Introduction
The microbiota and microbiome-derived metabolites in
the colon have been recognized as an alternative therapy for colon
cancer treatment (1). The
microbiota in the colon produce short-chain fatty acids (SCFAs;
butyrate, propionate and acetate) via dietary fiber fermentation to
maintain human health (2). Several
in vivo and in vitro studies have reported that
SCFAs, mainly butyrate, exert anticancer effects, such as
suppressing cell growth, migration and invasion, on colon cancer
(3,4). Recently, sodium butyrate treatment
was shown to upregulate miR-3935 expression, which prohibited the
proliferation and migration of A549 cells (5). However, despite studies on the roles
of SCFAs in the colon, the anticancer effects of SCFAs, especially
propionate, on lung cancer are not well understood. Therefore, the
present study examined the anticancer effects and molecular
mechanism of sodium propionate (SP) using lung cancer cell
lines.
Survivin, an antiapoptotic protein, is overexpressed
in several types of cancer, and knockdown of Survivin induces cell
apoptosis by increasing Bad and Bax expression and inducing G2/M
arrest (6). Additionally, in an
in vivo xenograft model of KRAS-mutant lung adenocarcinoma,
Survivin knockdown and trametinib treatment induced cell death
(7). Moreover, in hepatocellular
carcinoma cells, treatment with ATB-263, a novel Bcl-2 inhibitor,
and silencing of Survivin induced cell apoptosis; these results
implied that Survivin knockdown is an important method to overcome
the hurdle of drug resistance in cancer therapy (8), and the development of a method for
silencing Survivin is urgently needed.
Therefore, in the present study, cell cycle arrest
and apoptosis were investigated in lung cancer cell lines treated
with SP, and downregulated Survivin expression and upregulated p21
expression was found. Based on the results of this study, the novel
utilization of propionate for lung cancer treatment is proposed,
due to its anticancer effects.
Materials and methods
Cell culture and reagents
H1299 and H1703 are non-small cell lung carcinoma
(NSCLC) cell lines. NSCLC accounts for ~85% of all lung cancer
cases and is more insensitive to chemotherapy than small cell lung
carcinoma (SCLC). As NSCLCs are a main lung cancer type and are
difficult to treat, NSCLC cell lines were selected to assess the
activity of propionate. The human lung cancer cell lines H1299 and
H1703 were purchased from the Korean Cell Line Bank and cultured in
RPMI supplemented with 10% FBS and 1% penicillin/streptomycin in a
humidified atmosphere with 5% CO2 at 37°C. The normal
human lung cell line MRC5 was purchased from the Korean Cell Line
Bank and cultured in MEM supplemented with 10% FBS and 1%
penicillin/streptomycin in a humidified atmosphere with 5%
CO2 at 37°C. SP (cat. no. P5436) was purchased from
Sigma-Aldrich; Merck KGaA. H1299 and H1703 cells were treated with
10 mM SP for 48 h. Distilled water was used for the control
treatments (9).
Cell viability assay
For crystal violet staining (10,11),
cells treated with 0 mM (DW), and 10 mM SP for 48 h were washed
twice with PBS and fixed with cold 100% methanol for 5 min at 20°C.
After being washed twice with PBS, the cells were stained with a
0.1% crystal violet solution (cat. no. C0775; Sigma Aldrich; Merck
KGaA) for 5 min at room temperature. The cells were then washed
five times with distilled water and observed under a light
microscope (magnification, ×100; OLYMPUS 1X71; Olympus
Corporation).
Fluorescence-activated cell sorting
(FACS) analysis
After treatment with SP for 48 h, the cells were
collected and incubated with Muse Annexin V & Dead Cell Reagent
(Merck KGaA; cat. no. MCH100105) for 20 min at room temperature.
After incubation, approximately 5×103 cells were
analyzed with a Muse cell analyzer (Merck KGaA) (12). FACS analysis with propidium iodide
staining was performed. For cell cycle analysis after treatment
with SP, cells treated with SP for 48 h were fixed with 70% ethanol
and incubated with Muse® Cell Cycle Assay reagent (Merck
KGaA; cat. no. MCH1001060) for 30 min at room temperature,
according to the manufacturer's instructions. To measure the
activity of caspase 3/7, the Muse Caspase-3/7 kit (Merck KGaA; cat.
no. MCH100108) was used. According to the user's guide, cells
treated with SP for 48 h were treated with Muse Caspase-3/7 working
solution and incubated for 30 min in a 37°C incubator with 5%
CO2. After incubation, ~5×103 cells were
analyzed with a Muse cell analyzer (EMD Millipore). The FACS
results were analyzed using Muse 1.5 Analysis software (Merck
KGaA).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was isolated from the indicated cell lines
using a Qiagen RNeasy Mini kit (Qiagen, Inc.), according to the
manufacturer's instructions. RNA aliquots of 1 µg were then reverse
transcribed using the iScript™ cDNA synthesis kit (Bio-Rad
Laboratories, Inc.), according to standard protocols: 5 min at
25°C, 20 min at 46°C and 1 min at 95°C. qPCRs were performed using
the AriaMx Real-Time PCR instrument (Agilent Technologies, Inc.),
according to the manufacturer's instructions. qPCR was performed on
cDNA samples using Brilliant III Ultra-Fast SYBR® Green
qPCR Master Mix (Agilent Technologies, Inc.), and the signal was
detected by the AriaMx Real-time PCR System (Agilent Technologies).
The thermocycling conditions were as follows: 95°C for 3 min,
followed by 40 cycles of 95°C for 5 sec and 55°C for 10 sec. The
fluorescence threshold value was calculated using Agilent Aria 1.6
software (Agilent Technologies, Inc.) (13,14).
Using the Agilent Aria 1.6 software, ΔCq values were calculated and
normalized to β-actin (ACTB; http://www.agilent.com/cs/library/applications/application-fod-cannabis_5994-0430en-agilent.pdf).
The following PCR primers were used: E2F transcription factor
(E2F)1 (forward, 5′-GGACCTTCGTAGCATTGCAG-3′ and reverse,
5′-CTGATCCCACCTACGGTCTC-3′), E2F2 (forward,
5′-AGGAGCTGAAGGAGCTGATG-3′ and reverse,
5′-TCTTGTTGGCCTTGTCCTCA-3′), E2F4 (forward,
5′-CGGGAGCAAGAACTAGACCA-3′ and reverse,
5′-TCCTCATGAGTGACGTAGGC-3′), CDK4 (forward,
5′-ACAGCTACCAGATGGCACTT-3′ and reverse,
5′-GTCGGCTTCAGAGTTTCCAC-3′), cyclin A2 (CCNA2; forward,
5′-CATGGACCTTCACCAGACCT-3′ and reverse,
5′-AGTGTCTCTGGTGGGTTGAG-3′), cyclin B2 (CCNB2; forward,
5′-AGTTCCAGTTCAACCCACCA-3′ and reverse,
5′-GCAGAGCAAGGCATCAGAAA-3′), p21 (forward,
5′-CTTTGTCACCGAGACACCAC-3′ and reverse,
5′-CAGGTCCACATGGTCTTCCT-3′), Survivin (forward,
5′-AGGACCACCGCATCTCTACAT-3′ and reverse,
5′-AAGTCTGGCTCGTTCTCAGTG-3′) and ACTB (forward,
5′-ACTCTTCCAGCCTTCCTTCC-3′ and reverse,
5′-CAATGCCAGGGTACATGGTG-3′).
Western blot analysis
Cells were washed once with PBS and then lysed in
cold lysis buffer [50 mM Tris HCl (pH 7.4), 150 mM NaCl, 1% Triton
X-100, 0.1% SDS, 1 mM EDTA, 1 mM Na3VO4, 1 mM
NaF and 1X protease inhibitor cocktail]. Cell lysates were
centrifuged at 14,000 × g for 15 min at 4°C, boiled in 5X sample
buffer, and subjected to protein determination (BSA; cat. no.
23208; Thermo Fisher Scientific, Inc.). Protein samples (10 µg)
were subjected to western blot analysis as follows: Nitrocellulose
membranes (cat. no. 1620145; Bio-Rad Laboratories, Inc.), blocking
reagent (5% skim milk; 1 h at room temperature) and precast gels
(4–20% Mini-PROTEAN® TGX™; cat. no. 456 1094; Bio-Rad
Laboratories, Inc.) were used with the indicated antibodies at a
1:1,000 dilution ratio (15,16).
Samples were stained with anti-Survivin (cat. no. 2803S), anti-poly
(ADP-ribose) polymerase (PARP; cat. no. 9542S), and anti-Caspase 3
(cat. no. 9662S) antibodies from Cell Signaling Technology, Inc.,
and anti-p21 (cat. no. SC-6246) and anti-ACTB (cat. no. SC-47778)
antibodies from Santa Cruz Biotechnology Inc. at 4°C (overnight).
Secondary antibodies at a 1:5,000 dilution (rabbit, cat. no.
SC-2357; mouse, cat. no. SC-2031; Santa Cruz Biotechnology, Inc.)
were incubated at room temperature for 1 h, and an ECL solution
(cat. no. 170-5060; Bio-Rad Laboratories, Inc.) was used for
visualization. The results were analyzed using ImageJ software
(version 1.8.0; National Institutes of Health).
Statistical analysis
Results are expressed as the mean ± SD of three
independent experiments. Student's t-test was used to assess
significance using Microsoft Excel 2013 (Microsoft Corporation).
P<0.05 was considered to indicate a statistically significant
difference.
Results
SP suppresses the growth of lung
cancer cell lines
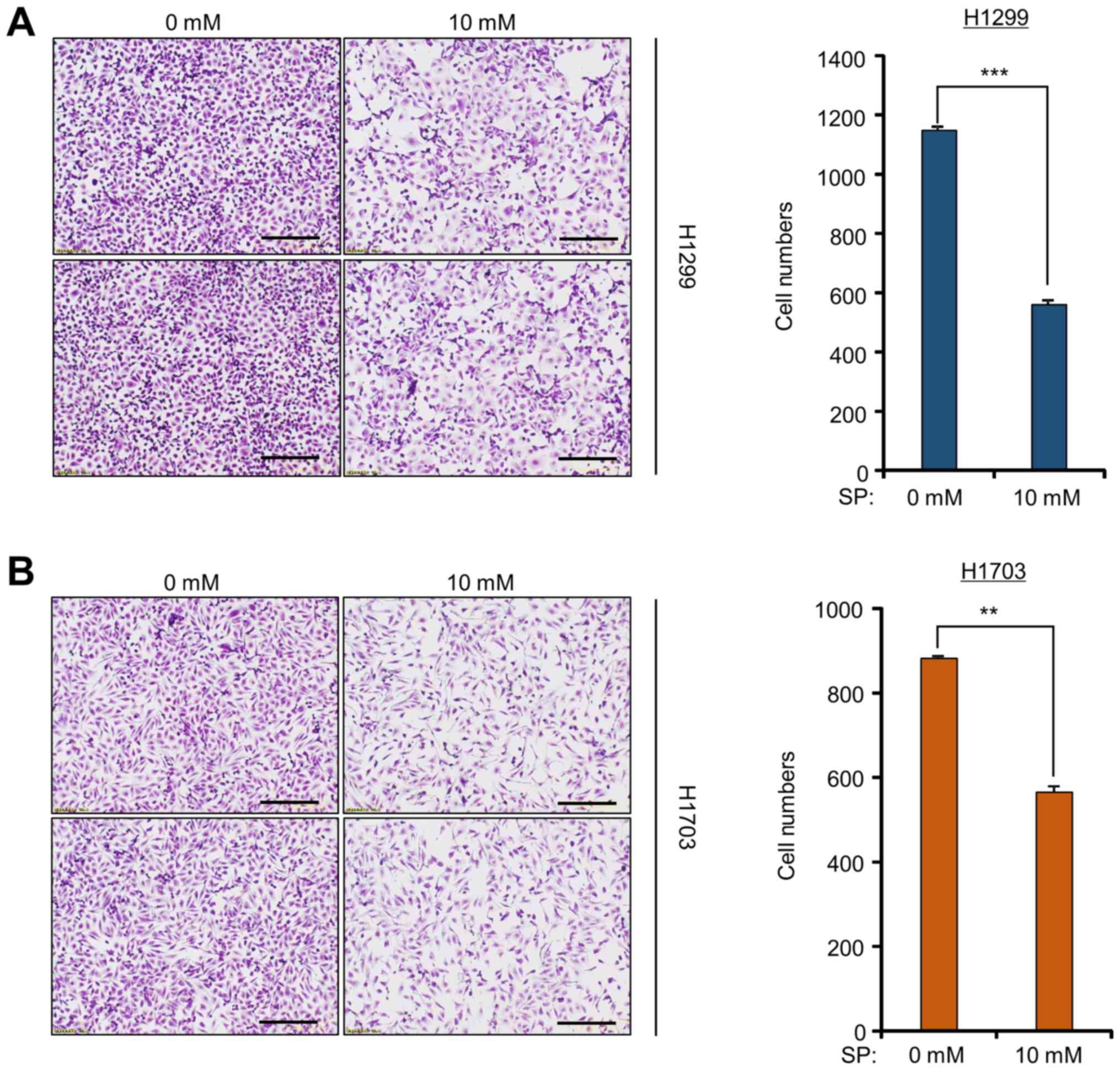
To investigate the anticancer effect of SP against
lung cancer, cell growth assays were performed on H1299 and H1703
cell lines after SP treatment. After 10 mM SP treatment, cell
growth was significantly lower than that after control treatment,
as determined by crystal violet staining (Fig. 1). However, treatment of a normal
lung cell line (MRC5) with SP had no effect on cell growth compared
to that observed in the lung cancer cell lines, as determined by
the cell growth assay (data not shown). Although several studies
have recently shown that SCFAs, including propionate, suppress
growth in colon cancer cell lines (17–19),
it was observed in the present study that SP also has anticancer
effects against lung cancer cell lines. Thus, an alternative
therapeutic method using SP is proposed for lung cancer
treatment.
SP induces G2/M arrest in lung cancer
cell lines
SCFA treatment initiates cell cycle arrest and
apoptosis in colon cancer cell lines (18,20).
Thus, the present study also assessed the effects of SP on the cell
cycle in lung cancer cell lines. To verify the relationship between
the cell cycle and SP treatment in the H1299 and H1703 cell lines,
FACS analysis with propidium iodide staining was performed. As
shown in Fig. 2A and B,
dose-dependent G2/M arrest was clearly observed after treatment of
the H1299 and H1703 cell lines with SP, implying that SP-induced
G2/M arrest inhibited cell growth in these lung cancer cell
lines.
Next, to evaluate the role of SP treatment at the
molecular level, primers were designed for the amplification of
various cell cycle-related genes (E2F1, E2F2, E2F4, CDK4, CCNA2,
CCNB2, p21 and Survivin) (6,21–23).
The RT-qPCR results clearly showed that the expression levels of
the E2F family members, CDK4, CCNA2, CCNB2 and Survivin were
significantly decreased after SP treatment in the H1299 and H1703
cell lines. Moreover, the expression of p21, a cyclin-dependent
kinase inhibitor (22), was
clearly induced by SP treatment in both lung cancer cell lines. In
particular, G2/M arrest-related genes (CCNA2, CCNB2, Survivin and
p21) were significantly reduced or induced in the SP-treated groups
compared to those in the control groups (Fig. 2C). Thus, it may be concluded that
SP treatment reduced the transcription of cell cycle-related genes,
especially those related to the G2/M phase, to inhibit the cell
growth of lung cancer cell lines.
SP treatment induces the apoptosis of
H1299 and H1703 cells
Li et al (6)
reported that silencing Survivin expression by siRNA treatment
resulted in cell apoptosis and G2/M arrest in the Heal and MCF7
cell lines. In addition, the upregulation of p21 expression induced
cell apoptosis and cell cycle arrest (24). Thus, to confirm the regulation of
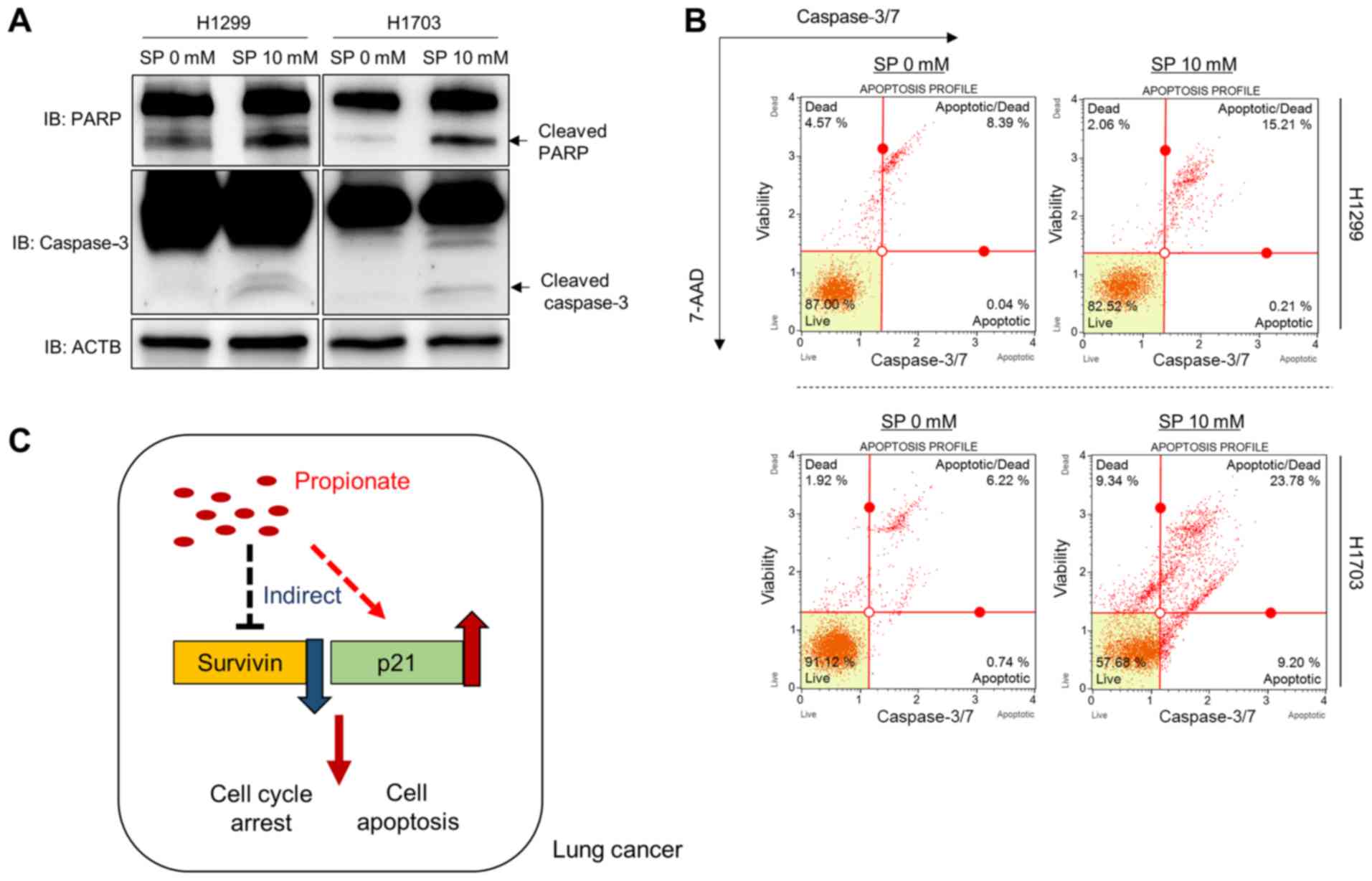
Survivin and p21 expression by SP treatment in more detail, western
blot analysis was performed with anti-Survivin and anti-p21
antibodies. After treatment with SP (10 mM), the Survivin and p21
protein expression levels were significantly decreased and
increased, respectively (Fig. 3A).
Next, to determine whether SP-induced growth suppression was
related to cell apoptosis, FACS analysis was performed using
Annexin V. The proportions of cells in early and late apoptosis
were greater in the SP treatment group than in the control group
(Fig. 3B). Moreover, the induction
of the apoptosis markers cleaved PARP and cleaved caspase 3 by SP
treatment was investigated in the H1299 and H1703 cell lines,
revealing that SP treatment induced cell apoptosis by controlling
Survivin and p21 expression in these lung cancer cell lines
(Fig. 4A). In addition, to confirm
the western blotting results, FACS analysis was performed to
measure the activity of caspase 3/7. As shown in Fig. 4B, the activity of caspase 3/7 was
increased by SP treatment. Taken together, the present data show
that SP treatment induced apoptosis and cell cycle arrest by
regulating Survivin and p21 expression in lung cancer cell lines,
similar to its effects on colon cancer; these molecular mechanism
data provide useful information on the potential role of the
anticancer effector propionate in lung cancer treatment (Fig. 4C).
Discussion
SCFAs derived from the microbiome affect colon
cancer proliferation by modulating histone hyperacetylation and
dysregulating Bcl-2, Bax, p21 and proliferating cell nuclear
antigen expression (18,25). In addition, sodium butyrate can
also suppress the proliferation of other cancer types, such as lung
and prostate cancer, by regulating p21 expression (26, 27).
However, as most studies on SCFAs have focused on butyrate, the
functionality of propionate in lung cancer is not widely known.
Therefore, the present study established a hypothesis regarding the
relationship between lung cancer and propionate based on several
previous studies (25–27), and provided novel functional
information regarding the role of propionate in lung cancer
treatment. To assess the function of propionate in lung cancer, it
was necessary to choose between two types of propionate, SP and
propionic acid (PA), because propionate cannot exist solely in
nature. However, when PA is added to cell culture media, the acidic
effects on cancer cells must be negated. Thus, to assess the
anticancer activity of propionate, SP was selected, although its
general use is as a mold inhibitor in baked goods.
SP inhibited lung cancer cell proliferation by
inducing cell cycle arrest, especially in the G2/M phase. Moreover,
to identify the relationship between cell cycle-related genes and
SP treatment, RT-qPCR analysis was performed, and it was found that
several cell cycle-related genes were up- or downregulated by SP
treatment. Although downregulation of the E2F family and CDK4 was
previously shown to be mainly involved in G1 arrest (21), the reduction rates of the E2F
family and CDK4 were broadly lower than those of CCNA2, CCNB2 and
Survivin as determined by RT-qPCR analysis. Thus, it was
hypothesized that in the lung cancer cell lines, SP treatment may
affect the entire cell cycle machinery, particularly G2/M arrest,
and that this regulation may suppress cell proliferation.
The anticancer drug epigallocatechin-3-gallate
(EGCG) augments the anticancer effect of leptomycin B (LMB)
treatment on lung cancer cells by upregulating p21 and
downregulating Survivin. Although the clinical usage of EGCG is a
subject of debate, combination therapy with LMB exerts a
synergistic effect on lung cancer cell lines (28). In the present study, the induction
of caspase 3/7 activity and apoptosis by SP treatment was observed
in lung cancer cell lines. Western blot analysis also showed that
SP treatment increased cleaved PARP and caspase 3 expression by
down- and upregulating Survivin and p21, respectively. Thus, the
novel suggestion is proposed that SP may function as a sensitizer
of LMB treatment and could be combined with LMB in lung cancer
therapy. However, to translate this hypothesis to the clinic, the
activity of SP must be evaluated in vivo, and an accurate
mode-of-action study on propionate treatment for the regulation of
Survivin and p21 expression is required.
In conclusion, although propionate treatment is
known to suppress cell growth and induce apoptosis in colon cancer
cell lines, the results of the present study suggested that it
could also function as an anticancer drug for the treatment of lung
cancer, by inducing cell apoptosis and cell cycle arrest by down-
and upregulating Survivin and p21 expression, respectively.
Therefore, although many hurdles must be cleared for SCFAs to be
applicable in lung cancer treatment, the present results suggest
that SCFAs derived from the microbiome and dietary therapy may help
patients with lung cancer; however, in vivo studies are
needed to validate the effectiveness of SP.
Acknowledgements
Not applicable.
Funding
This study was supported by a grant from the
National Research Foundation of Korea (NRF), funded by the Ministry
of Science, ICT and Future Planning (grant no.
NRF-2018M3A9H3023077).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MYS, DSK, JK and HSC were involved in the conception
and design of the study. KK, OK and TYR were involved in the
development of the methodology, and the analysis and interpretation
of the data. CRJ, JK, JKM, DSK, MYS and HSC were involved in the
writing, reviewing and/or revision of the manuscript. CRJ, JKM, JK
and MYS were responsible for administrative, technical or material
support. CRJ and JKM were involved in the design of the study,
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rea D, Coppola G, Palma G, Barbieri A,
Luciano A, Del Prete P, Rossetti S, Berretta M, Facchini G, Perdonà
S, et al: Microbiota effects on cancer: From risks to therapies.
Oncotarget. 9:17915–17927. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tan J, McKenzie C, Potamitis M, Thorburn
AN, Mackay CR and Macia L: The role of short-chain fatty acids in
health and disease. Adv Immunol. 121:91–119. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tian Y, Xu Q, Sun L, Ye Y and Ji G:
Short-chain fatty acids administration is protective in
colitis-associated colorectal cancer development. J Nutr Biochem.
57:103–109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu X, Wu Y, He L, Wu L, Wang X and Liu Z:
Effects of the intestinal microbial metabolite butyrate on the
development of colorectal cancer. J Cancer. 9:2510–2517. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xiao X, Cao Y and Chen H: Profiling and
characterization of microRNAs responding to sodium butyrate
treatment in A549 cells. J Cell Biochem. 119:3563–3573. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li Y, Liu D, Zhou Y, Li Y, Xie J, Lee RJ,
Cai Y and Teng L: Silencing of Survivin Expression Leads to Reduced
Proliferation and Cell Cycle Arrest in Cancer Cells. J Cancer.
6:1187–1194. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sumi T, Hirai S, Yamaguchi M, Tanaka Y,
Tada M, Yamada G, Hasegawa T, Miyagi Y, Niki T, Watanabe A, et al:
Survivin knockdown induces senescence in TTF 1-expressing,
KRAS-mutant lung adenocarcinomas. Int J Oncol. 53:33–46.
2018.PubMed/NCBI
|
|
8
|
Zhao X, Ogunwobi OO and Liu C: Survivin
inhibition is critical for Bcl-2 inhibitor-induced apoptosis in
hepatocellular carcinoma cells. PLoS One. 6:e219802011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim K, Son MY, Jung CR, Kim DS and Cho HS:
EHMT2 is a metastasis regulator in breast cancer. Biochem Biophys
Res Commun. 496:758–762. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim SK, Kim K, Ryu JW, Ryu TY, Lim JH, Oh
JH, Min JK, Jung CR, Hamamoto R, Son MY, et al: The novel
prognostic marker, EHMT2, is involved in cell proliferation via
HSPD1 regulation in breast cancer. Int J Oncol. 54:65–76.
2018.PubMed/NCBI
|
|
11
|
Ryu TY, Kim K, Kim SK, Oh JH, Min JK, Jung
CR, Son MY, Kim DS and Cho HS: SETDB1 regulates SMAD7 expression
for breast cancer metastasis. BMB Rep. 52:139–144. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ryu TY, Kim K, Son MY, Min JK, Kim J, Han
TS, Kim DS and Cho HS: Downregulation of PRMT1, a histone arginine
methyltransferase, by sodium propionate induces cell apoptosis in
colon cancer. Oncol Rep. 41:1691–1699. 2019.PubMed/NCBI
|
|
13
|
Kim DS, Ryu JW, Son MY, Oh JH, Chung KS,
Lee S, Lee JJ, Ahn JH, Min JS, Ahn J, et al: A liver-specific gene
expression panel predicts the differentiation status of in vitro
hepatocyte models. Hepatology. 66:1662–1674. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Son MY, Jung CR, Kim DS and Cho HS:
Comparative in silico profiling of epigenetic modifiers in human
tissues. Mol Biol Rep. 45:309–314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jung KB, Lee H, Son YS, Lee MO, Kim YD, Oh
SJ, Kwon O, Cho S, Cho HS, Kim DS, et al: Interleukin-2 induces the
in vitro maturation of human pluripotent stem cell-derived
intestinal organoids. Nat Commun. 9:30392018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ryu JW, Kim SK, Son MY, Jeon SJ, Oh JH,
Lim JH, Cho S, Jung CR, Hamamoto R, Kim DS, et al: Novel prognostic
marker PRMT1 regulates cell growth via downregulation of CDKN1A in
HCC. Oncotarget. 8:115444–115455. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Davie JR: Inhibition of histone
deacetylase activity by butyrate. J Nutr. 133 (Suppl
7):2485S–2493S. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hinnebusch BF, Meng S, Wu JT, Archer SY
and Hodin RA: The effects of short-chain fatty acids on human colon
cancer cell phenotype are associated with histone hyperacetylation.
J Nutr. 132:1012–1017. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ooi CC, Good NM, Williams DB, Lewanowitsch
T, Cosgrove LJ, Lockett TJ and Head RJ: Structure-activity
relationship of butyrate analogues on apoptosis, proliferation and
histone deacetylase activity in HCT-116 human colorectal cancer
cells. Clin Exp Pharmacol Physiol. 37:905–911. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heerdt BG, Houston MA and Augenlicht LH:
Short-chain fatty acid-initiated cell cycle arrest and apoptosis of
colonic epithelial cells is linked to mitochondrial function. Cell
Growth Differ. 8:523–532. 1997.PubMed/NCBI
|
|
21
|
Bertoli C, Skotheim JM and de Bruin RA:
Control of cell cycle transcription during G1 and S phases. Nat Rev
Mol Cell Biol. 14:518–528. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
El-Deiry WS: p21(WAF1) Mediates Cell-Cycle
Inhibition, Relevant to Cancer Suppression and Therapy. Cancer Res.
76:5189–5191. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park AM, Tsunoda I and Yoshie O: Heat
shock protein 27 promotes cell cycle progression by down-regulating
E2F transcription factor 4 and retinoblastoma family protein p130.
J Biol Chem. 293:15815–15826. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karimian A, Ahmadi Y and Yousefi B:
Multiple functions of p21 in cell cycle, apoptosis and
transcriptional regulation after DNA damage. DNA Repair (Amst).
42:63–71. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Emenaker NJ, Calaf GM, Cox D, Basson MD
and Qureshi N: Short-chain fatty acids inhibit invasive human colon
cancer by modulating uPA, TIMP-1, TIMP-2, mutant p53, Bcl-2, Bax,
p21 and PCNA protein expression in an in vitro cell culture model.
J Nutr. 131 (Suppl 11):3041S–3046S. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rephaeli A, Blank-Porat D, Tarasenko N,
Entin-Meer M, Levovich I, Cutts SM, Phillips DR, Malik Z and
Nudelman A: In vivo and in vitro antitumor activity of
butyroyloxymethyl-diethyl phosphate (AN-7), a histone deacetylase
inhibitor, in human prostate cancer. Int J Cancer. 116:226–235.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Edmond V, Brambilla C, Brambilla E,
Gazzeri S and Eymin B: SRSF2 is required for sodium
butyrate-mediated p21(WAF1) induction and premature senescence in
human lung carcinoma cell lines. Cell Cycle. 10:1968–1977. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cromie MM and Gao W:
Epigallocatechin-3-gallate enhances the therapeutic effects of
leptomycin B on human lung cancer a549 cells. Oxid Med Cell Longev.
2015:2173042015. View Article : Google Scholar : PubMed/NCBI
|