Introduction
The microtubule cytoskeleton is essential for almost
all cellular activities. Microtubules themselves are the main
components of important structures, such as spindles, axons and
centrosomes, and mutations in tubulin-encoding genes cause severe
human diseases, with their common cause being severe defects in
microtubule organization (1–3). A
key feature of the microtubule, upon which all its functions rely,
is its dynamicity. For example, during interphase or each stage of
mitosis in somatic cells, microtubules remain highly dynamic
(growth, shrinkage, pause, catastrophe) at either or both ends to
achieve diverse processes, such as vesicle and organelle transport,
chromosome segregation, centrosome orientation and protein turnover
(4–6). The dynamicity of microtubules is
primarily determined by the cooperation of different families of
microtubule-associated proteins (MAPs), thus, mutations or
abnormalities of these MAPs are also the primary cause of certain
severe human diseases (7–9). Among the various MAPs,
microtubule-severing proteins (MTSPs), a meiotic subfamily of the
AAA superfamily (containing ATP-associated proteins with diverse
cellular activities), are crucial for microtubule (MT) dynamicity
(10,11). MTSPs primarily use their
microtubule-severing activities to sever MTs near their plus or
minus ends to remove the protective caps, thereby exposing the free
ends to MT polymerizer or depolymerizer (12–15).
Another proposed major function of MTSPs is ‘nucleating’, which
entails severing a long microtubule into multiple short segments so
that each short part can polymerize into a new long microtubule to
promote efficient MT organization (16). Notably, the mutation or deletion of
MTSPs causes severe hereditary disease in humans (17), neuronal disorders, developmental
problems and subfertility in mouse models (18–23).
Although MTSPs share a conserved AAA domain, their other regions
are entirely different, so they localize distinctly and function
cooperatively but also individually (13).
The mechanism of cell division, either mitosis or
meiosis, is a core issue in the study of the cell cycle. Abnormal
cell division can cause aneuploidy, cancer cell-like features or
infertile gametes. In normal meiosis, especially in female meiosis,
gene stability is critical, and meiotic abnormalities are closely
related to many human genetic diseases (3,24–26).
Compared to mitosis, meiosis is a more unique process. Oocytes and
spermatocytes do not have a typical centrosome, but some key
centrosome components are found at the spindle poles and are
essential for the organization of meiotic spindle poles. The
division apparatus (regardless of the fact that its function is not
limited to division), the spindle, is constructed from dynamic
microtubules and all types of MAPs including MTSPs. Katanin p60
ATPase-containing subunit A-like 1 (p60 katanin-like 1) is an MTSP
that exhibits typical microtubule-severing activity, and a study
revealed that p60 katanin-like 1 is essential to maintain normal
microtubule intensity at the poles of mitotic somatic cells
(27). Additionally, p60
katanin-like 1 was revealed to regulate terminal dendrite stability
and dendrite pruning in sensory neurons, and missense mutations
caused intellectual disability and microcephaly in humans and
defects in neuronal migration and morphology (18,19,21).
However, no studies have been performed on the function of p60
katanin-like 1 in mammalian female meiosis. In the present study,
it was revealed that p60 katanin-like 1, a member of the MTSP
family, was predominantly enriched within oocytes and ovaries and
essential for oocyte meiosis and maturation.
Materials and methods
General chemicals, reagents cells and
animals
Chemicals and reagents were obtained from
Sigma-Aldrich; Merck KGaA unless otherwise stated. The NIH3T3 cell
line was purchased from the American Type Culture Collection. A
total of 265 3 week old female specific pathogen free ICR mice
(weighing 18–20 g) used in this study were obtained from Vital
River Experimental Animal Technical Co., Ltd. Animals were housed
at a temperature of 20–26°C and a humidity of 40–70% with a 12 h
light/dark cycle. The mice were fed in feeding boxes, food was
replaced 2 times a week and the water bottle was replaced 3 times a
week. All animal experiments were approved by the Animal Care and
Use Committee of Nanjing Medical University (Nanjing, China) and
were performed in accordance with institutional guidelines.
Antibodies
Mouse monoclonal anti-β-actin (cat. no. A5316-100)
antibody was obtained from Sigma-Aldrich; Merck KGaA. Mouse
monoclonal anti-katanin p60 AL1 (A-10) (cat. no. sc-373814) and
mouse monoclonal anti-β-tubulin (cat. no. sc-5274) antibodies were
purchased from Santa Cruz Biotechnology, Inc. Human anti-centromere
CREST antibody (cat. no. 15-234) was purchased from Antibodies
Incorporated. Mouse monoclonal anti-EGFP (F56-6A1.2.3) (cat. no.
ab184601) was purchased from Abcam. Cy2-conjugated donkey
anti-mouse IgG (code no. 715-225-150), rhodamine (TRITC)-conjugated
donkey anti-goat IgG (code no. 705-025-147), and Alexa Fluor
647-conjugated donkey anti-human IgG (code no. 709-605-149) were
purchased from Jackson ImmunoResearch Laboratories, Inc.
Horseradish peroxidase (HRP)-conjugated rabbit anti-goat IgG (cat.
no. 31402) and HRP-conjugated goat anti-mouse IgG (cat. no. 31430)
were purchased from Invitrogen; Thermo Fisher Scientific, Inc.
Oocyte collection and culture
Immature oocytes arrested in prophase I [germinal
vesicle (GV) oocytes] were obtained from the ovaries of 3 to
4-week-old female ICR mice. The mice were first euthanized with
CO2 and then sacrificed by cervical dislocation, and the
ovaries were isolated and placed in operation medium (HEPES) with
2.5 nM milrinone and 10% fetal bovine serum (FBS) (Gibco; Thermo
Fisher Scientific, Inc.). Oocytes were released from the ovary by
puncturing the follicles with a hypodermic needle. Cumulus cells
were washed off the cumulus-oocyte complexes, and 50 isolated
denuded oocytes were placed in 100-µl droplets of culture medium
under mineral oil in plastic dishes (BD Biosciences). The culture
medium was MEM+ (MEM with 0.01 mM EDTA, 0.23 mM Na-pyruvate, 0.2 mM
pen/strep, 3 mg/ml BSA and 20% FBS). The oocytes were cultured at
37.0°C, 5% O2, and 5% CO2 in a humidified
atmosphere. Prior to in vitro maturation (IVM), all culture
medium included 2.5 nM milrinone to prevent the resumption of
meiosis.
siRNA production and transfection
Sequences of all DNA templates used for siRNA
production are listed in Table I.
The sequence of the control templates was a mock sequence that did
not specifically bind to any mRNA from the mouse genome. DNA
templates against four different DNA coding (sequence coding for
the amino acids in a protein, CDS) regions of KATNAL1 siRNA were
designed online through BLOCK-iT™ RNAi Designer (http://rnaidesigner.invitrogen.com/rnaiexpress/) with
some modifications. The sequence specificity was verified through a
BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) homology
search.
 | Table I.DNA oligos for siRNA production. |
Table I.
DNA oligos for siRNA production.
| Target site | DNA templates |
|---|
| KATNAL1 CDS
58–76a | Oligo1:
GGATCCTAATACGACTCACTATAGGAAGAATATGAACAGGTTb |
|
|
Oligo2:AAAACCTGTTCATATTCTTCCTATAGTGAGTCGTATTAGGATCCb |
|
| Oligo3:
GGATCCTAATACGACTCACTATAAACCTGTTCATATTCTTCCb |
|
|
Oligo4:AAGGAAGAATATGAACAGGTTTATAGTGAGTCGTATTAGGATCCb |
| KATNAL1 CDS
581–599a | Oligo1:
GGATCCTAATACGACTCACTATAGGGACATTGTGTCCAGGAAb |
|
|
Oligo2:AATTCCTGGACACAATGTCCCTATAGTGAGTCGTATTAGGATCCb |
|
| Oligo3:
GGATCCTAATACGACTCACTATATTCCTGGACACAATGTCCCb |
|
|
Oligo4:AAGGGACATTGTGTCCAGGAATATAGTGAGTCGTATTAGGATCCb |
| KATNAL1 CDS
701–719a | Oligo1:
GGATCCTAATACGACTCACTATAGGATTAGAAGGCCATGGAAb |
|
|
Oligo2:AATTCCATGGCCTTCTAATCCTATAGTGAGTCGTATTAGGATCCb |
|
| Oligo3:
GGATCCTAATACGACTCACTATATTCCATGGCCTTCTAATCCb |
|
|
Oligo4:AAGGATTAGAAGGCCATGGAATATAGTGAGTCGTATTAGGATCCb |
| KATNAL1 CDS
922–940a | Oligo1:
GGATCCTAATACGACTCACTATAGATTCTATCTGCAGTCGAAb |
|
|
Oligo2:AATTCGACTGCAGATAGAATCTATAGTGAGTCGTATTAGGATCCb |
|
| Oligo3:
GGATCCTAATACGACTCACTATATTCGACTGCAGATAGAATCb |
|
|
Oligo4:AAGATTCTATCTGCAGTCGAATATAGTGAGTCGTATTAGGATCCb |
|
Controlc | Oligo1:
GGATCCTAATACGACTCACTATACCTACGCCACCAATTTCGTTTb |
|
|
Oligo2:AAAAACGAAATTGGTGGCGTAGGTATAGTGAGTCGTATTAGGATCCb |
|
| Oligo3:
GGATCCTAATACGACTCACTATAAAACGAAATTGGTGGCGTAGGb |
|
|
Oligo4:AACCTACGCCACCAATTTCGTTTTATAGTGAGTCGTATTAGGATCCb |
siRNAs were produced using the T7 RiboMAX™ Express
RNAi System (Promega Corporation), according to the manufacturer's
instructions. Briefly, for each double-stranded siRNA against one
of the four KATNAL1 CDS regions, two pairs of synthesized
complementary single-stranded DNA oligonucleotides were first
annealed to form two double-stranded DNA templates. Subsequently,
two complementary single-stranded siRNAs were separately
synthesized in accordance with these two templates and then
annealed to form a final double-stranded siRNA. Next, the siRNA was
purified by conventional phenol/chloroform/isopropanol
precipitation and then aliquoted and stored at −80°C after a
quality check on an agarose gel. A ready-to-use siRNA mixture was
prepared by mixing the siRNAs against four target regions together
at an equal molar ratio to a final concentration of 5 µM.
For siRNA transfection, the N-TERM™ Nanoparticle
siRNA Transfection System (Sigma-Aldrich; Merck KGaA) was used.
Briefly, two tubes, one containing 1.1 µl N-TERM nanoparticles in
5.15 µl nuclease-free water (Acros Organics) and the other
containing 1.625 µl of the siRNA mixture (5 µM) in 4.625 µl of
siRNA dilution buffer (provided by the kit) were prepared; they
were then gently mixed and incubated at room temperature (RT) for
20 min. Next, the siRNA-nanoparticle complex solution was added
into a 100-µl drop of medium containing 50 oocytes. After treatment
for 12–14 h, the oocytes were washed to remove the
nanoparticle-containing medium. After a period of 1–2 h, another
round or two rounds of siRNA treatment were performed, depending on
how difficult the target was to significantly knock down. During
the whole siRNA treatment, which was typically 36–44 h long, 2.5 nM
milrinone was included to prevent the resumption of meiosis. Next,
the oocytes were transferred into milrinone-free MEM+ and cultured
for 8 or 16 h. They were then used for the phenotype analysis
experiment described below.
Plasmid construction and mRNA
synthesis
Total RNA was extracted from 100 mouse oocytes using
an Arcturus PicoPure RNA Isolation kit (Applied Biosystems; Thermo
Fisher Scientific, Inc.), and cDNA was generated with a QIAquick
PCR Purification kit (Qiagen GmBH). The following primers were used
to amplify the CDS sequence of KATNAL1: Forward primer,
5′-CGCGGATCCGCCACCATGAATTTGGCGGAGATTTGTG-3′ and reverse primer,
5′-AAGGAAAAAAGCGGCCGCTCATGCAGACCCAAACTCAAC-3′. The PCR products
were purified, digested with BamHI and NotI (New
England Biolabs, Inc.), and then cloned into the pCS2+
vector with EGFP tags.
To synthesize EGFP-KATNAL1 mRNA, the
KATNAL1-pCS2+ plasmids were linearized using
EcoRI. Capped cRNAs were produced using in vitro
transcription with an SP6 mMessage mMachine kit (Ambion; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions and then purified by an RNeasy Micro kit (Qiagen
GmBH). The synthesized RNA was portioned into aliquots and stored
at −80°C.
Microinjections of mRNA with a Narishige
microinjector (Narishige Group) were used to knock down or
overexpress specific proteins in mouse oocytes. Ten picoliters of
the mRNA solution (10 ng/ml) was injected into the oocyte cytoplasm
for overexpression analysis. The same amount of RNase-free PBS was
injected as a control.
After the injections, the oocytes were arrested at
the GV stage in M2 medium containing 2.5 mM milrinone for 20 h to
either facilitate the knockdown of mRNA translation or permit
overexpression. Following three washes, the oocytes were cultured
in milrinone-free medium for different time-points to evaluate the
cellular events during maturation.
Immunofluorescence
The oocytes were briefly washed in PBS with 0.05%
polyvinylpyrrolidone (PVP), permeated in 0.5% Triton X-100/PHEM (60
mM PIPES, 25 mM HEPES pH 6.9, 10 mM EGTA, 8 mM MgSO4) for 5 min and
washed three times rapidly in PBS/PVP. Next, the oocytes were fixed
in 3.7% paraformaldehyde (PFA)/PHEM for 20 min at room temperature,
washed three times (10 min each) in PBS/PVP and blocked with
blocking buffer (1% BSA/PHEM with 100 mM glycine) at room
temperature for 1 h. Then, the oocytes were in sequence incubated
at 4°C overnight with a primary antibody diluted in blocking
buffer, washed three times (10 min each) in PBS with 0.05% Tween-20
(PBST), incubated at room temperature for 45 min with a secondary
antibody diluted in blocking buffer (1:750 in all cases), and
washed three times (10 min each) in PBST. Finally, the DNA was
stained with 10 µg/ml Hoechst 33258 (Sigma-Aldrich; Merck KGaA) at
room temperature for 10 min, and the oocytes were mounted onto a
slide with mounting medium (0.5% propyl gallate, 0.1 M Tris-HCl, pH
7.4, 88% glycerol) and covered with a cover glass (thickness,
0.13–0.17 µm). To maintain the dimension of the oocytes, two strips
of double-stick tape (thickness, 90 µm) were placed between the
slide and cover glass. The primary antibodies were diluted as
follows: Anti-p60 katanin-like 1, 1:200; anti-tubulin, 1:500;
anti-human centromere, 1:500. The oocytes were examined with an
Andor Revolution spinning disk confocal workstation (Oxford
instruments).
Western blotting
A total of 100 oocytes were lysed in Laemmli sample
buffer (Bio-Rad Laboratories, Inc.) containing a protease inhibitor
and boiled for 5 min before being subjected to 10% SDS-PAGE. The
separated proteins were transferred to a PVDF membrane and then
blocked in TBST (TBS containing 0.05% Tween-20) with 5% nonfat milk
at room temperature for 1 h. Then, the PVDF membrane was separated
and incubated overnight at 4°C with primary antibodies as follows:
Mouse monoclonal anti-β-actin (cat. no. A5316-100) was diluted with
a blocking buffer (TBS containing 0.05% Tween-20) at a ratio of
1:1,000; mouse monoclonal anti-katanin p60 AL1 (A-10) (cat. no.
sc-373814; Santa Cruz Biotechnology, Inc.) was diluted with a
blocking buffer at a ratio of 1:500; mouse monoclonal
anti-γ-tubulin (cat. no. T3559-.2ML; Sigma-Aldrich; Merck KGaA) was
diluted with a blocking buffer at a ratio of 1:2,000. After being
washed in TBST, the membranes were incubated with HRP-conjugated
rabbit anti-goat IgG or HRP-conjugated goat anti-mouse IgG (diluted
with a blocking buffer to 1:1,000) for 1 h at room temperature and
then processed using an ECL Plus Western Blotting Detection System
(Vazyme). ImageJ 1.8.0 (National Institutes of Health) was used for
data analysis.
Immunoprecipitation
For immunoprecipitation experiments, 5 µg control
IgG or anti-p60 katanin-like 1 antibody was first coupled to 30 µl
protein-A/G beads [M&C Gene Technology (Beijing), Ltd] for 4 h
at 4°C on a rotating wheel in 250 µl IP buffer (20 mM Tris-HCl, pH
8.0, 10 mM EDTA, 1 mM EGTA, 150 mM NaCl, 0.05% Triton X-100, 0.05%
Nonidet P-40, and 1 mM phenylmethylsulfonyl fluoride) with 1:100
protease inhibitor and 1:500 phosphatase inhibitor (both from
Sigma-Aldrich; Merck KGaA). Meanwhile, 600 ZP-free GV oocytes were
lysed and ultrasonicated in 250 IP buffer before being incubated
with 30 µl protein-A/G beads for 4 h at 4°C. Then, a protein
A/G-coupled control IgG or anti-p60 katanin-like 1 antibody was
incubated overnight at 4°C with 250 µl pre-cleaned oocyte lysate
supernatant. Finally, the beads were washed the next morning three
times for 10 min each with 1 ml IP buffer, and the resulting beads
with bound immunocomplexes were subjected to 10% SDS-PAGE and
silver staining by incubating with 0.1% AgNO3 solution at room
temperature for 30 min.
Mitochondrial staining
For mitochondrial staining, the oocytes were stained
in HEPES containing 100 nM MitoTracker (cat. no. M7521; Invitrogen;
Thermo Fisher Scientific, Inc.) and 10 µg/ml Hoechst 33342
(Sigma-Aldrich; Merck KGaA) for 30 min. Subsequently, the oocytes
were examined with an Andor Revolution spin disk confocal
workstation.
Data analysis and statistics
All experiments were repeated at least three times.
Measurements on confocal images was performed with ImageJ (National
Institutes of Health). Data are presented as the average ± sem.
Statistical comparisons were performed with Student's t-test in
Excel. P<0.05 was considered to indicate a statistically
significant difference.
Results and Discussion
p60 katanin-like 1 is predominant in
mouse ovaries and oocytes
The expression and localization patterns of p60
katanin-like 1 were first examined in mouse ovaries and oocytes.
Western blot analysis revealed that the expression of KATNAL1 in
oocytes was significantly higher than that in somatic cells
(Fig. 1A), indicating that p60
katanin-like 1 may be more critical in oocytes. p60 katanin-like 1
was more pronounced in oocytes than in granular cells (Fig. 1B), indicating that the role of p60
katanin-like 1 may be primarily in oocytes. Additionally, it was
also determined that the abundance of p60 katanin-like 1 in the
ovary and testis was markedly high (Fig. 1C), indicating that p60 katanin-like
1 may be closely related to the activities of germ cells.
p60 katanin-like 1 is located at the
spindle poles of MI and MII oocytes and co-localizes with
γ-tubulin
The abundance of p60 katanin-like 1 in mouse oocytes
indicated that p60 katanin-like 1 participates in germ cell
activity. Next, the localization of p60 katanin-like 1 in oocytes
was assessed by immunofluorescence. p60 katanin-like 1 was located
at the spindle poles of MI and MII mouse oocytes (Fig. 2A and B). In addition, p60
katanin-like 1 and γ-tubulin co-localized at the spindle poles of
MI and MII oocytes (Fig. 2C),
indicating that the function of p60 katanin-like 1 may be related
to the organization of spindle poles.
p60 katanin-like 1 knockdown leads to
severe maturation abnormalities in oocytes
To explore the function of p60 katanin-like 1 in
oocyte development, specific siRNAs for p60 katanin-like 1 were
designed and the siRNA mixture was transfected into GV oocytes with
nanoparticle reagent. Then, the effect of siRNA knockdown was
examined. RT-PCR (Fig. 3B, lower
panel) and western blot (Fig. 3A)
revealed that siRNA knockdown reduced the p60 katanin-like 1 mRNA
or protein level to 20% of that in the control. RT-PCR also
revealed that the siRNAs could specifically target p60 katanin-like
1 but had no detectable effect on p60 katanin, another member of
MTSP family (Fig. 3B, upper
panel). Next, oocyte maturation after p60 katanin-like 1 knockdown
was examined. Compared with that in the control, p60 katanin-like
1-depleted cells did not exhibit a significantly lower rate of
germinal vesicle breakdown (GVBD) after 3 h of in vitro
maturation (IVM) (Fig. 3C upper
panel). However, the percentage of cells exhibiting a first polar
body (1 Pb) extrusion was significantly lower than that of the
control group after 16 h of IVM (Fig.
3C, lower panel). Furthermore, in vitro fertilization
(IVF) with normal sperm revealed that the p60 katanin-like
1-depleted group had a decreased fertility rate and significantly
more fertilized eggs with multiple pronuclei (less fertilized eggs
with two pronuclei) than the control group (Fig. 3D). These results indicated that p60
katanin-like 1 is essential for the potential of oocytes.
p60 katanin-like 1 knockdown causes
abnormal pole-correlated spindle organization in metaphase I (MI)
or metaphase II (MII) oocytes
Next, the spindle phenotype was characterized after
p60 katanin-like 1 knockdown. p60 katanin-like 1 knockdown caused
different extents of spindle irregularities. The ratio of
width:length of the spindles was assessed and it was revealed that,
compared with the control, the spindles in the p60 katanin-like
1-knockdown group were longer and thinner (Fig. 4A). In addition, p60 katanin-like 1
knockdown resulted in more aster-like microtubule structures in the
cytoplasm and more multipolar spindles than control cells (Fig. 4B). Since p60 katanin-like 1 was
concentrated at the spindle poles and these phenotypes were
pole-related, we next examined whether p60 katanin-like 1 interacts
with crucial pole components. Co-immunoprecipitation experiments
revealed that p60 katanin-like 1 and γ-tubulin interacted well with
each other (Fig. 4C).
Collectively, these results revealed that p60 katanin-like 1 may
interact with γ-tubulin and that p60 katanin-like 1 knockdown
caused pole-correlated spindle defects.
In addition, EGFP-KATNAL1 mRNA was expressed in
vitro and injected into oocytes, which inhibited development.
After 8 h of development in vitro, immunofluorescence of the
EGFP tags was performed and it was revealed that the localization
of the EGFP tags was consistent with that of p60 katanin-like 1 in
the oocytes, which demonstrated the expression of EGFP-KATNAL1 mRNA
in oocytes (Fig. S1A). In
addition, it was revealed that oocytes overexpressing KATNAL1 had a
wide spindle shape at the two poles (Fig. S1B). After 16 h of in vitro
development, 1 Pb extrusion in oocytes decreased significantly
compared to that in control cells (Fig. S1C). It was speculated that the
overexpression of KATNAL1 may lead to disorder in the polar
structure of the spindle by severing the microtubules excessively,
thus affecting the morphology and function of the spindle, thereby
affecting oocyte development.
p60 katanin-like 1 knockdown causes
the abnormal subcellular distribution of mitochondria in
oocytes
To further study how p60 katanin-like 1 knockdown
caused severe defects in oocyte maturation and spindle
organization, its effects on the mitochondria were investigated in
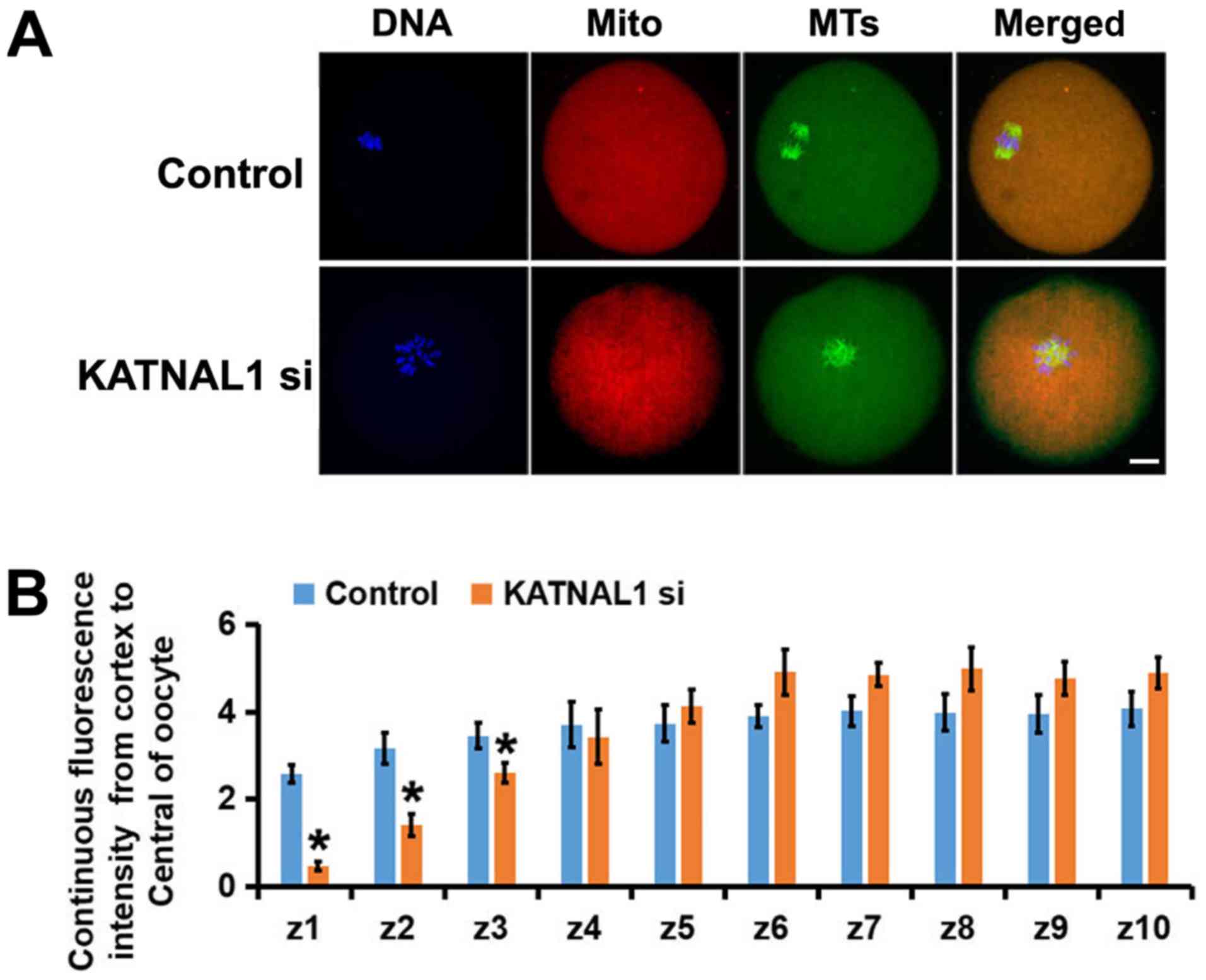
oocytes. The deletion of KATNAL1 resulted in an uneven
distribution of mitochondria in oocytes compared with their
distribution in the control group (Fig. 5A). Through careful assessment of
the mitochondrial intensity in each subregion, it was determined
that the mitochondria in the KATNAL1-depleted oocytes tended to
gather in the center; accordingly, there were fewer mitochondria
near the membrane (Fig. 5B). These
results indicate that KATNAL1 knockdown markedly affected the
distribution of mitochondria.
Normal meiosis ensures the euploidy of oocytes,
subsequent regular fertilization, and early embryo development
until the birth of healthy pups. Meiotic spindles undergo marked
changes throughout meiosis, and markedly abnormal organization of
the spindle microtubules frequently causes defective meiosis,
consequently affecting all the following processes (3,16,22–26).
Typical abnormal spindle organization includes improper
kinetochore-MT attachment (merotelic, syntelic), spindles with
significantly decreased MT intensity, an abnormal width:length
ratio (which usually indicates that MT dynamics are abnormal), a
monopolar spindle, and an extra aster-like microtubule structure.
In the present study, it was first revealed that p60 katanin-like
1, a member of the MTSP family, is predominant in the ovaries and
more enriched in oocytes than granular cells, indicating that p60
katanin-like 1 may play essential roles in oocyte growth and
maturation. It was revealed in the present study, that p60
katanin-like 1 localizes at the spindle poles and interacts with
γ-tubulin, indicating that the function of p60 katanin-like 1 may
be correlated with the organization of the spindle poles. Next, p60
katanin-like 1 was depleted with specific siRNAs and mainly three
typical meiotic spindle phenotypes were revealed: An abnormal
width:length ratio, multipolar microtubules, and an extra
aster-like microtubule structure. Previous studies revealed that
the depletion of the spindle pole matrix caused an abnormal
width:length ratio, multipolarity or additional asters when diverse
centrosome or pole components were knocked down or depleted
(28–32). These results mostly correspond with
the results of our study, suggesting that p60 katanin-like 1 is
essential for meiotic spindle integrity by regulating pole
organization. A previous study reported that in U2OS cells, p60
katanin-like 1 was restricted to the spindle poles and absent from
centrosomes, and the siRNA depletion of p60 katanin-like 1 from
U2OS cells caused a significant reduction in the density of the
spindle poles and increased the spindle length (27). Thus, p60 katanin-like 1 functions
somewhat similarly in meiosis and mitosis. However, it was also
observed that p60 katanin-like 1 depletion caused multipolar
spindles and extra asters, which could cause even severe meiotic
defects. Therefore, it appears that p60 katanin-like 1 is more
indispensable in meiosis than in mitosis. Numerous studies have
demonstrated that defective spindle organization could severely
affect the progression of meiosis and subsequent fertilization. In
the present study, p60 katanin-like 1 knockdown significantly
reduced the percentage of GVBD or MII oocytes, which further
indicates its essential roles in meiosis.
Mitochondria are the factory for adenosine
triphosphate (ATP) generation, and the abnormal subcellular
distribution of mitochondria could lead to insufficient ATP
generation in some regions and excessive ATP generation in others;
either situation is unfavorable for normal meiosis since the
regular cell cycle, either mitosis or meiosis, requires the
cooperation of multiple enzymes that bind and hydrolyze the proper
level of ATP to perform appropriately. Insufficient or excessive
ATP generation could have an inhibitory or activating effect that
could inhibit or overactivate these enzymes. In fact, multiple
previous studies revealed that abnormal mitochondrial distribution
caused significantly reduced cellular ATP levels (33–36).
Mitochondrial malfunction is closely related to a reduction in
reproductivity (37–40). Mitochondria are also highly dynamic
and motile during the cell cycle, and their movement is
microtubule-dependent (41–44);
therefore, the abnormal mitochondrial distribution in our study
could be caused by irregular spindle organization. This eventually
disrupted spindle organization and caused the uneven distribution
of mitochondria, which were both caused by p60 katanin-like 1
knockdown; together, these retarded meiosis and reduced
fertilization.
In conclusion, the present study, revealed for the
first time that p60 katanin-like 1, an MTSP that is enriched in
oocytes, is concentrated at the spindle poles and essential for
pole organization during mouse oocyte meiosis. p60 katanin-like 1
knockdown caused severe spindle defects characterized by an
abnormal width:length ratio, multipolarity, and extra aster
microtubules. p60 katanin-like 1 knockdown also caused irregular
mitochondrial distribution. These factors collectively retarded
meiosis and reduced fertilization. Further investigation is
required to determine how p60 katanin-like 1 organizes the spindle
poles and what regulates its activity.
Supplementary Material
Supporting Data
Acknowledgements
The authors would like to thank Dr Zhang (Hangzhou
Medical College) for providing us with a mitochondrial probe.
Funding
This present study was supported by the Grant for
Zhejiang Provincial Science and Technology Department and the
Department of Health (2017KY199 and 2018KY240).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JWL and LLG conceived and designed the experiments.
LLG, FX, ZJ and XYY performed the experiments. LLG analyzed the
data. JWL and LLG contributed the reagents/materials/analysis
tools. JWL and LLG wrote the paper. All authors read and approved
the manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All animal experiments were approved by the Animal
Care and Use Committee of Nanjing Medical University (Nanjing,
China) and performed in accordance with institutional
guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sferra A, Fattori F, Rizza T, Flex E,
Bellacchio E, Bruselles A, Petrini S, Cecchetti S, Teson M,
Restaldi F, et al: Defective kinesin binding of TUBB2A causes
progressive spastic ataxia syndrome resembling sacsinopathy. Hum
Mol Genet. 27:1892–1904. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Luscan R, Mechaussier S, Paul A, Tian G,
Gerard X, Defoort-Dellhemmes S, Loundon N, Audo I, Bonnin S,
LeGargasson JF, et al: Mutations in TUBB4B cause a distinctive
sensorineural disease. Am J Hum Genet. 101:1006–1012. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Feng R, Sang Q, Kuang Y, Sun X, Yan Z,
Zhang S, Shi J, Tian G, Luchniak A, Fukuda Y, et al: Mutations in
TUBB8 and human oocyte meiotic arrest. N Engl J Med. 374:223–232.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Martin M and Akhmanova A: Coming into
focus: Mechanisms of microtubule minus-end organization. Trends
Cell Biol. 28:574–588. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Aher A and Akhmanova A: Tipping
microtubule dynamics, one protofilament at a time. Curr Opin Cell
Biol. 50:86–93. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Akhmanova A and Steinmetz MO: Control of
microtubule organization and dynamics: Two ends in the limelight.
Nat Rev Mol Cell Biol. 16:711–726. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huynh W and Vale RD: Disease-associated
mutations in human BICD2 hyperactivate motility of dynein-dynactin.
J Cell Biol. 216:3051–3060. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lewis WR, Malarkey EB, Tritschler D, Bower
R, Pasek RC, Porath JD, Birket SE, Saunier S, Antignac C, Knowles
MR, et al: Mutation of growth arrest specific 8 reveals a role in
motile cilia function and human disease. PLoS Genet.
12:e10062202016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Decker JM, Kruger L, Sydow A, Dennissen
FJ, Siskova Z, Mandelkow E and Mandelkow EM: The Tau/A152T
mutation, a risk factor for frontotemporal-spectrum disorders,
leads to NR2B receptor-mediated excitotoxicity. EMBO Rep.
17:552–569. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Frickey T and Lupas AN: Phylogenetic
analysis of AAA proteins. J Struct Biol. 146:2–10. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Vale RD: AAA proteins. Lords of the ring.
J Cell Biol. 150:F13–E19. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Buster D, McNally K and McNally FJ:
Katanin inhibition prevents the redistribution of gamma-tubulin at
mitosis. J Cell Sci. 115:1083–1092. 2002.PubMed/NCBI
|
|
13
|
Sharp DJ and Ross JL: Microtubule-severing
enzymes at the cutting edge. J Cell Sci. 125:2561–2569. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang D, Rogers GC, Buster DW and Sharp
DJ: Three microtubule severing enzymes contribute to the
‘Pacman-flux’ machinery that moves chromosomes. J Cell Biol.
177:231–242. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang D, Grode KD, Stewman SF,
Diaz-Valencia JD, Liebling E, Rath U, Riera T, Currie JD, Buster
DW, Asenjo AB, et al: Drosophila katanin is a microtubule
depolymerase that regulates cortical-microtubule plus-end
interactions and cell migration. Nat Cell Biol. 13:361–370. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Srayko M, O'Toole ET, Hyman AA and
Muller-Reichert T: Katanin disrupts the microtubule lattice and
increases polymer number in C. elegans meiosis. Curr Biol.
16:1944–1949. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Errico A, Ballabio A and Rugarli EI:
Spastin, the protein mutated in autosomal dominant hereditary
spastic paraplegia, is involved in microtubule dynamics. Hum Mol
Genet. 11:153–163. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Banks G, Lassi G, Hoerder-Suabedissen A,
Tinarelli F, Simon MM, Wilcox A, Lau P, Lawson TN, Johnson S,
Rutman A, et al: A missense mutation in Katnal1 underlies
behavioural, neurological and ciliary anomalies. Mol Psychiatry.
23:713–722. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mishra-Gorur K, Caglayan AO, Schaffer AE,
Chabu C, Henegariu O, Vonhoff F, Akgümüş GT, Nishimura S, Han W, Tu
S, et al: Mutations in KATNB1 cause complex cerebral malformations
by disrupting asymmetrically dividing neural progenitors. Neuron.
84:1226–1239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mao CX, Xiong Y, Xiong Z, Wang Q, Zhang YQ
and Jin S: Microtubule-severing protein Katanin regulates
neuromuscular junction development and dendritic elaboration in
Drosophila. Development. 141:1064–1074. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stewart A, Tsubouchi A, Rolls MM, Tracey
WD and Sherwood NT: Katanin p60-like1 promotes microtubule growth
and terminal dendrite stability in the larval class IV sensory
neurons of Drosophila. J Neurosci. 32:11631–11642. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
O'Donnell L, Rhodes D, Smith SJ, Merriner
DJ, Clark BJ, Borg C, Whittle B, O'Connor AE, Smith LB, McNally FJ,
et al: An essential role for katanin p80 and microtubule severing
in male gamete production. PLoS Genet. 8:e10026982012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Smith LB, Milne L, Nelson N, Eddie S,
Brown P, Atanassova N, O'Bryan MK, O'Donnell L, Rhodes D, Wells S,
et al: KATNAL1 regulation of sertoli cell microtubule dynamics is
essential for spermiogenesis and male fertility. PLoS Genet.
8:e10026972012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen B, Zhang Z, Sun X, Kuang Y, Mao X,
Wang X, Yan Z, Li B, Xu Y, Yu M, et al: Biallelic mutations in
PATL2 cause female infertility characterized by oocyte maturation
arrest. Am J Hum Genet. 101:609–615. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nguyen AL, Marin D, Zhou A, Gentilello AS,
Smoak EM, Cao Z, Fedick A, Wang Y, Taylor D, Scott RT Jr, et al:
Identification and characterization of Aurora kinase B and C
variants associated with maternal aneuploidy. Mol Hum Reprod.
23:406–416. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Caburet S, Arboleda VA, Llano E, Overbeek
PA, Barbero JL, Oka K, Harrison W, Vaiman D, Ben-Neriah Z,
García-Tuñón I, et al: Mutant cohesin in premature ovarian failure.
N Engl J Med. 370:943–949. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sonbuchner TM, Rath U and Sharp DJ: KL1 is
a novel microtubule severing enzyme that regulates mitotic spindle
architecture. Cell Cycle. 9:2403–2411. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pimenta-Marques A, Bento I, Lopes CA,
Duarte P, Jana SC and Bettencourt-Dias M: A mechanism for the
elimination of the female gamete centrosome in Drosophila
melanogaster. Science. 353:aaf48662016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Connolly AA, Osterberg V, Christensen S,
Price M, Lu C, Chicas-Cruz K, Lockery S, Mains PE and Bowerman B:
Caenorhabditis elegans oocyte meiotic spindle pole assembly
requires microtubule severing and the calponin homology domain
protein ASPM-1. Mol Biol Cell. 25:1298–1311. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim JS, Kim EJ, Oh JS, Park IC and Hwang
SG: CIP2A modulates cell-cycle progression in human cancer cells by
regulating the stability and activity of Plk1. Cancer Res.
73:6667–6678. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Patel H, Zich J, Serrels B, Rickman C,
Hardwick KG, Frame MC and Brunton VG: Kindlin-1 regulates mitotic
spindle formation by interacting with integrins and Plk-1. Nat
Commun. 4:20562013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Eot-Houllier G, Venoux M, Vidal-Eychenie
S, Hoang MT, Giorgi D and Rouquier S: Plk1 regulates both ASAP
localization and its role in spindle pole integrity. J Biol Chem.
285:29556–29568. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhou CX, Shi LY, Li RC, Liu YH, Xu BQ, Liu
JW, Yuan B, Yang ZX, Ying XY and Zhang D: GTPase-activating protein
Elmod2 is essential for meiotic progression in mouse oocytes. Cell
Cycle. 16:852–860. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang XL, Liu P, Yang ZX, Zhao JJ, Gao LL,
Yuan B, Shi LY, Zhou CX, Qiao HF, Liu YH, et al: Pnma5 is essential
to the progression of meiosis in mouse oocytes through a chain of
phosphorylation. Oncotarget. 8:96809–96825. 2017.PubMed/NCBI
|
|
35
|
Dinkelmann MV, Zhang H, Skop AR and White
JG: SPD-3 is required for spindle alignment in Caenorhabditis
elegans embryos and localizes to mitochondria. Genetics.
177:1609–1620. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kong XW, Wang DH, Zhou CJ, Zhou HX and
Liang CG: Loss of function of KIF1B impairs oocyte meiotic
maturation and early embryonic development in mice. Mol Reprod Dev.
83:1027–1040. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Maccarinelli F, Regoni M, Carmona F, Poli
M, Meyron-Holtz EG and Arosio P: Mitochondrial ferritin deficiency
reduces male fertility in mice. Reprod Fertil Dev. 29:2005–2010.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang M, Huang YP, Wu H, Song K, Wan C, Chi
AN, Xiao YM and Zhao XY: Mitochondrial complex I deficiency leads
to the retardation of early embryonic development in Ndufs4
knockout mice. Peer J. 5:e33392017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
May-Panloup P, Boucret L, Chao de la Barca
JM, Desquiret-Dumas V, Ferre-L'Hotellier V, Moriniere C, Descamps
P, Procaccio V and Reynier P: Ovarian ageing: The role of
mitochondria in oocytes and follicles. Hum Reprod Update.
22:725–743. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ben-Meir A, Burstein E, Borrego-Alvarez A,
Chong J, Wong E, Yavorska T, Naranian T, Chi M, Wang Y, Bentov Y,
et al: Coenzyme Q10 restores oocyte mitochondrial function and
fertility during reproductive aging. Aging Cell. 14:887–895. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bartolak-Suki E, Imsirovic J, Nishibori Y,
Krishnan R and Suki B: Regulation of mitochondrial structure and
dynamics by the cytoskeleton and mechanical factors. Int J Mol Sci.
18(pii): E18122017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fu L, Dong Q, He J, Wang X, Xing J, Wang
E, Qiu X and Li Q: SIRT4 inhibits malignancy progression of NSCLCs,
through mitochondrial dynamics mediated by the ERK-Drp1 pathway.
Oncogene. 36:2724–2736. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang C, Du W, Su QP, Zhu M, Feng P, Li Y,
Zhou Y, Mi N, Zhu Y, Jiang D, et al: Dynamic tubulation of
mitochondria drives mitochondrial network formation. Cell Res.
25:1108–1120. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fu C, Jain D, Costa J, Velve-Casquillas G
and Tran PT: mmb1p binds mitochondria to dynamic microtubules. Curr
Biol. 21:1431–1439. 2011. View Article : Google Scholar : PubMed/NCBI
|