Introduction
Immunoglobulin A (IgA) nephropathy (IgAN) is the
most common chronic glomerular disease worldwide, and ~20% IgANs
develop into end-stage renal disease (ESRD) within 20 years of
treatment (1). The incidence of
IgAN varies in different regions, being much higher (30–50%) in
Asia than in Europe and the United States (5–25%). Compared to
other ethnicities, in Asian patients, IgAN has been revealed to be
more likely to progress to end-stage renal disease (ESRD) (2). IgAN accounts for >40% of primary
glomerular diseases in China (3),
being the top cause of ESRD there. IgAN has diverse clinical
manifestations, such as macroscopic or microscopic hematuria,
microscopic hematuria with proteinuria, nephrotic syndrome, and
renal dysfunction. Currently, there is no effective treatment
method for IgAN. Although cytotoxic drugs such as glucocorticoids
and cyclophosphamide can reduce proteinuria in some patients, their
long-term protective effect on renal function is not known
(4). Owing to a lack of auxiliary
diagnostic methods, the definitive diagnosis of IgAN relies mainly
on pathology. The etiology and pathogenesis of IgAN are not clear,
but the incidence and progression are mainly related to mucosal
infections such as those of the mouth, throat, and intestine
(5). Therefore, clarifying the
relationship between the composition of microflora in the digestive
tract and IgAN may provide insights on its pathogenesis, early
diagnosis, and treatment.
As the largest micro-ecological system of the human
body, the intestinal tract mainly includes intestinal bacteria, in
addition to a very small amount of viruses, mycoplasmas and fungi.
The intestinal tract harbors as many as 1,014 bacterial species,
constituting over 10 times the total number of human cells
(6,7). Therefore, homeostasis of the
intestinal flora is physiologically significant in promoting the
digestion and absorption of host nutrients, maintaining normal
physiological functions of the intestine, regulating body immunity,
and antagonizing the colonization of pathogenic microbes (8–10).
Changes in the intestinal environment disrupt the homeostasis of
the intestinal flora, causing autoimmune diseases such as
inflammatory bowel diseases, type I diabetes, cardiovascular
diseases, central nervous system diseases, allergic diseases,
rheumatoid arthritis, and systemic lupus erythematosus (11).
In addition, the oral microbial flora comprises one
of the most complex microbial communities known. Microbes in the
mouth and intestines interact with each other to some extent.
Bacteria in the mouth enter the stomach by food ingestion,
eventually reaching the intestines. At least 700 microbe species
have been identified to occur in the oral bacterial community or
biofilms (12). Similar to
intestinal flora, a disruption in oral flora is closely associated
with the occurrence of many malignant tumors and autoimmune
diseases (13,14).
The relationship between oral microflora and the
occurrence of IgAN is unclear at present; therefore, the salivary
flora of IgAN patients and healthy people was investigated and
compared by high-throughput sequencing to discover saliva-specific
microbes associated with IgAN. Furthermore, preliminary modeling in
combination with clinical indicators was performed to facilitate
the early diagnosis of IgAN and provide novel insights for its
treatment.
Materials and methods
Research subjects
Twenty-eight patients who were definitively
diagnosed with IgAN through renal biopsy at the Shenzhen Longhua
District Central Hospital (Shenzhen, China), were randomly selected
as the experimental group, while 25 healthy volunteers from the
same area served as normal control subjects. Each subject
volunteered to participate in the experiment and signed an informed
consent form. The clinical information of all the subjects is
listed in Table I. The study
protocol was approved by the Ethics Committee of Guangdong Medical
University, and informed consent was obtained from each patient and
healthy subject enrolled in the study.
 | Table I.General information of research
subjects. |
Table I.
General information of research
subjects.
| Parameters | Healthy control
group | IgAN group |
|---|
| Age | 36.12±6.71 | 36.14±8.87 |
| Male | 13 | 15 |
| Female | 12 | 13 |
| BMI | 21.72±2.13 | 22.31±3.29 |
| Proteinuria
(g/day) | 1.978±1.719 | 0.068±0.032 |
| Systolic blood
pressure (mmHg) | 112.2±11.55 | 139.04±22.72 |
| Diastolic blood
pressure (mmHg) | 71.76±10.3 | 86.14±14.33 |
| sCR (µmol/l) | 65.16±15.17 | 127.7±60.81 |
| eGFR (ml/min/1.73
m2) | 106.5±17.93 | 64.6±30.62 |
Subject exclusion criteria
Patients with diarrhea or other intestinal diseases
who had taken antibiotics, probiotics, or laxatives within 4 weeks
before sampling were excluded.
Collection of saliva samples
Subjects underwent comprehensive dental and
periodontal examinations by specialists who performed clinical
status assessments. Two hours after the subjects brushed their
teeth in the morning, they were required to spit 4–5 ml of saliva
directly into sterile collection containers over a time span of 30
min. The collected saliva was naturally produced, without any
stimulation. After collection, the samples were immediately mixed
with RNAlater (Sigma-Aldrich; Merck KGaA) and stored at −80°C until
use.
Nucleic acid extraction
Salivary microbial genomic DNA was extracted using
the HiPure Bacterial DNA kit (Magen) according to the
manufacturer's protocol. The integrity of the extracted DNA was
verified by agarose gel electrophoresis, and the extracted DNA was
stored at −20°C until use.
16S rRNA-based NGS library
construction
Primers were designed for the V4 region in the
microbial 16S rRNA gene, and the primers used for PCR included
Illumina Bridge PCR-compatible primers, barcode primers, and the V4
universal primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R
(5′-GGACTACHVGGGTWTCTAAT-3′). PCR products were purified with
magnetic beads, and the sizes were verified by an Agilent 2100
Bioanalyzer (Agilent Technologies, Inc.). This was followed by
quantification with Qubit 3.0 (Thermo Fisher Scientific, Inc.),
qPCR, and sequencing with the Illumina HiSeq 4000 platform
(Illumina, Inc.).
Raw data quality control and sequence
splicing
Quality filtering of paired-end raw read data was
performed using Trimmomatic software (version 0.33; http://www.usadellab.org/cms/?page=trimmomatic).
Concurrently, according to the barcode and primer information at
both ends of each sequence, sequences were assigned to the
corresponding samples with Mothur software (version 1.35.1;
http://www.mothur.org), after which the barcode
and primers were removed to obtain the paired-end clean reads after
quality control. Each pair of clean reads was spliced with FLASH
software (version 1.2.11; http://ccb.jhu.edu/software/FLASH/) to obtain the
original splicing sequence (raw tags). The spliced sequences were
subjected to quality control and were filtered to obtain effective
splicing fragments (clean tags).
Bioinformatics analysis
All the clean tags of the samples were clustered
with URESEARCH software (version 8 10.0.240; http://www.drive5.com/usearch/). The sequences were
clustered into operational taxonomic units (OTUs) as per 97%
identity. An OTU was considered to represent a species. The default
clustering method was set as UPARSE (http://www.drive5.com/uparse/). Using QIIME
(http://qiime.org), a sequence with the highest
frequency of occurrence was extracted from the sequence to which
each OTU belongs as a representative sequence of the OTU. Singleton
OTUs usually result from sequencing errors or the chimeras
generated during PCR. Therefore, singleton OTUs were removed with
USEARCH (http://www.drive5.com/usearch/) after clustering, and
chimeric sequences were removed using UCHIME (EdiTar Bio-Tech
Ltd.).
The representative sequence of each OTU was compared
with the Human Oral Microbiome Database (HOMD; http://www.homd.org) to obtain species annotation
information, thereby obtaining community composition information
for each sample. The OTUs and their tags that were annotated as
chloroplast or mitochondrion (16S amplicon) and those that could
not be annotated to genus level were excluded; thus, the valid tag
sequence and OTU classification for each sample were obtained.
Finally, the data of each sample were homogenized with the sample
with the minimum amount of data as the standard. Differences were
analyzed using GraphPad Prism version 6.0 (GraphPad Software, Inc.)
for nonparametric Mann-Whitney test analysis (two-tailed, 95% CI);
P<0.05 was considered to indicate a statistically significant
result.
Results
Characterization of sequencing
results
The salivary microbes of IgAN patients and healthy
control individuals were analyzed by sequencing the 16S rRNA gene
with Illumina HiSeq 4000. After quality control, the total number
of salivary microbial sequences from the 53 subjects was 2,842,648
reads, which included 1,386,638 from 25 healthy control subjects
(mean age, 36.12 years), at an average of 55,467 reads per sample,
and 1,455,965 from 28 IgAN patients (mean age, 36.14 years), at an
average of 51,998 reads per sample, with an average length of 500
bp each. Shannon-Wiener curves plateaued, indicating that the
sequencing depth could fully analyze the composition of salivary
microbes (Fig. 1).
Taxonomic analysis of salivary
microbes
The composition of salivary bacterial colonies was
analyzed in terms of a richness estimator (Chao1) and diversity
index (Shannon). IgAN patients (7,194 OTUs) harbored higher
microbial diversity, with an average of 256 OTUs, while the healthy
control group (6,305 OTUs) had an average of 252 OTUs. According to
the mean of the OTUs, the Shannon index was higher in the healthy
control group, while Chao 1 was higher in IgAN patients (Fig. 2). However, Wilcoxon analysis
revealed no significant difference in α-diversity (Chao1, Shannon)
between the groups. In the salivary microbes, 12 phyla were
identified [Absconditabacteria (SR1), Actinobacteria,
Bacteroidetes, Chloroflexi, Cyanobacteria, Firmicutes,
Fusobacteria, Gracilibacteria (GN02), Proteobacteria,
Saccharibacteria (TM7), Spirochaetes, and
Synergistetes]. There were no significant differences in the
above 12 phyla between the healthy control subjects and IgAN
patients (P<0.05; data not shown). In the experimental group,
Firmicutes, Bacteroidetes, Proteobacteria, and
Fusobacteria accounted for 92.05% of the total sequencing
data, while in the control group, these four phyla accounted for
93.49% of the total sequencing data (Fig. 3).
Ten genera with the highest relative abundance
(relative abundance >3%) were selected to compare the difference
between the experimental group and the control group. The abundance
of Granulicatella was significantly lower in the healthy
individuals than in the IgAN patients (P<0.05), while
Prevotella and Veillonella were significantly more
abundant in the healthy population than in IgAN patients (P<0.05
and P<0.01, respectively) (Fig.
4).
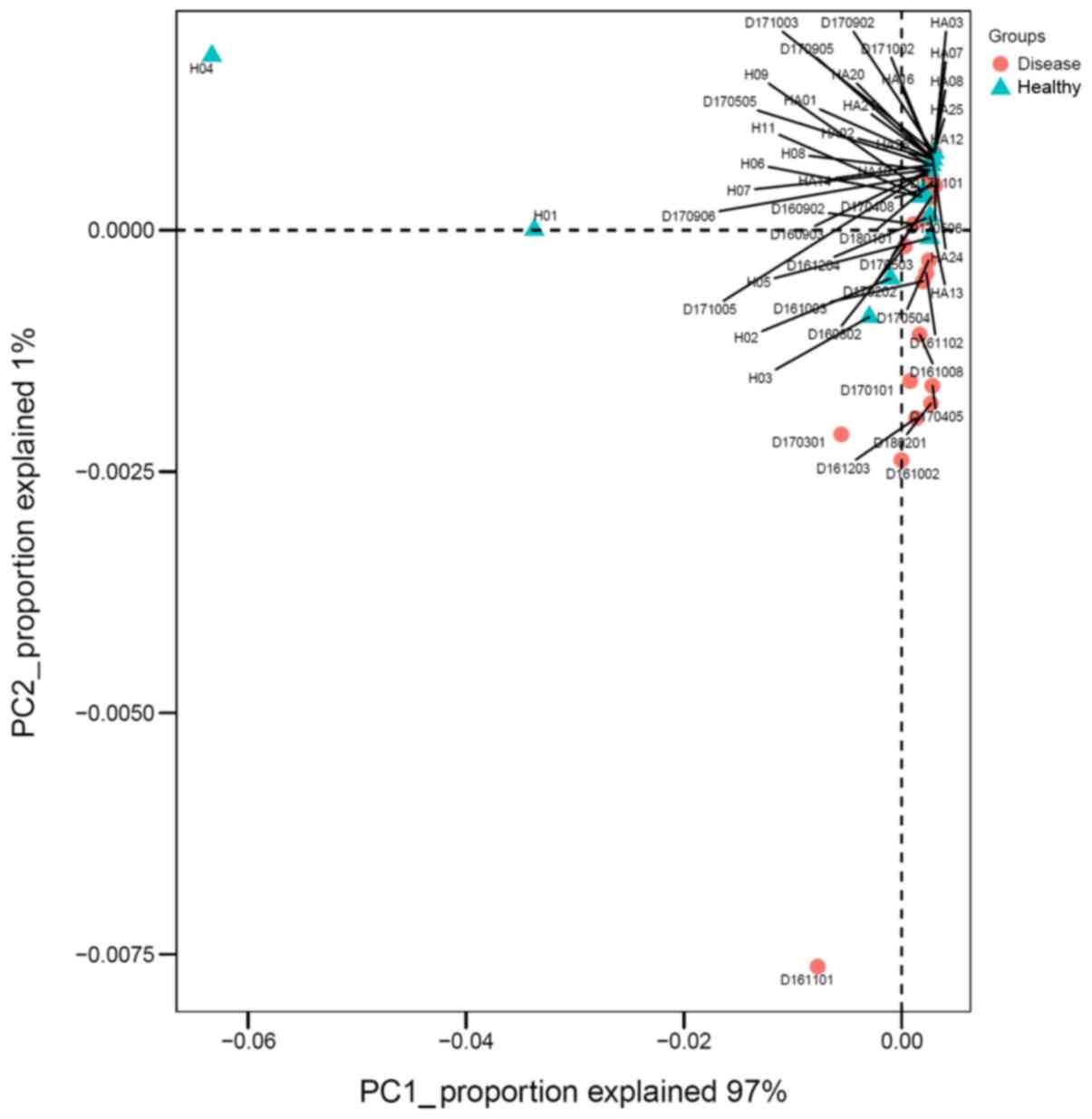
Principal component analysis
Principal component analysis (PCA) was conducted at
the genus level between the IgAN group and the control group.
Fig. 5 reveals the main components
1 and 2, which distinguished the IgAN group from the healthy
control group. Principal component 1 could explain 97% of
differences between the two groups.
Correlation analysis between
biochemical indicators and OTUs
The correlation between biochemical indicators,
Lee's grading system, and OTUs based on available data was
analyzed. The results revealed that the glomerular filtration rate
(eGFR) was positively correlated with OTU86 and OTU287 at
P<0.05, positively correlated with OTU165 at P<0.001, and
negatively correlated with OTU455 at P<0.05. The serum
creatinine (sCR) index was negatively correlated with OTU287 and
OTU165 at P<0.05 and P<0.001, respectively. Lee's grading
system was positively correlated with OTU255 at P<0.05, with
OTU200 at P<0.01, and with OTU455 and OTU75 at P<0.001, while
it was negatively correlated with OTU86, OTU287, and OTU788 at
P<0.05 and negatively correlated with OTU165 at P<0.01
(Fig. 6).
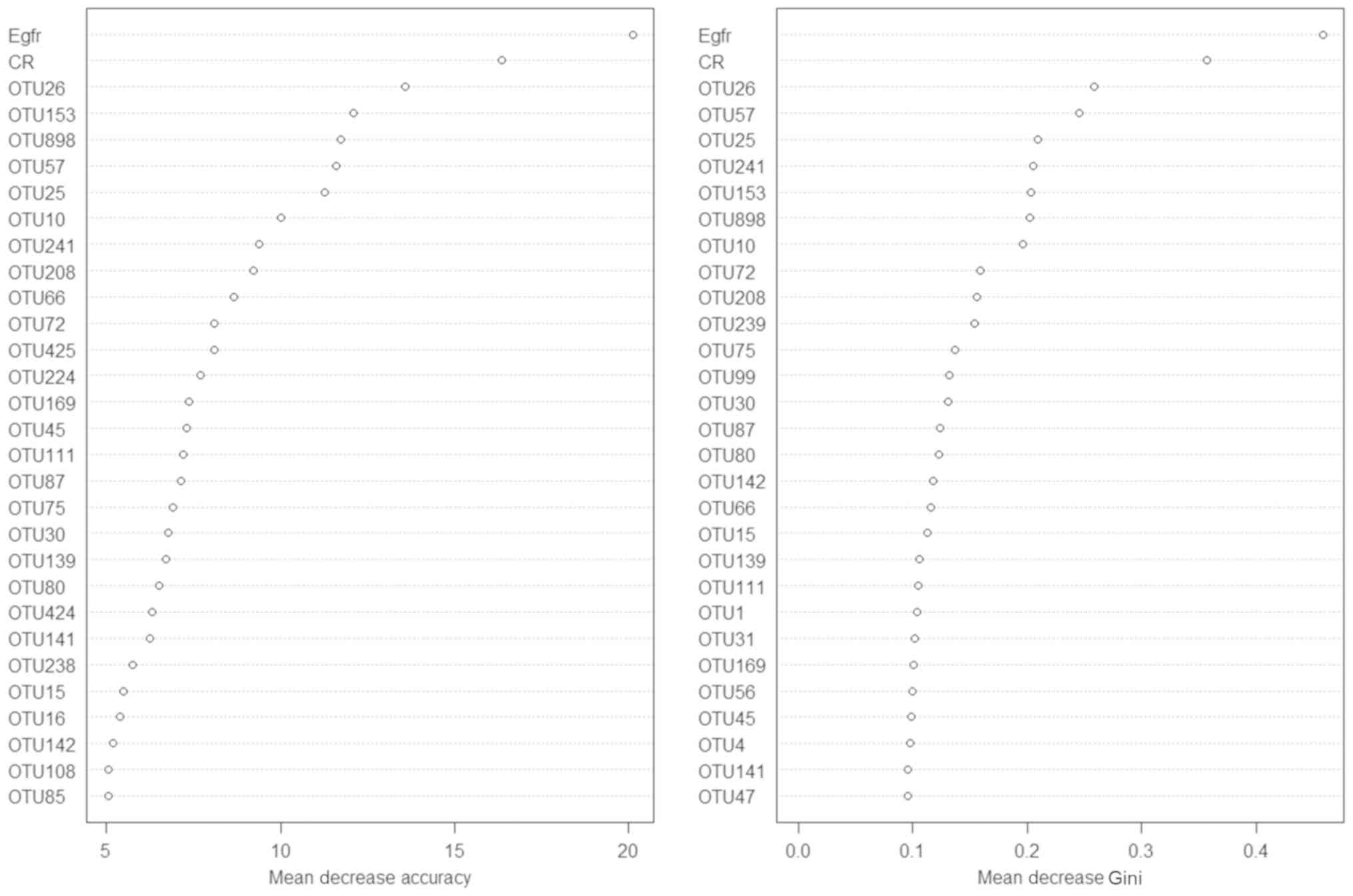
Random forest modeling
To distinguish the IgAN condition from the healthy
status, a random forest model based on the data of the existing
samples was established. First, 70% of the samples were used as the
training set to build the model. Next, the remaining 30% were used
as the test set to verify the model built. Two renal function test
biochemical indicators, sCR and eGFR, were then added for modeling.
The model without the biochemical indicators revealed a prediction
accuracy of 71.25%. After the addition of sCR and eGFR, the model
revealed a prediction accuracy of 79.49% (Fig. 7).
Discussion
The salivary microbes in IgAN patients and control
individuals were comprehensively explored using the Illumina HiSeq
4000 sequencing platform and the microbes with a non-culture
method, which is commonly used to analyze microbes that are
difficult to culture, were directly analyzed (15). Although the species level can be
identified under the condition of 3% OTU accuracy based on the full
length of 16S rDNA (16), the
number of species may be underestimated (17). Therefore, the microbial composition
of the V4 region in the 16S rRNA gene was only identified and the
microbial composition at the genus level based on the 3% OTU gap
rather than the species level was analyzed.
It was revealed that the overall microbial
composition of the healthy individuals and IgAN patients was very
similar, but there were some significant differences between them
at the genus level, consistent with previous studies (18). At the phylum level, IgAN patients
and healthy subjects did not differ: Both groups exhibited a high
abundance of Firmicutes, Bacteroidetes, Proteobacteria, and
Fusobacteria. Piccolo et al (18) revealed that the
Firmicutes/Proteobacteria ratio was significantly
lower in IgAN patients than in healthy individuals, however in our
experiment this ratio was determined to be 1, with no significant
decrease in IgAN. Nevertheless, this finding requires confirmation
in a larger sample size.
Further analysis at the genus level revealed that
levels of Veillonella of Firmicutes and
Prevotella of Bacteroidetes were significantly higher
in the control subjects than in the IgAN patients, while the levels
of Granulicatella of Firmicutes were significantly
higher in the IgAN patients than in the control subjects. Several
Prevotella spp., including P. melaninogenica, P.
intermedia, and P. loescheii, are present in the human
oral cavity (19,20). In the salivary microbiome, the
relative abundances of Prevotella spp. (P. nigrescens, P.
intermedia, P. pallens, and P. salivae) have been
revealed to be higher in healthy control individuals compared to
IgAN patients, and the only exception is P. aurantiaca
(18); the present results were
consistent with these previous findings.
The genus Veillonella is common in saliva.
Veillonella is present in different parts of the mouth
(tongue dorsum, lateral part of the tongue, buccal epithelium, hard
palate, soft palate, supragingival plaque of tooth surfaces,
subgingival plaque, maxillary anterior vestibule, and tonsillar
region) (12). Veillonella,
Granulicatella, and Streptococcus can produce
antimicrobial compounds (such as bacteriocins and hydrogen
peroxide) and inhibit pathogen growth; hence, they are associated
with oral health (21). The
present results revealed that the abundance of the genus
Granulicatella was significantly lower in healthy
individuals than in IgAN patients, which is inconsistent with the
result of Piccolo et al (18). This inconsistency may have been
caused by differences in the environment and diet. Further research
is warranted to determine the effect of Granulicatella on
the development of IgAN.
Oral dysbacteriosis has been revealed to be closely
associated with the occurrence of many malignant tumors and
autoimmune diseases (13,14,22).
Oral and throat infections are closely related to the occurrence
and development of IgAN. For example, nearly 30% of IgAN patients
may manifest with pharyngitis-related gross hematuria (23). A study of the tonsils excised from
IgAN patients revealed that the proliferation of microbes such as
Haemophilus, Campylobacter, and Treponema was
abnormally high and closely related to relief from proteinuria
(5). The present findings are
consistent with studies revealing that the salivary microbial flora
is very complex, irrespective of IgAN. Although there was no
significant difference between healthy control individuals and IgAN
patients at the phylum level, both groups significantly differed in
Veillonella, Granulicatella, and Prevotella
abundance. Because microbes are affected by many factors such as
diet and environment, some OTUs that had a relatively low content
were removed and the top 10 genera with high relative abundance
were only selected to compare the significant differences between
the two groups. Some research has been performed on the composition
and identification of microbes in saliva, however varying results
have been obtained due to the small sample size. Although the
present study revealed significant differences in the three genera
between IgAN patients and healthy subjects, the relationship
between these differential floras and IgAN remains unclear.
Therefore, the role of these differential floras in IgAN requires
further studying.
eGFR is an important indicator of renal filtration
function, indicating the amount of ultrafiltrate produced by two
kidneys per unit time (per minute). sCR, generally considered to be
endogenous serum creatinine, is a product of human muscle
metabolism. Clinically, detecting sCR is one of the main methods
used to understand renal function. Based on the data of the current
53 samples, the correlation between biochemical indicators and OTUs
(Fig. 6) was preliminarily
analyzed to establish a link between them and to explore the
possibility of applying the OTU-assisted clinical indicators sCR
and eGFR to diagnose and treat IgAN.
Lee's glomerular grading system is useful to compare
biopsy specimens and to predict the natural course of disease in
IgAN. The typical pathological change in IgAN is the deposition of
IgA in the glomerular mesangium. IgAN can be divided into five
stages according to the severity of mesangial proliferation,
crescent formation, and tubulointerstitial changes. In the present
study, the correlation of pathological changes and OTUs (Fig. 6) was investigated to explore the
possible relationship between them.
Although IgAN is the most common chronic glomerular
disease, few clinical auxiliary diagnostic methods are available.
The poor accuracy of sCR and eGFR in the clinical setting results
in some IgAN patients failing to receive timely diagnosis and
treatment (24–26) or relying on invasive pathological
diagnosis, thereby causing greater physical injury. Therefore,
additional auxiliary diagnostic methods need to be developed. In
the present study, the existing data was fully utilized and a
mathematical model was created, that is expected to distinguish
healthy people from IgAN patients to assist clinical diagnosis.
When this model is combined with the biochemical indicators sCR and
eGFR, its accuracy can reach up to nearly 80% (Fig. 7). However, the availability of more
samples is likely to improve the accuracy of the model.
Nevertheless, the present study provides the landscape of oral
microbiota in IgAN patients; the difference in genera between the
IgAN group and control group provides possible therapeutic targets
for IgAN, beyond diagnostic applications.
In conclusion, the salivary microbiota landscape in
IgAN patients and healthy subjects was compared, the differences
were preliminarily analyzed, the correlation of OTUs with
biochemical indicators (eGFR and sCR) and pathological changes was
explored, and a mathematical model was established to assist in the
clinical diagnosis of IgAN. This provides a promising approach for
the early diagnosis of IgAN patients, which has implications in the
early and effective treatment of this condition.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from The
Shenzhen Foundation of Science and Technology (grant/award nos.
JCYJ20160428142040945, JCYJ20180306172449376 and
JCYJ20180306172459580), The National Natural Science Foundation of
China (grant/award no. 81502410), the Natural Science Foundation of
Guangdong Province (grant/award no. 2016A030313028), The Medical
Scientific Research Foundation of Guangdong Province (grant/award
no. B2018003), and The Shenzhen Longhua District Foundation of
Science and Technology (grant/award no. 2017013).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. All sequencing data related to present study can be found
at https://submit.ncbi.nlm.nih.gov/subs/sra/SUB5024679/files.
Authors' contributions
SL and HG supervised the study, wrote the manuscript
and analyzed the data. SZ, HZ, RL, CL, PZ, XW and WL collected the
samples from patients. YZ and XW revised the manuscript and helped
analyze the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study protocol was approved by The Ethics
Committee of Guangdong Medical University. Informed consent was
obtained from each patient and healthy subject enrolled in the
study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Moriyama T, Tanaka K, Iwasaki C, Oshima Y,
Ochi A, Kataoka H, Itabashi M, Takei T, Uchida K and Nitta K:
Prognosis in IgA nephropathy: 30-year analysis of 1,012 patients at
a single center in Japan. PLoS One. 9:e917562014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Barbour SJ, Cattran DC, Kim SJ, Levin A,
Wald R, Hladunewich MA and Reich HN: Individuals of Pacific Asian
origin with IgA nephropathy have an increased risk of progression
to end-stage renal disease. Kidney Int. 84:1017–1024. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Salvadori M and Rosso G: Update on
immunoglobulin A nephropathy, Part I: Pathophysiology. World J
Nephrol. 4:455–467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Floege J and Eitner F: Current therapy for
IgA nephropathy. J Am Soc Nephrol. 22:1785–1794. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nagasawa Y, Iio K, Fukuda S, Date Y,
Iwatani H, Yamamoto R, Horii A, Inohara H, Imai E, Nakanishi T, et
al: Periodontal disease bacteria specific to tonsil in IgA
nephropathy patients predicts the remission by the treatment. PLoS
One. 9:e816362014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sekirov I, Russell SL, Antunes LC and
Finlay BB: Gut microbiota in health and disease. Physiol Rev.
90:859–904. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Arumugam M, Raes J, Pelletier E, Le
Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto
JM, et al: Enterotypes of the human gut microbiome. Nature.
473:174–180. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bron PA, van Baarlen P and Kleerebezem M:
Emerging molecular insights into the interaction between probiotics
and the host intestinal mucosa. Nat Rev Microbiol. 10:66–78. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maynard CL, Elson CO, Hatton RD and Weaver
CT: Reciprocal interactions of the intestinal microbiota and immune
system. Nature. 489:231–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Slack E, Hapfelmeier S, Stecher B,
Velykoredko Y, Stoel M, Lawson MA, Geuking MB, Beutler B, Tedder
TF, Hardt WD, et al: Innate and adaptive immunity cooperate
flexibly to maintain host-microbiota mutualism. Science.
325:617–620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kronbichler A, Kerschbaum J and Mayer G:
The influence and role of microbial factors in autoimmune Kidney
diseases: A systematic review. J Immunol Res. 2015:8580272015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Aas JA, Paster BJ, Stokes LN, Olsen I and
Dewhirst FE: Defining the normal bacterial flora of the oral
cavity. J Clin Microbiol. 43:5721–5732. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mager DL, Haffajee AD, Devlin PM, Norris
CM, Posner MR and Goodson JM: The salivary microbiota as a
diagnostic indicator of oral cancer: A descriptive, non-randomized
study of cancer-free and oral squamous cell carcinoma subjects. J
Transl Med. 3:272005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Farrell JJ, Zhang L, Zhou H, Chia D,
Elashoff D, Akin D, Paster BJ, Joshipura K and Wong DT: Variations
of oral microbiota are associated with pancreatic diseases
including pancreatic cancer. Gut. 61:582–588. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Watanabe H, Goto S, Mori H, Higashi K,
Hosomichi K, Aizawa N, Takahashi N, Tsuchida M, Suzuki Y, Yamada T,
et al: Comprehensive microbiome analysis of tonsillar crypts in IgA
nephropathy. Nephrol Dial Transplant. 32:2072–2079. 2017.PubMed/NCBI
|
|
16
|
Acinas SG, Klepac-Ceraj V, Hunt DE,
Pharino C, Ceraj I, Distel DL and Polz MF: Fine-scale phylogenetic
architecture of a complex bacterial community. Nature. 430:551–554.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pedrós-Alió C: Marine microbial diversity:
Can it be determined? Trends Microbiol. 14:257–263. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Piccolo M, De Angelis M, Lauriero G,
Montemurno E, Di Cagno R, Gesualdo L and Gobbetti M: Salivary
microbiota associated with immunoglobulin A nephropathy. Microb
Ecol. 70:557–565. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gmur R and Thurnheer T: Direct
quantitative differentiation between Prevotella intermedia
and Prevotella nigrescens in clinical specimens.
Microbiology. 148:1379–1387. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kononen E: Pigmented Prevotella
species in the periodontally healthy oral cavity. FEMS Immunol Med
Microbiol. 6:201–205. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar PS, Griffen AL, Moeschberger ML and
Leys EJ: Identification of candidate periodontal pathogens and
beneficial species by quantitative 16S clonal analysis. J Clin
Microbiol. 43:3944–3955. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Finegold SM: Host factors predisposing to
anaerobic infections. FEMS Immunol Med Microbiol. 6:159–163. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Moreno JA, Martin-Cleary C, Gutierrez E,
Toldos O, Blanco-Colio LM, Praga M, Ortiz A and Egido J: AKI
associated with macroscopic glomerular hematuria: Clinical and
pathophysiologic consequences. Clin J Am Soc Nephrol. 7:175–184.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shemesh O, Golbetz H, Kriss JP and Myers
BD: Limitations of creatinine as a filtration marker in
glomerulopathic patients. Kidney Int. 28:830–838. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Caregaro L, Menon F, Angeli P, Amodio P,
Merkel C, Bortoluzzi A, Alberino F and Gatta A: Limitations of
serum creatinine level and creatinine clearance as filtration
markers in cirrhosis. Arch Intern Med. 154:201–205. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rule AD, Rodeheffer RJ, Larson TS, Burnett
JC Jr, Cosio FG, Turner ST and Jacobsen SJ: Limitations of
estimating glomerular filtration rate from serum creatinine in the
general population. Mayo Clinic Proc. 81:1427–1434. 2006.
View Article : Google Scholar
|