chiang GG and Abraham RT: Targeting the
mTOR signaling network in cancer. Trends Mol Med. 13:433–442. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sarbassov DD, Ali SM and Sabatini DM:
Growing roles for the mTOR pathway. Curr Opin Cell Biol.
17:596–603. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Long X, Lin Y, Ortiz-Vega S, Yonezawa K
and Avruch J: Rheb binds and regulates the mTOR kinase. Curr Biol.
15:702–713. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Julien LA, Carriere A, Moreau J and Roux
PP: mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135
and regulates mTORC2 signaling. Mol Cell Biol. 30:908–921. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rattan R, Giri S, Hartmann LC and Shridhar
V: Metformin attenuates ovarian cancer cell growth in an AMP-kinase
dispensable manner. J Cell Mol Med. 15:166–178. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meijer AJ and Codogno P: AMP-activated
protein kinase and autophagy. Autophagy. 3:238–240. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Divac Rankov A, Ljujic M, Petric M,
Radojkovic D, Pesic M and Dinic J: Targeting autophagy to modulate
cell survival: A comparative analysis in cancer, normal and
embryonic cells. Histochem Cell Biol. 148:529–544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian X, Song HS, Cho YM, Park B, Song YJ,
Jang S and Kang SC: Anticancer effect of Saussurea lappa extract
via dual control of apoptosis and autophagy in prostate cancer
cells. Medicine (Baltimore). 96:e76062017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kon M, Kiffin R, Koga H, Chapochnick J,
Macian F, Varticovski L and Cuervo AM: Chaperone-mediated autophagy
is required for tumor growth. Sci Transl Med. 3:109ra1172011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cheong H: Integrating autophagy and
metabolism in cancer. Arch Pharm Res. 38:358–371. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang W, Li Q, Song C and Lao L: Knockdown
of autophagy-related protein 6, Beclin-1, decreases cell growth,
invasion, and metastasis and has a positive effect on
chemotherapy-induced cytotoxicity in osteosarcoma cells. Tumour
Biol. 36:2531–2539. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ni HM, Bockus A, Boggess N, Jaeschke H and
Ding WX: Activation of autophagy protects against
acetaminophen-induced hepatotoxicity. Hepatology. 55:222–232. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Athamneh K, Hasasna HE, Samri HA, Attoub
S, Arafat K, Benhalilou N, Rashedi AA, Dhaheri YA, AbuQamar S, Eid
A and Iratni R: Rhus coriaria increases protein ubiquitination,
proteasomal degradation and triggers non-canonical
Beclin-1-independent autophagy and apoptotic cell death in colon
cancer cells. Sci Rep. 7:116332017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kenific CM, Thorburn A and Debnath J:
Autophagy and metastasis: Another double-edged sword. Current
opinion in cell biology. 22:241–245. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kabeya Y, Mizushima N, Ueno T, Yamamoto A,
Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T: LC3, a
mammalian homologue of yeast Apg8p, is localized in autophagosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu WJ, Ye L, Huang WF, Guo LJ, Xu ZG, Wu
HL, Yang C and Liu HF: p62 links the autophagy pathway and the
ubiqutin-proteasome system upon ubiquitinated protein degradation.
Cell Mol Biol Lett. 21:292016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Park JM, Huang S, Wu TT, Foster NR and
Sinicrope FA: Prognostic impact of Beclin 1, p62/sequestosome 1 and
LC3 protein expression in colon carcinomas from patients receiving
5-fluorouracil as adjuvant chemotherapy. Cancer Biol Ther.
14:100–107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ngabire D and Kim GD: Autophagy and
inflammatory response in the tumor microenvironment. Int J Mol Sci.
18(pii): E20162017. View Article : Google Scholar : PubMed/NCBI
|