Introduction
Colorectal cancer (CRC) is the third most common
cancer worldwide. Although there have been marked improvements in
clinical treatment methods, CRC still causes many deaths every
year, especially in developed countries (1,2).
Great efforts to understand the complicated pathogenesis of CRC
have been made, however it is still largely unknown, and critical
regulatory molecules that are important for monitoring CRC tumor
progression and detecting efficient therapeutic targets for future
clinical treatments are required.
Rapid growth, migration and invasion are critical
characteristics that induce malignant tumor genesis and development
(3–5). Many signaling pathways have been
reported to be closely related to these biological processes. For
example, the upregulation of Sox2 in breast cancer tissues was
associated with lymph node metastasis, pathological grade and TNM
classification (6). Kisspeptin
suppressed cancer growth and metastasis by activating EIF2AK2 in
CRC (7). Cyclin D2/miR-1297
signaling was associated with the growth and metastasis of CRC
(8). miR-184 inhibited cell
proliferation and metastasis in CRC by targeting IGF-1R (9). MACC1 was identified as a biomarker of
metastasis and disease prognosis (10). BNIPL-2 was revealed to interact
with Bcl-2 and Cdc42GAP to regulate apoptosis in cells (11). Additionally, BNIPL-2 promoted cell
migration, invasion and metastasis in hepatocellular carcinoma
(HCC) cells (12). However,
whether BNIPL-2 is also a potential critical regulator that is
associated with CRC progression and prognosis remains largely
unknown.
CD44 is reported to be an important regulator of
cell proliferation. CD44 regulates endothelial cell proliferation
by modulating CD31 and VE-cadherin (13). Knockdown of CD44 suppressed breast
cancer cell proliferation (14).
CD44 is also a potential biomarker for predicting hepatic
metastases and survival (15).
Upregulation of CD44 is associated with a metastatic CRC phenotype
(16). However, whether CD44
directly regulates CRC proliferation and the related upstream
regulators remains largely unknown.
It was hypothesized that BNIPL-2 and CD44 may
regulate CRC proliferation, and the present study aimed to identify
whether BNIPL-2 is associated with CRC prognosis and can be a
potential biomarker by bioinformatics methods. The effects of
BNIPL-2 and CD44 on cell cycle regulation and their regulatory
relationship were also determined. In the present study,
transcriptome data analysis of patient samples from the Cancer
Genome Atlas (TCGA) dataset was performed to investigate potential
gene expression correlations with CRC prognosis and progression. It
was revealed that compared with that in adjacent tissues, BNIPL-2
was upregulated in CRC tissues. A higher expression level of
BNIPL-2 was associated with greater malignant cancer progression
and a poorer prognosis and was also closely correlated with the
activation of signaling pathways involved in tumor growth, invasion
and migration. The present study not only revealed the potential
relationship between BNIPL-2 and CRC during CRC proliferation but
also suggested the potential diagnostic value of BNIPL-2 as a
biomarker and treatment target for CRC in future clinical
therapy.
Materials and methods
Gene expression analysis of TCGA
data
Data for the analysis of BNIPL-2 expression
differences and the RSEM value of BNIPL-2-related regulatory
signaling pathways in clinical CRC samples with related genome
transcriptomic profiles were obtained online from TCGA (http://cancergenome.nih.gov/). All data regarding
patients was obtained from TCGA.
Stratification of high or low levels
of BNIPL-2 expression in patients
To determine the relationship between clinical
malignancy stage, related signaling, and survival rate and BNIPL-2
expression in CRC patients, the CRC tissue samples were categorized
according to high and low BNIPL-2 expression, as related to the
median. Patients with higher BNIPL-2 expression than the median
were classified into the high expression group, and those with
lower BNIPL-2 expression than the median were classified into the
low expression group.
Gene Set Enrichment Analysis (GSEA)
using CRC tissues
GSEA was performed with the
msigdb.v6.0.symbols.gmtgene set. A total of 10,000 permutations
were used for P-value statistics.
Cell culture
The CRC cell lines SW480 and HCT116 were obtained
from the American Type Culture Collection and cultured in DMEM
(HyClone; GE Healthcare Life Sciences) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.). Cells were cultured at
37°C in an atmosphere with 5% CO2.
Overexpression of BNIPL-2
Total RNA was isolated from SW480 cells and used to
synthesize cDNA with a reverse transcription PCR kit (Takara Bio,
Inc.). The CDS sequence of BNIPL-2 mRNA was amplified from the cDNA
and inserted into the Fugw vector (Addgene, Inc.). The primer
sequences were as follows: forward,
5′-GGCGGATCCATGCGCAAGCGTCTTTCTGC-3′ (BamH1 site) and
reverse, 5′-GGCGAATTCCTATGTCCCTCCTGAGCCATGGAG-3′ (EcoR1
site). The vector was transfected into cells to overexpress BNIPL-2
using Lipofectamine 2000 (Thermo Fisher Scientific, Inc.).
Knockdown of BNIPL-2
The siRNA sequences were as follows: Control siRNA,
5′-UUCUCCGAACGUCUCACGU-3′;
siRNA-1-BNIPL-2,5′-CGCGUAGACAUGACUGUCAUU-3′;
siRNA-2-BNIPL-2,5′-CCUUUGCAUGACCCUACUUUC-3′; and negative siRNA
control, 5′-UUCUCCGAACGUGUCACGUTT-3′.
Knockdown of p53
siRNA-p53: Sense sequences
5′-GACUCCAGUGGUAAUCUACdTT-3′ [sequence was referenced by a previous
study (17)] was transfected into
HCT116 cells to downregulated p53 expression.
Overexpression of CD44
The CDS sequence of BNIPL-2 was amplified to insert
into the Fugw vector. The primer sequences were as follows:
Forward, 5′-GGCACCGGTATGGACAAGTTTTGGTGGCACG-3′ (Age1 site)
and reverse, 5′-GGCGAATTCTTACACCCCAATCTTCATGTCCACA-3′ (EcoR1
site).
Knockdown of CD44
For CD44 knockdown, two siRNAs were used. The sense
sequences for CD44 siRNA were as follows: siRNA control,
5′UUCUCCGAACGUCUCACGU-3′; siRNA-1-CD44, 5′-UGCCUUUGAUGGACCAAUU-3′;
and siRNA-2-CD44, 5′-UAUUCCACGUGGAGAAAAATT-3′.
Flow cytometric assay
Cells were digested into a cell suspension and
washed with PBS. Then, the cells were fixed with 70% alcohol at 4°C
for 2 h. RNase A (20 mg/ml final concentration) was used to degrade
whole RNA for 30 min at room temperature. Then, the cells were
incubated with a propidium iodide solution (final concentration 50
mg/ml) for 15 min in the dark to stain the DNA. The cell cycle was
analyzed using a Cytomics FC 500 instrument (Beckman Coulter,
Inc.). The data were analyzed by FlowJo 7.6.1 software (FlowJo
LLC).
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was isolated by RNAiso plus (Takara
Biotechnology Co., Ltd.). cDNA was subsequently reverse-transcribed
with M-MLV reverse transcriptase (Takara Biotechnology Co., Ltd.).
RT-PCR included 40 cycles of amplification using SYBR Green qPCR
mix (BioRad Laboratories, Inc.). Relative gene expression
(2−ΔΔCq) (18) was
normalized to GAPDH. The primer sequences used were as follows:
GAPDH (forward, 5′-CTGGGCTACACTGAGCACC-3′ and reverse,
5′-AAGTGGTCGTTGAGGGCAATG-3′); cyclin E (forward,
5′-CGCCTGCCGGGACTGGAG-3′ and reverse, 5′-TCTTCCTGGAGCGAGCCG-3′);
cyclin D (forward, 5′-GACCACCGAGGAGTTTAATCG-3′ and reverse,
5′-GGGTGATCCCCTGATCCTTTG-3′); p21 (forward,
5′-TGTCCGTCAGAACCCATGC-3′; and reverse,
5′-AAAGTCGAAGTTCCATCGCTC-3′); BNIPL-2 (forward,
5′-GAGTCTGACTAAGGGGCCTG-3′ and reverse,
5′-CTCCGAGTCTGAAGGTGTCT-3′); and CD44 (forward,
5′-TTGCTGCACAGATGGAGTTGG-3′ and reverse,
5′-GAAAGCTCTGAGCATCGGATTTG-3′). Thermocycling conditions were:
Initial denaturation: 95°C for 30 sec, 95°C for 5 sec, 60 °C for 34
sec for 30 cycles.
Western blot analysis
Cells were lysed with SDS Lysis buffer (Beyotime
Institute of Biotechnology, Inc.) and heated at 95°C for 10 min. A
BCA Protein Assay Kit (Beyotime Institute of Biotechnology, Inc.)
was used for protein amount determination. 10 µg protein was used
for electrophoresis in agarose gel (12.5%; Epizyme, Inc.) Protein
was transferred onto the PVDF membrane (Bio-Rad Laboratories,
Inc.). The membrane was blocked by using 3% BSA (Epizyme, Inc.) for
1 h at room temperature and then incubated with primary antibodies
against CD44 (Abcam, ab157107, Rabbit polyclonal antibody, 1:1,000
dilution), BNIPL-2 (Novus biologicals, NBP1-79502, Rabbit
polyclonal antibody, 1:1,000 dilution) and GAPDH (Cell signaling
technology, 14C10 2118, Rabbit polyclonal antibody, 1:2,000
dilution) respectively at room temperature for 2 h. Membrane was
then incubated by secondary antibodies (Thermo, Inc, G-21234,
1:3,000 dilution) at room temperature for 1 h. Results for
signaling visualization was detected by enhanced chemiluminescence
(ECL) western blotting substrate (Thermo, Inc.) and analyzed using
the Bio-Rad ChemiDoc system (Bio-Rad Laboratories, Inc.).
Cell proliferation analysis
Cells were seeded at a density of 3×103
cells/well) in 96-well plates and used to perform a proliferation
assay according to the Cell Titer 96 AQueous One Solution Cell
Proliferation Assay instructions (MTS) (Promega Corporation). A
microplate reader was used to detect the absorbance at 490 nm.
Statistical analysis
T-tests were used to determine statistical
significance. Fisher's exact test was used to determine TMN stage
differences. Prognosis analysis was performed using the
Kaplan-Meier method. The values are presented as the mean ±
standard deviation. P<0.05, P<0.01 and P<0.001 were
considered to indicate statistically significant differences.
Results
BNIPL-2 is upregulated in CRC tissues
and correlated with poor prognosis
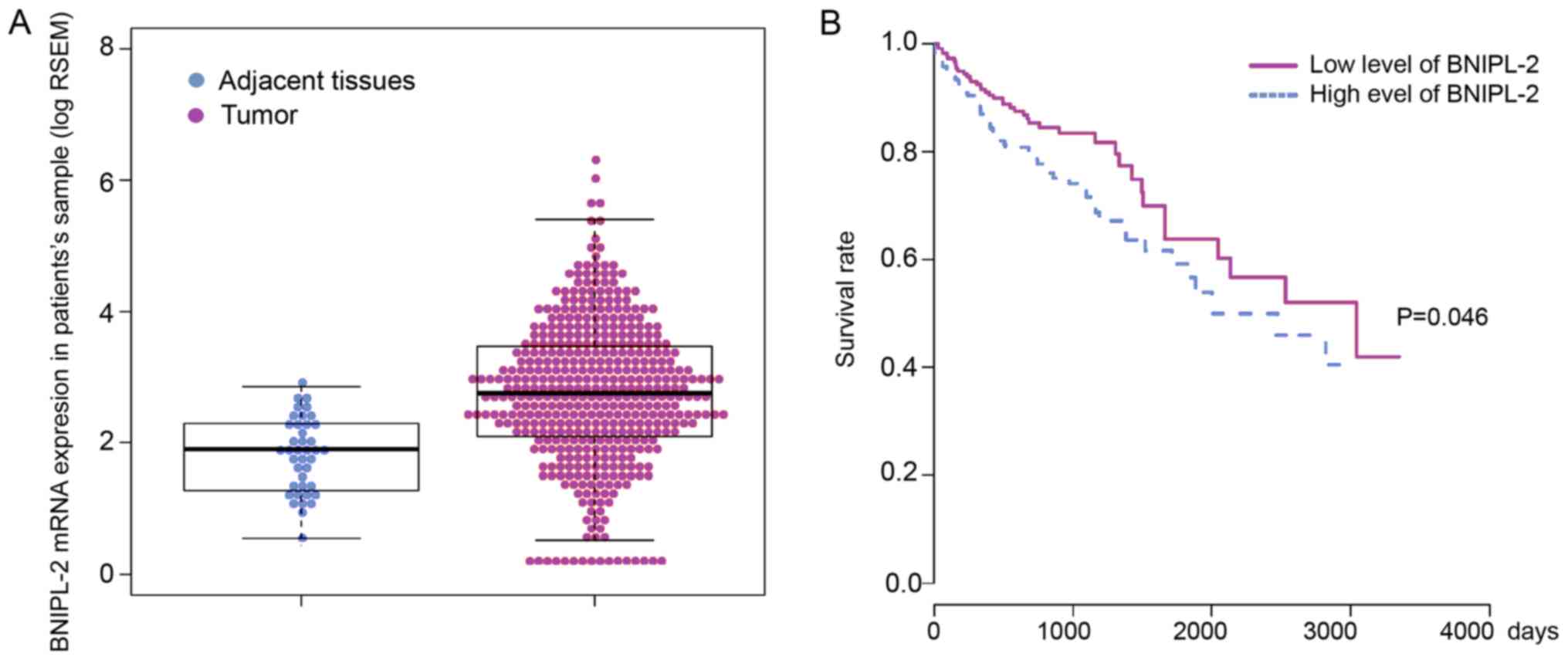
To determine whether BNIPL-2 expression was
associated with CRC, the expression level of BNIPL-2 was analyzed
in 41 adjacent tissue samples and 459 cancer samples from TCGA and
significantly higher BNIPL-2 levels were revealed in CRC samples
than in adjacent tissues (Fig.
1A). A Kaplan-Meier survival curve was used to analyze whether
the different levels of BNIPL-2 expression were related to the
prognosis of CRC patients. Patients were assigned into two groups
corresponding to low and high BNIPL-2 expression, which was defined
according to the median patient expression, and it was revealed
that there was a significantly poorer survival rate for the high
BNIPL-2 expression group than for the low BNIPL-2 expression group.
Additionally, the five-year survival rate of patients with low
expression of BNIPL-2 was approximately 10% higher than that of
patients with high BNIPL-2 expression (Fig. 1B).
High expression of BNIPL-2 is
associated with the malignant stage characteristics of CRC
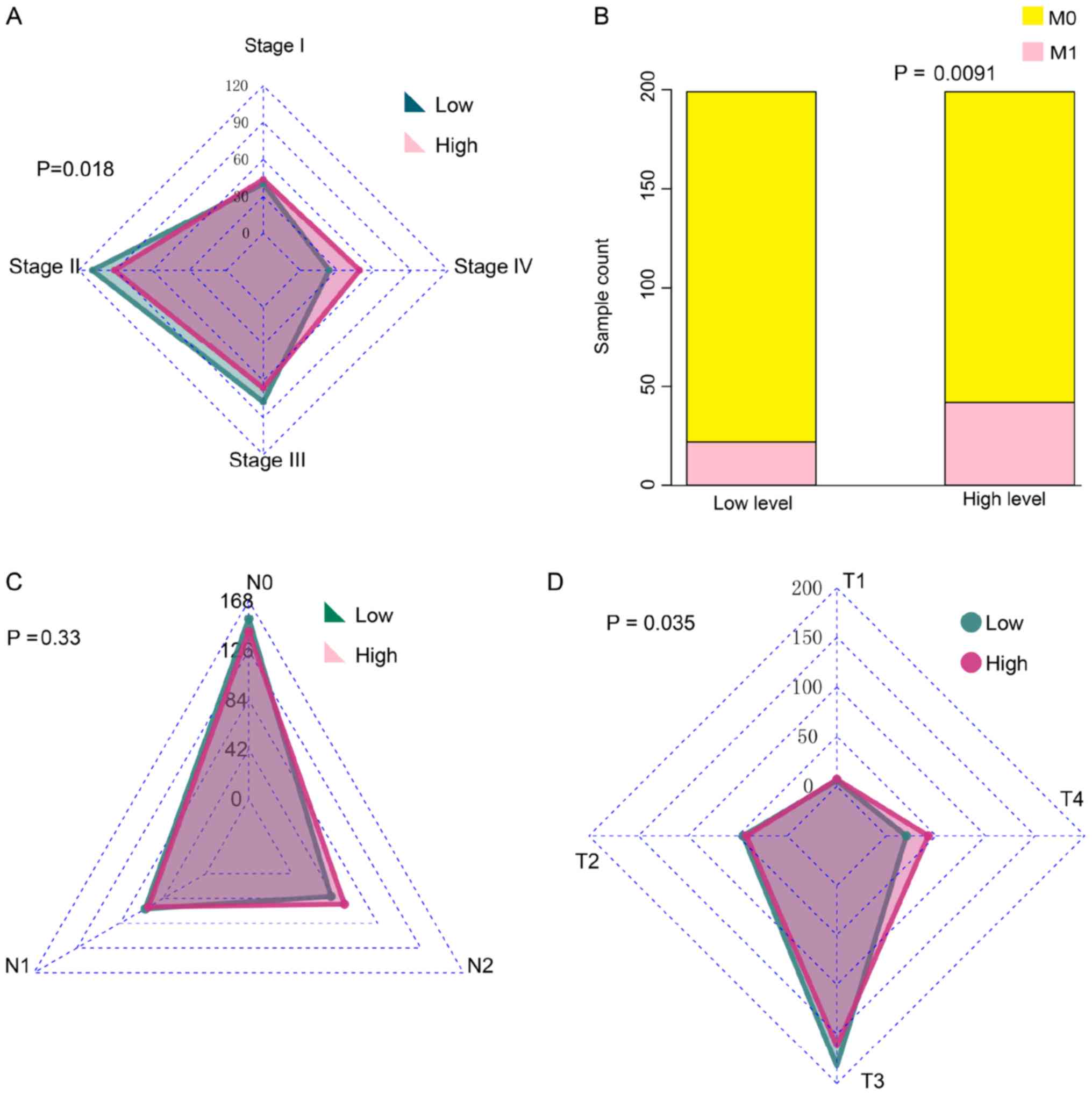
Due to the relationship between high BNIPL-2
expression and poor prognosis, it was hypothesized that BNIPL-2
expression would be associated with malignant stage
characteristics. It was revealed that tissues with higher
expression of BNIPL-2 stratified at the more adverse stage IV,
whereas tissues with lower levels of BNIPL-2 stratified at stages
II and III (Fig. 2A). TNM staging
was further analyzed and it was revealed that there were more
tissues with high BNIPL-2 expression classified as M1 than as M0
compared to low BNIPL-2 expression (Fig. 2B). There was no significant
difference between BNIPL-2 expression and N staging classification
(Fig. 2C). However, T staging
analysis revealed that more cancer tissues with high BNIPL-2
expression were in the T4 stage than CRC tissues with low BNIPL-2
expression (Fig. 2D).
BNIPL-2 promotes the proliferation of
the CRC cell line SW480
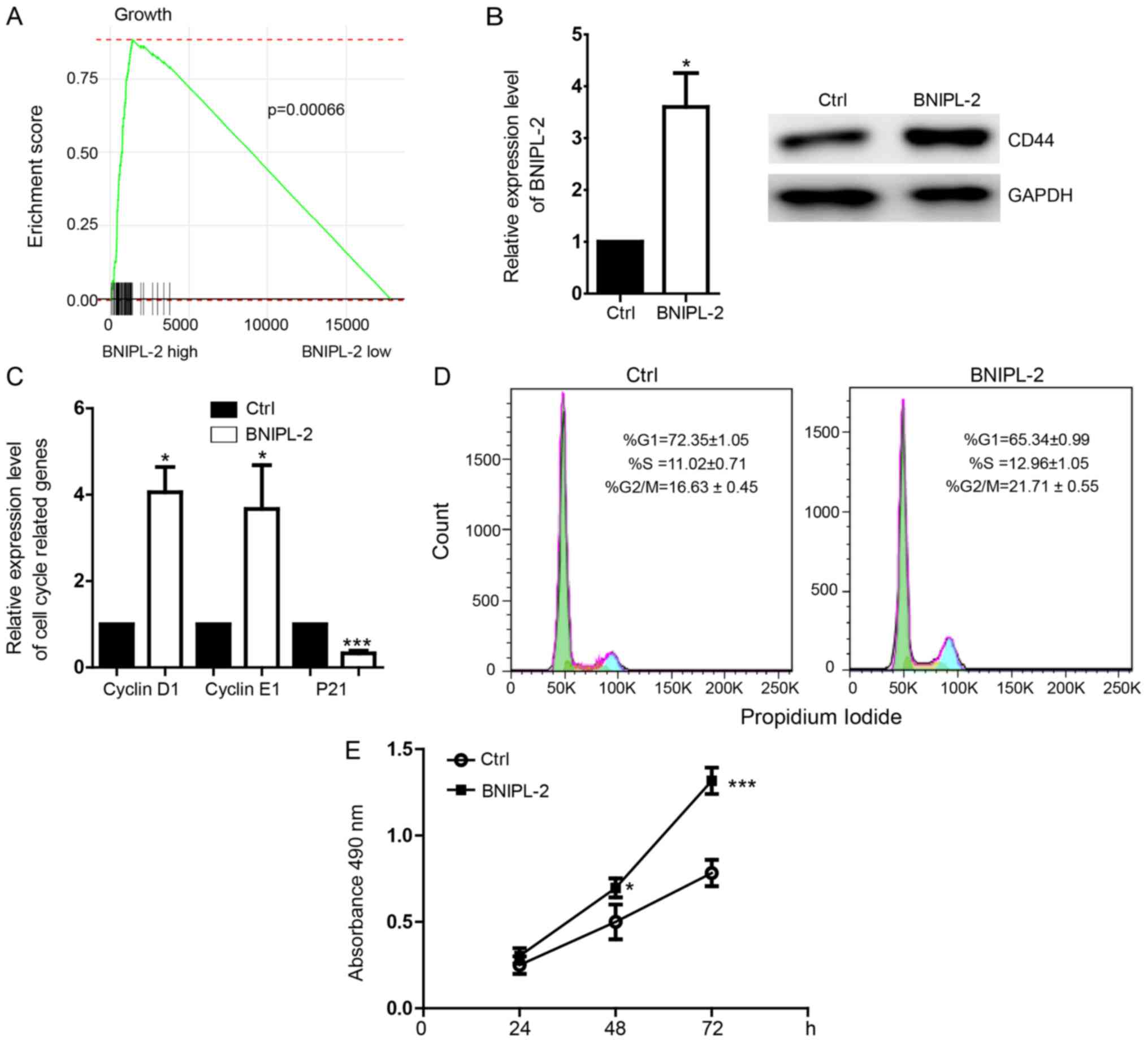
The tumor genesis- and development-related
regulatory pathways associated with BNIPL-2 expression were
detected in CRC transcriptomic profiles by gene set enrichment
analysis (GSEA) and the results revealed that the cell
migration-related signaling pathway was activated in BNIPL-2
high-expression tissues but not in BNIPL-2 low-expression tissues
(Fig. 3A). Overexpression of
BNIPL-2 (Fig. 3B) in SW480 cells
upregulated the expression of cyclin D1 and cyclin E1 and
downregulated the expression of the cell cycle inhibitor p21
(Fig. 3C). The proportion of cells
was decreased in the G1 phase and increased in the S and G2/M
phases by the overexpression of BNIPL-2 (Fig. 3D). An MTS proliferation assay
revealed that cell proliferation was promoted by BNIPL-2
overexpression (Fig. 3E).
BNIPL-2 also promotes the
proliferation of the CRC cell line HCT116
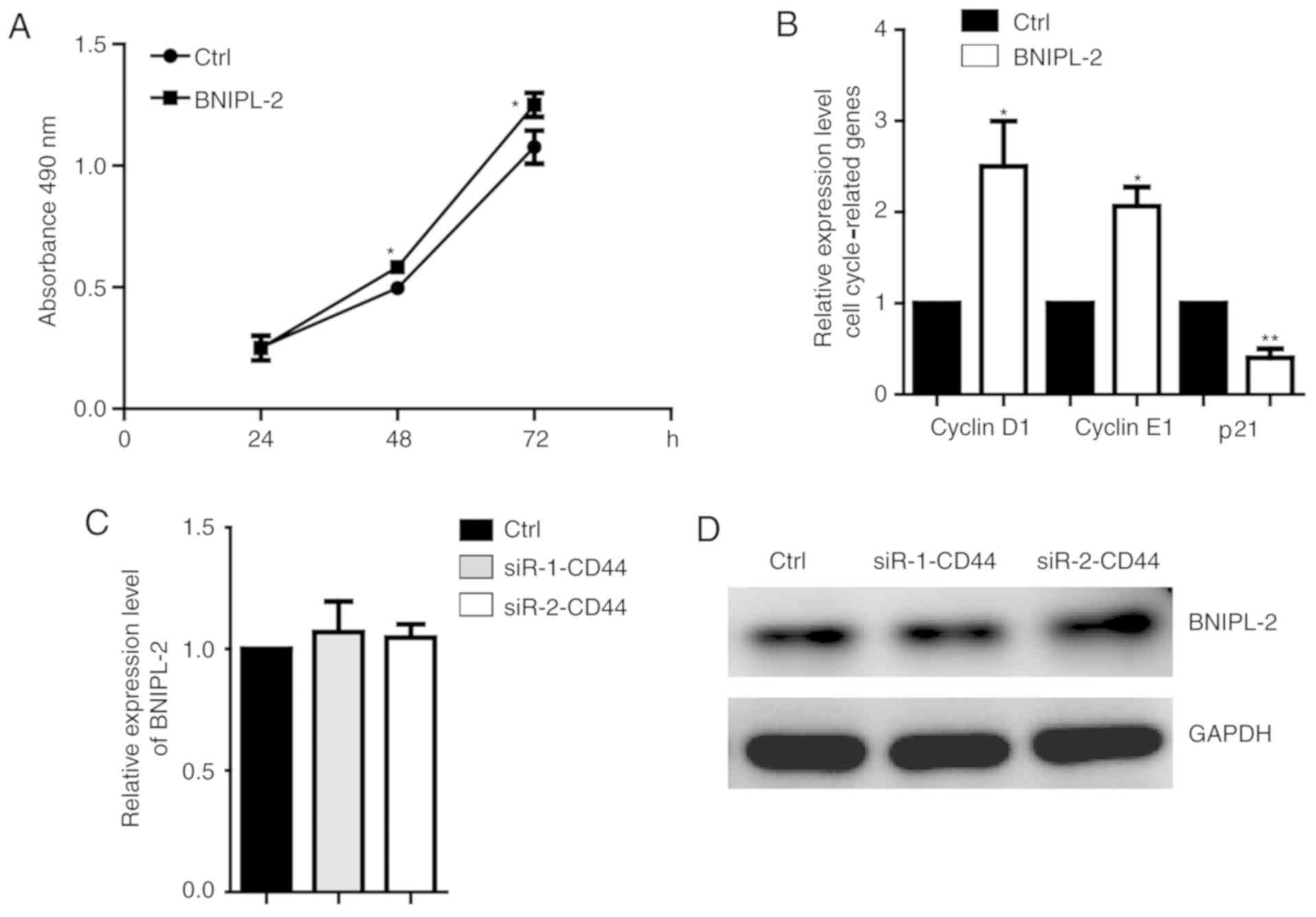
To further confirm the function of BNIPL-2 in
regulating CRC proliferation, BNIPL-2 was overexpressed in another
CRC cell line, HCT116, and the results revealed that proliferation
was also promoted (Fig. 4A). The
critical cell cycle-related genes cyclin D1 and cyclin E1 were
higher, and p21 was lower in BNIPL-2-overexpressing cells than in
control cells (Fig. 4B).
Downregulation of CD44 did not induce changes in BNIPL-2 mRNA
(Fig. 4C) and protein (Fig. 4D) expression, which suggested that
there was no reverse regulation of BNIPL-2 and CD44.
CD44 is regulated by BNIPL-2 and
promotes CRC proliferation
Overexpression of BNIPL-2 increased the protein
level of CD44 in SW480 cells (Fig.
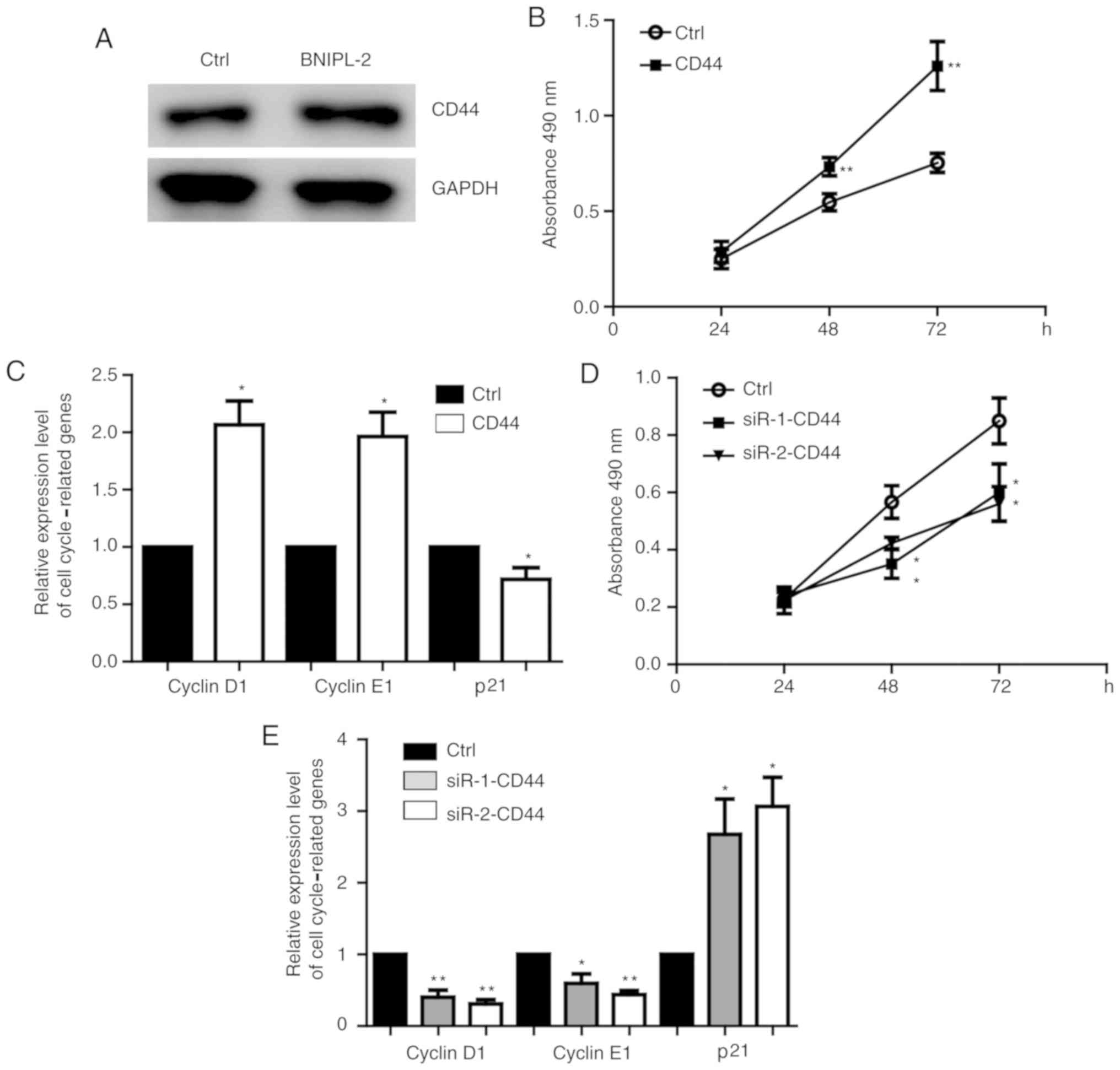
5A). Overexpression of CD44 (Fig.
S1A) promoted SW480 cell proliferation (Fig. 5B) and increased the expression of
the cell cycle-related genes cyclin D1 and E1 and decreased the
expression of p21 (Fig. 5C). CD44
knockdown (Fig. S1B) inhibited
proliferation (Fig. 5D), increased
p21 expression and decreased cyclin D1 and E1 expression (Fig. 5E). In order to demonstrate the
effect of transfection, a positive siRNA experimental group was
also employed. siRNA-p53 [sequence was referenced by a previous
study (17)] was transfected into
HCT116 cells and the downregulation of p53 was detected to confirm
the effective transfection system (Fig. S1C). It was further determined that
CD44 overexpression (Fig. S1D)
promoted HCT116 cell proliferation (Fig. S1E), and CD44 knockdown (Fig. S1F) suppressed HCT116 cell
proliferation (Fig. S1G).
CD44 mediates the function of BNIPL-2
in regulating CRC proliferation
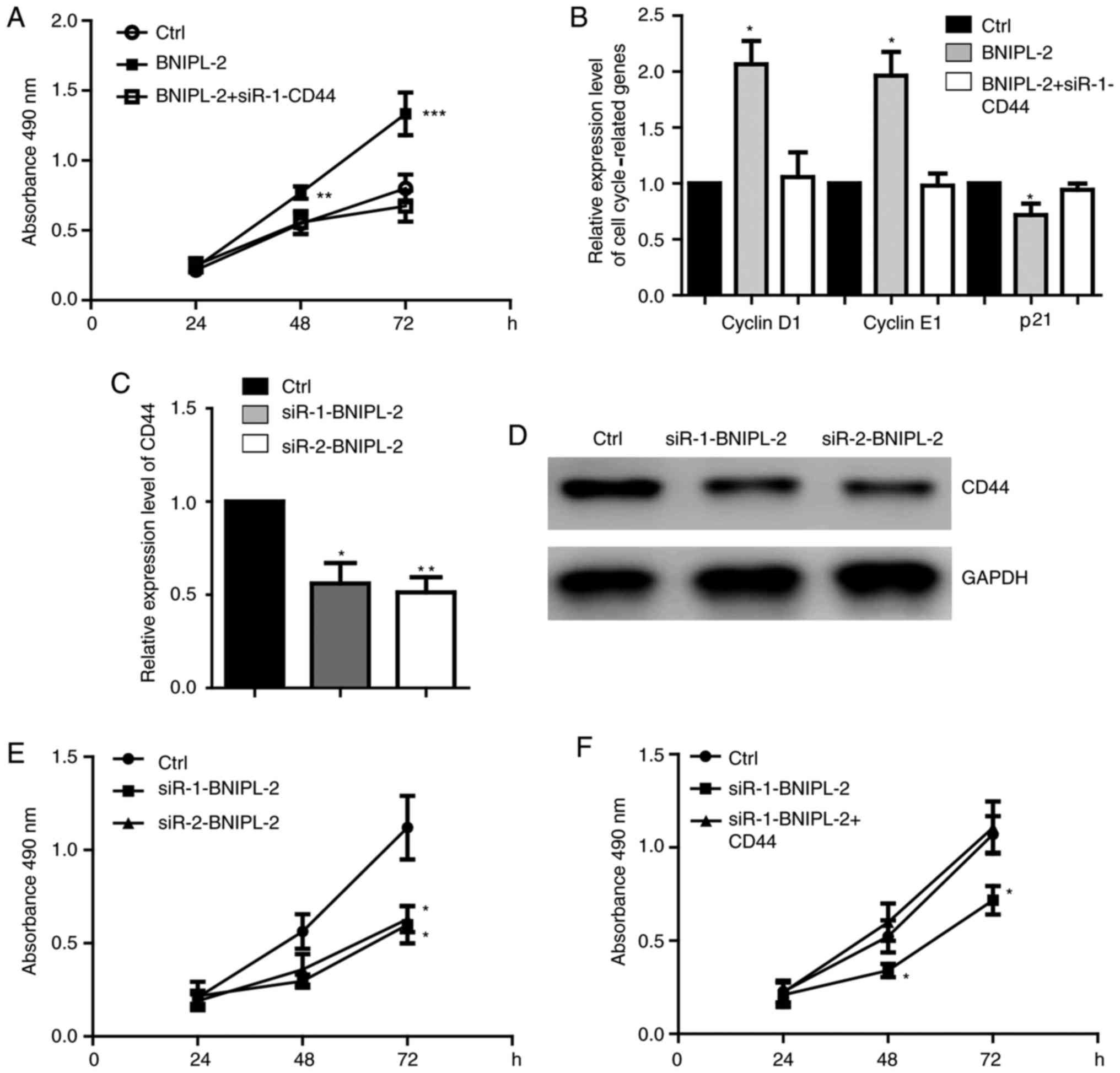
A rescue experiment was then performed to detect
whether BNIPL-2 promotes cell proliferation through CD44. It was
revealed that CD44 knockdown significantly blocked the promotion of
proliferation caused by overexpressing BNIPL-2 in SW480 cells
(Figs. 6A and S2A). The expression of cell
cycle-regulated genes was also restored by CD44 downregulation
(Fig. 6B). BNIPL-2 knockdown
downregulated CD44 at the mRNA (Fig.
6C) and protein levels (Fig.
6D) and inhibited cell proliferation (Fig. 6E). CD44 overexpression rescued the
proliferation caused by BNIPL-2 knockdown in both SW480 cells
(Fig. 6F) and HCT116 cells
(Fig. S2B and C).
Discussion
Colorectal cancer has become one of the most
frequent types of malignant cancers worldwide (2). An improved understanding of the risk
factors of CRC is of great importance to improve the future
diagnosis and treatment of the disease. In the present study, it
was revealed that a high level of BNIPL-2 is a critical factor
involved in the adverse T and M stages and poor prognosis and is
correlated with cell growth, migration, and invasion regulatory
signaling pathways.
BNIPL-2 has been reported to promote the invasion
and metastasis of human hepatocellular carcinoma cells (12). However, apoptosis and growth
inhibition-related genes were upregulated, and cellular
proliferation was downregulated in Hep3B cells overexpressing
BNIPL-2 (19). These studies
indicated the complicated function of BNIPL-2 in regulating cancer
physiology. It was revealed that upregulation of BNIPL-2 in CRC
tissues suggested a poor prognosis. These results are the first to
indicate the critical relationship between BNIPL-2 and CRC.
BNIPL-2 interacts with Bcl-2 and Cdc42GAP (11), which are both important apoptosis-
and proliferation-related genes in many types of cancers (20,21).
These studies indicated the critical regulatory function of BNIPL-2
in cancer cell growth. TMN stage analysis was performed and a
correlation between higher BNIPL-2 expression levels and adverse T
and M stages was revealed, which suggested the potential function
of BNIPL-2 in the regulation of cell growth involved in
proliferation and apoptosis. It was further detected that gene
enrichment for tumor genesis and development, including cell
growth, migration, and invasion, were all stratified in the BNIPL-2
high expression tissues. BNIPL-2 was also critically involved in
regulating CRC cell proliferation. BNIPL-2 upregulated the
expression of cyclin D1 and cyclin E1 and downregulated the
expression of p21. The proportion of cells was increased in the G1
phase and decreased in the S and G2/M phases upon overexpression of
BNIPL-2. The present results indicated the critical function of
BNIPL-2 in regulating CRC proliferation and cell cycle
processes.
CD44 has been reported to promote cell proliferation
in non-small cell lung cancer (16). νCD44 regulated proliferation in
lung cancer PDL cells, possibly through BMP-2, FGF-1 and ICAM-1
(22). In cutaneous squamous
cells, miR-199a targeted CD44 and reduced proliferation (23). These studies indicated the function
of CD44 in regulating cell proliferation. In the present study, it
was confirmed that CD44 mediated the function of BNIPL-2 in
regulating CRC proliferation. BNIPL-2/CD44 signaling plays an
important role during CRC growth.
Additionally, there are some limitations to this
study. It will further be determined whether BNIPL-2 can be a
transcriptional factor that regulates the expression of CD44 by
ChIP and luciferase reporter gene assays in the future.
Additionally, the BNIPL-2/CD44 signaling axis regulation of CRC
cell proliferation in vivo will be verified. The present
results not only indicated the important roles of BNIPL-2 in CRC
genesis and development and cell cycle and proliferation regulation
but also suggested the potential function of BNIPL-2 as an
efficient biomarker or treatment target for future therapy.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81801047 and
U1404808).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LG and HL performed most of the cell and molecular
experiments. NY, SZ and GJ conceived and performed the experiments
and analyzed the data. YH, DH and YL performed the bioinformatics
analysis. QS and XF conceived and supervised the project and wrote
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Farinetti A, Zurlo V, Manenti A, Coppi F
and Mattioli AV: Mediterranean diet and colorectal cancer: A
systematic review. Nutrition. 43-44:83–88. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu C, He T, Li Z, Liu H and Ding B:
Regulation of HOXA11-AS/miR-214-3p/EZH2 axis on the growth,
migration and invasion of glioma cells. Biomed Pharmacother.
95:1504–1513. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang XQ, Zhang XF, Xia JH, Chao J, Pan
QZ, Zhao JJ, Zhou ZQ, Chen CL, Tang Y, Weng DS, et al: Tripartite
motif-containing 3 (TRIM3) inhibits tumor growth and metastasis of
liver cancer. Chin J Cancer. 36:772017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao Z, Jia Q, Wu MS, Xie X, Wang Y, Song
G, Zou CY, Tang Q, Lu J, Huang G, et al: Degalactotigonin, a
natural compound from Solanum nigrum L., inhibits growth and
metastasis of osteosarcoma through GSK3β inactivation-mediated
repression of the Hedgehog/Gli1 pathway. Clin Cancer Res.
24:130–144. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li XT, Liang Z, Wang TT, Yang JW, Ma W,
Deng SK, Wang XB, Dai YF, Guo JH and Li LY: Brain-derived
neurotrophic factor promotes growth of neurons and neural stem
cells possibly by triggering the phosphoinositide
3-kinase/AKT/glycogen synthase kinase-3β/β-catenin pathway. CNS
Neurol Disord Drug Targets. 16:828–836. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim TH and Cho SG: Kisspeptin inhibits
cancer growth and metastasis via activation of EIF2AK2. Mol Med
Rep. 16:7585–7590. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Xue J, Kuang H, Zhou X, Liao L and
Yin F: microRNA-1297 inhibits the growth and metastasis of
colorectal cancer by suppressing cyclin D2 expression. DNA Cell
Biol. 36:991–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu G, Liu J, Wu Z, Wu X and Yao X:
MicroRNA-184 inhibits cell proliferation and metastasis in human
colorectal cancer by directly targeting IGF-1R. Oncol Lett.
14:3215–3222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lu H, Liufu N, Dong Y, Xu G, Zhang Y, Shu
L, Soriano SG, Zheng H, Yu B and Xie Z: Sevoflurane acts on
ubiquitination-proteasome pathway to reduce postsynaptic density 95
protein levels in young mice. Anesthesiology. 127:961–975. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qin W, Hu J, Guo M, Xu J, Li J, Yao G,
Zhou X, Jiang H, Zhang P, Shen L, et al: BNIPL-2, a novel homologue
of BNIP-2, interacts with Bcl-2 and Cdc42GAP in apoptosis. Biochem
Biophys Res Commun. 308:379–385. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie L, Qin W, Li J, He X, Zhang H, Yao G,
Shu H, Yao M, Wan D and Gu J: BNIPL-2 promotes the invasion and
metastasis of human hepatocellular carcinoma cells. Oncol Rep.
17:605–610. 2007.PubMed/NCBI
|
|
13
|
Tsuneki M and Madri JA: CD44 regulation of
endothelial cell proliferation and apoptosis via modulation of CD31
and VE-cadherin expression. J Biol Chem. 289:5357–5370. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nam K, Oh S, Lee KM, Yoo SA and Shin I:
CD44 regulates cell proliferation, migration, and invasion via
modulation of c-Src transcription in human breast cancer cells.
Cell Signal. 27:1882–1894. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jing F, Kim HJ, Kim CH, Kim YJ, Lee JH and
Kim HR: Colon cancer stem cell markers CD44 and CD133 in patients
with colorectal cancer and synchronous hepatic metastases. Int J
Oncol. 46:1582–1588. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Elliott VA, Rychahou P, Zaytseva YY and
Evers BM: Activation of c-Met and upregulation of CD44 expression
are associated with the metastatic phenotype in the colorectal
cancer liver metastasis model. PLoS One. 9:e974322014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang S, Wang K, Zhang C, Zhang W, Xu Q,
Wang Y, Zhang Y, Li Y, Zhang Y, Zhu H, et al: Overaccumulation of
p53-mediated autophagy protects against betulinic acid-induced
apoptotic cell death in colorectal cancer cells. Cell Death Dis.
8:e30872017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xie L, Qin WX, He XH, Shu HQ, Yao GF, Wan
DF and Gu JR: Differential gene expression in human hepatocellular
carcinoma Hep3B cells induced by apoptosis-related gene BNIPL-2.
World J Gastroenterol. 10:1286–1291. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Q, Peng J, Liu T and Zhang G: Effects
of celecoxib on cell apoptosis and Fas, FasL and Bcl-2 expression
in a BGC-823 human gastric cancer cell line. Exp Ther Med.
14:1935–1940. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shi C, Ren L, Sun C, Yu L, Bian X, Zhou X,
Wen Y, Hua D, Zhao S, Luo W, et al: miR-29a/b/c function as
invasion suppressors for gliomas by targeting CDC42 and predict the
prognosis of patients. Br J Cancer. 117:1036–1047. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yeh Y, Yang Y and Yuan K: Importance of
CD44 in the proliferation and mineralization of periodontal
ligament cells. J Periodontal Res. 49:827–835. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang SH, Zhou JD, He QY, Yin ZQ, Cao K and
Luo CQ: MiR-199a inhibits the ability of proliferation and
migration by regulating CD44-Ezrin signaling in cutaneous squamous
cell carcinoma cells. Int J Clin Exp Pathol. 7:7131–7141.
2014.PubMed/NCBI
|