Introduction
Gastric cancer (GC) is one of the most common types
of gastrointestinal cancer, being the second leading cause of
tumor-associated mortality in the world (1–4). It
is reported that the 5-year survival rate of patients with GC is
~20% (5). The high mortality rate
is primarily due to late diagnosis and local and systemic
metastasis (6–9). Surgery is the principal treatment,
and metastases are observed in the majority of patients with
advanced GC. Advances in diagnostic and therapeutic approaches have
led to improved expectations for the long-term survival of patients
with early GC. However, the prognosis for advanced GC with
extensive invasion and metastasis remains poor (10). Therefore, understanding the
potential molecular underpinnings of the GC metastatic process is
critical for the development of novel treatments.
MicroRNAs (miRs) are endogenous non-coding RNAs.
They are formed of 18–25 nucleotides and regulate gene expression
by binding to the untranslated region (UTR) of mRNAs (11,12).
miRs serve important and diverse roles in numerous biological
processes by controlling the translation of mRNA, including cell
growth and metabolism (13–15).
Furthermore, miRs serve important roles in multiple aspects of
tumor development, including cell proliferation, apoptosis,
invasion and migration (16).
Increasing evidence has indicated that miR-381 is abnormally
expressed in various types of tumors, including lung cancer,
prostate cancer, cervical carcinoma and GC (17–20),
and that miR-381 may regulate various biological aspects of
tumorigenesis (21). Previous
studies have reported miR-381 is upregulated in glioma and acts as
an oncogene (18); however, it is
downregulated in renal cancer cells, lung adenocarcinoma and
glioblastoma, and exhibits the characteristics of a tumor
suppressor in these types of cancer (1,17,19).
Liao et al (22) reported
that Twist-related protein 1 (TWIST1) was a direct target of
miR-381, and that miR-381 regulated the growth and migration of
pituitary adenoma cells by targeting TWIST. However, the biological
role of miR-381 in GC remains poorly investigated. In the present
study, the biological effect of miR-381 and the association between
miR-381 and TWIST1 in GC cells was investigated.
Materials and methods
Cell culture
The human gastric adenoma cell line BGC-823 (cat.
no. TCHu 11) and human gastric adenoma cell line SGC-7901 (cat. no.
TCHu 46) were purchased from the Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). The human gastric
epithelial cell line MKN-28 (cat. no. CL-0291) and the human
gastric epithelial mucosa cells (GES; cat. no. CL-0563) were
purchased from Procell Life Science & Technology Co., Ltd.
(Wuhan, China). The cells were cultured in RPMI 1640 medium
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 1:10 fetal bovine serum (FBS; Invitrogen; Thermo
Fisher Scientific, Inc.), penicillin (1:100; Invitrogen; Thermo
Fisher Scientific, Inc.) and streptomycin (1:100; Invitrogen;
Thermo Fisher Scientific, Inc.) in a humidified atmosphere
containing 5% CO2 at 37°C.
Transfection assay
The GC cell line SGC-7901 was cultured in medium
without antibiotics for 24 h. miR-381 mimic (cat. no. 4464066) or
miR-381 negative control (NC; cat. no. 4464059) (Ambion; Thermo
Fisher Scientific, Inc.) were transfected into the cultured
SGC-7901 cells (5,000 cells/well in a 96-well plate) at a final
concentration of 10 nM using the Lipofectamine® 3000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. The miR-381 NC was used as the
negative control. Protein and total RNA were extracted from the
transfected cells 72 h after transfection. TWIST1 small interfering
RNA (siRNA; cat. no. sc-38604) was purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). TWIST1 siRNA transfection
was performed using the Lipofectamine® 3000 reagent
according to the manufacturer's instructions. Cells were harvested
for further study 24 h post-transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total miRs and mRNAs were extracted from transfected
and mock GC cells using TRIzol® (Invitrogen; Thermo
Fisher Scientific, Inc.). miRs were converted to cDNAs via
PrimeScript miRNA cDNA Synthesis kit (cat. no. D350A; Takara Bio,
Inc., Otsu, Japan) and mRNAs were converted to cDNAs by using the
PrimeScript RT Master Mix kit (cat. no. DRR036A; Takara Bio, Inc.).
miRNA RT was conducted under the following conditions: 42°C for 60
min and 85°C for 15 min. mRNA RT was conducted under the following
conditions: 37°C for 15 min and 85°C for 1 min. miR-381
quantification was performed via TaqMan Universal PCR Master Mix
with a miRNA-specific TaqMan minor groove binder probe (cat. no.
4426961; Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. RNA U6 was used as internal
control for miR-381 quantification. The sequences for U6 were: F:
5′-GTGCTCGCTTCGGCAGCACATATAC-3′; R:
5′-AAAAATATGGAACGCTCACGAATTTG-3′. The sequences for GAPDH (cat. no.
qHsaCEP0041396; Bio-Rad Laboratories, Inc.) and miR-381 (cat. no.
4426961; Invitrogen; Thermo Fisher Scientific, Inc.) were
proprietary. The qPCR for TWIST1 was performed using PrimePCR™
SYBR® Green Assay (cat. no. qHsaCED0043959; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). GAPDH was the internal
control gene for TWIST1 quantification. qPCR was conducted under
the following conditions: 95°C for 10 min, followed by 40 cycles at
95°C for 10 sec, 57°C for 20 sec and 72°C for 10 sec. Relative
quantifications of miRs and mRNAs were calculated using the
2−ΔΔCq method as previously described (23).
miRNA target predictions
To further investigate the potential target of
miR-318, potential genes were predicted using TargetScan 7.2
(www.targetscan.org) and miRBase
(www.mirbase.org).
Dual luciferase assay
The wild-type (wt; GUUUUGUAAAUAUCUUUGUAUA) or mutant
(mut; GUUUUGUAAAUAUCUCCACGCA) TWIST1 genes were cloned downstream
of the luciferase gene in the pLUC Luciferase vector to construct
the TWIST1 luciferase reporter vector (Qcbio S&T Co., Ltd.).
miR-381 mimic or miR-381 NC were co-transfected with the TWIST1
luciferase reporter vector into the GC cell line SGC-7901 (10,000
cells/well in a 48-well plate) using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. The transfected cells were cultured at
37°C for 48 h, harvested and lysed for the measurement of
luciferase activity according to the manufacturer's instructions
(Dual-Luciferase® Reporter Assay system; Promega
Corporation). The relative luciferase activity was normalized to
Renilla luciferase activity.
Cell viability and colony formation
assays
An MTT assay was used to evaluate the effect of
miR-381 on GC cell viability. GC cells transfected with miR-381
mimic or miR-381 NC were cultured in 96-well plates (5,000
cells/well). Cell viability was measured by MTT assay every 24 h
according to the manufacturer's instructions. Following incubation
with MTT reagent, formazan crystals were dissolved in DMSO and
optical density was measured at 490 nm. For the colony formation
assay, GC cells transfected with miR-381 mimic or miR-381 NC were
cultured in 6-well plates for 2 weeks (500 cells/well), fixed with
100% methanol at room temperature (RTemp) for 15 min, and stained
with 0.5% crystal violet at 4°C for 30 min to count the number of
colonies using a light microscope (magnification, ×4).
Transwell cell migration and invasion
assays
Cell invasion and migration were assessed using a
Transwell chamber coated with or without Matrigel (EMD Millipore),
respectively, according to the manufacturer's protocol. miR-381
mimic or miR-381 NC transfected GC cells (100,000) were cultured in
serum free medium and seeded in the upper chamber (pore size, 8
µm). The lower chamber contained the chemoattractant medium with
10% FBS. After 48 h, the cultured cells in the upper chamber were
removed using a cotton swab, while the cells which had migrated to
the reverse face were fixed with 4% methanol for 20 min at RTemp,
and stained with giemsa for 15 min at RTemp to count the cell
numbers using a light microscope (magnification, ×200).
Cell apoptosis assay
miR-381 mimic or miR-381 NC transfected GC cells
were cultured for 2 days in RPMI1640 medium. Harvested cells were
stained with fluorescein isothiocyanate-Annexin V and propidium
iodide (PI; Beyotime Institute of Biotechnology). A flow cytometry
(FCM) assay was performed to evaluate cell apoptosis (BD
FACSCalibur; BD Biosciences), and the data were analyzed with
CellQuest Pro 7.5.3 software (BD Biosciences).
Western blotting
miR-381 mimic or miR-381 NC transfected GC cells
were cultured for 2 days in 6-well plates. Cells were harvested and
protein was extracted using a radioimmunoprecipitation assay buffer
containing protease inhibitors (Beyotime Institute of
Biotechnology). Protein concentration was measured using the
bicinchoninic acid assay. The extracted proteins (50 µg) were
separated by 10% SDS-PAGE and transferred onto a polyvinylidene
difluoride (PVDF) membrane. The PVDF membrane was blocked with 5%
milk in PBS-0.05% Tween 20 (PBST) for 2 h at RTemp, and incubated
with the following primary antibodies (1:2,000): TWIST1, (cat. no.
46702) and GAPDH (cat. no. 5174; both Cell Signaling Technology,
Inc.) for 2 h at RTemp. Following three washes with PBST, the PVDF
membrane was incubated with anti-rabbit secondary antibodies
(1:2,000; cat. no. 7074; Cell Signaling Technology, Inc.) for 1 h
at RTemp. Following three washes with PBST, the target proteins
were detected with an enhanced chemiluminescent kit (cat. no.
P0018; Beyotime Institute of Biotechnology) and exposed on x-ray
film.
Statistical analysis
Each experiment was repeated at least three times.
SPSS 16.0 (SPSS, Inc., Chicago, IL, USA) was used to perform
one-way analysis of variance and Tukey's HSD tests (post-hoc test)
to identify statistical differences. The data are expressed as the
mean ± standard deviation. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-381 is downregulated in GC
cells
In order to determine the expression of miR-381 in
human GC cells, total RNA was extracted from cultured cell lines,
and the expression levels of miR-381 were measured via RT-qPCR. The
relative expression of miR-381 was normalized to the endogenous U6
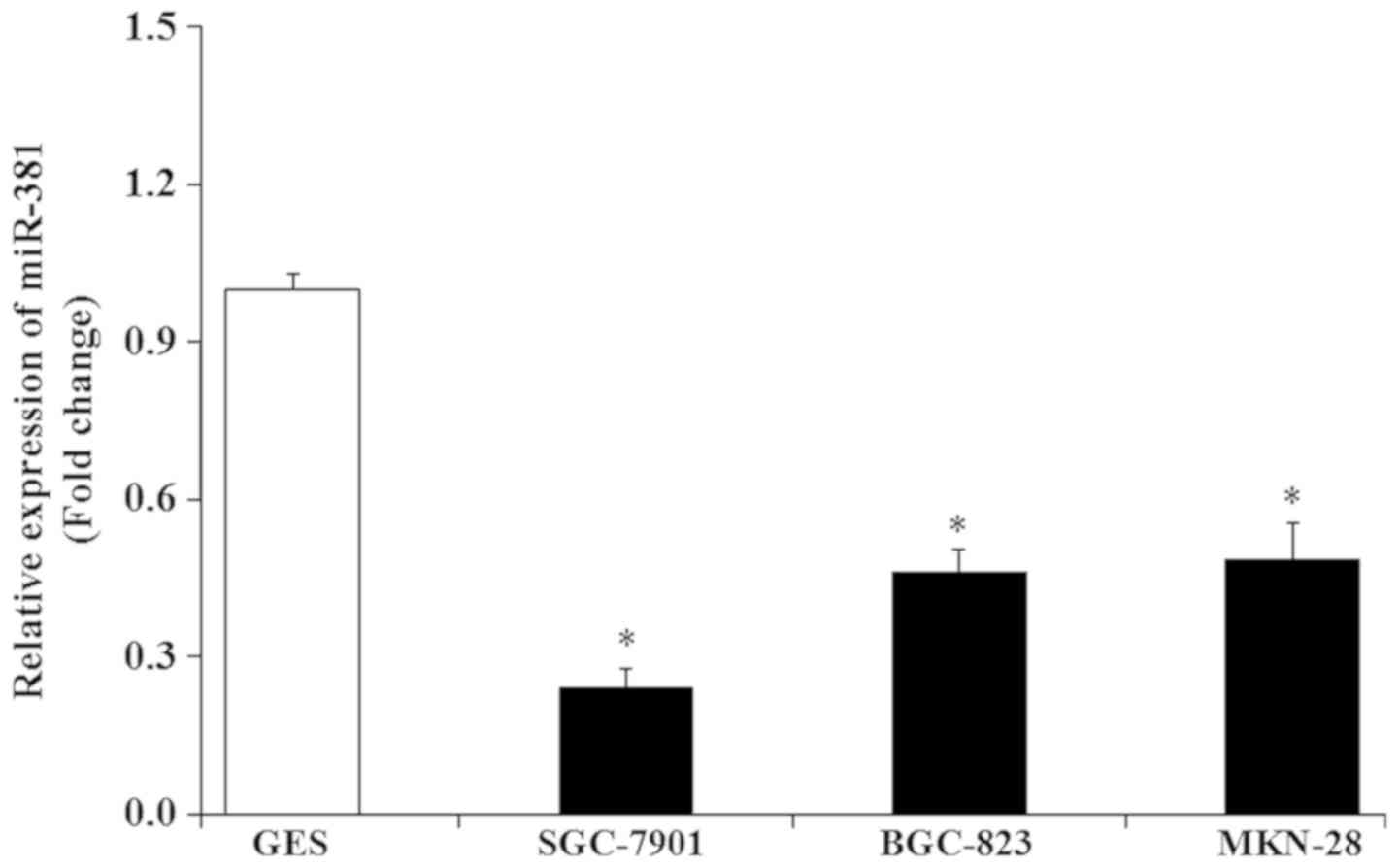
RNA expression. The expression of miR-381 (Fig. 1) was significantly decreased in GC
cell lines compared with the immortalized human gastric epithelial
GES cells (P<0.05). Notably, in SGC-7901 cells, the relative
expression of miR-381 was ~0.24 relative to GES cells; therefore,
SGC-7901 cells were used in the subsequent experiments.
TWIST1 is a direct target of miR-381
in GC cells
To investigate the biological function of miR-381 in
GC cells, the direct target gene of miR-381 was identified using
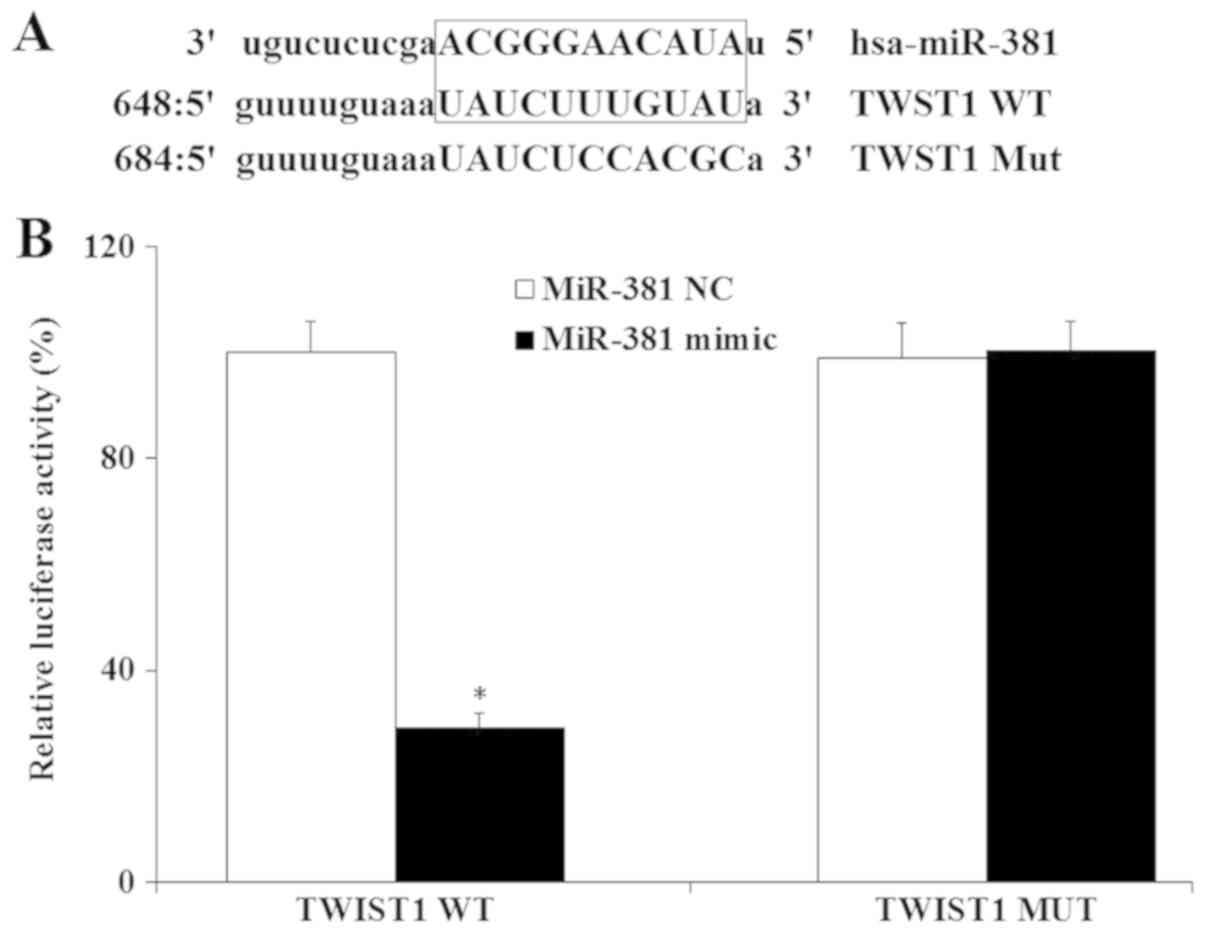
the miRBase database. The search results indicated that TWIST1 was
a potential target of miR-381 (Fig.
2A). Previous studies reported that TWIST1 is upregulated in
GC, and that TWIST1 promotes GC cell proliferation (24,25),
therefore miR-381 was hypothesized to regulate GC cells via TWIST1.
To confirm whether TWIST1 is a direct target gene of miR-381, a
dual luciferase assay was performed in SGC-7901 cells. Luciferase
reporter plasmids with either the wt or mut targeting sequence of
TWIST1 mRNA were created (Fig.
2A). These were co-transfected with miR-381 mimic or miR-381 NC
into SGC-7901 cells, cultured for 48 h, and the luciferase activity
was measured. As indicated in Fig.
2B, miR-381 significantly decreased the luciferase activity of
the wt TWIST1 UTR reporter (P<0.05), while the transfection with
the mut UTR of TWIST1 did not. These results demonstrated that
TWIST1 was a direct target gene of miR-381.
miR-381 inhibits GC cell
viability
To investigate the effect of miR-381 on GC SGC-7901
cell viability, an MTT assay was performed. To evaluate the effect
of miR-381 expression, the mimic transfection assay was used.
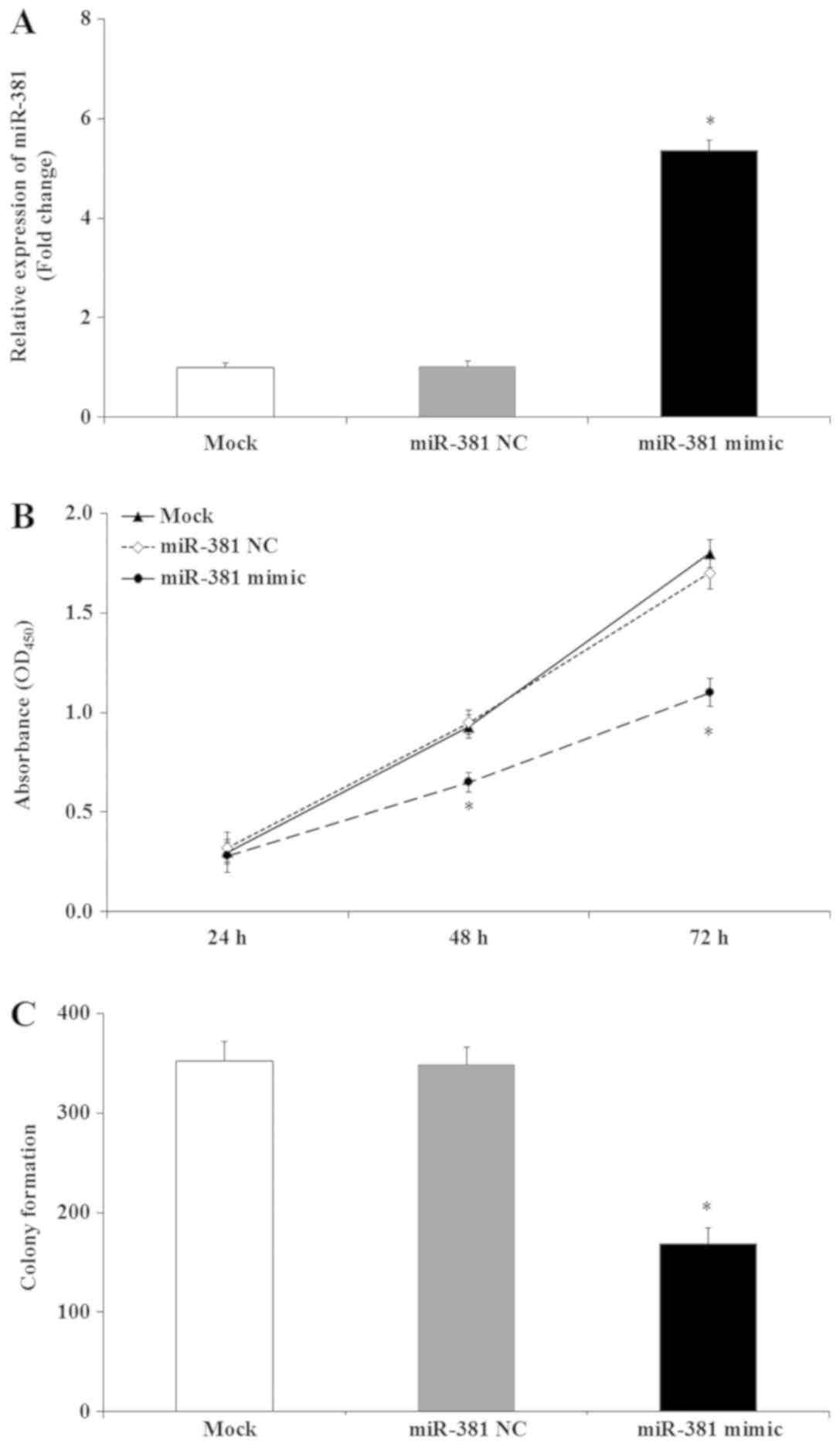
Transfection of miR-381 mimic significantly increased the
expression of miR-381 compared with the miR-381 NC and mock groups
(Fig. 3A). The MTT results
indicated that miR-381 significantly decreased the viability of
SGC-7901 cells compared with mock and miR-381 NC cells (P<0.05),
while there was no significant difference between miR-381 NC and
mock group (Fig. 3B). Further to
these results, colony formation assays demonstrated that miR-381
expression resulted in fewer SGC-7901 cell colonies compared with
mock and miR-381 NC (P<0.05; Fig.
3C). No significant differences in the number of colonies
between the miR-381 NC and mock group were observed (Fig. 3C). These results revealed that
miR-381 may serve an important role in GC cell viability, and that
induction of miR-381 expression may inhibit GC cell
proliferation.
miR-381 inhibits GC cell invasion and
migration
The effect of miR-381 on GC cell invasion and
migration was also verified to further characterize miR-381 in GC.
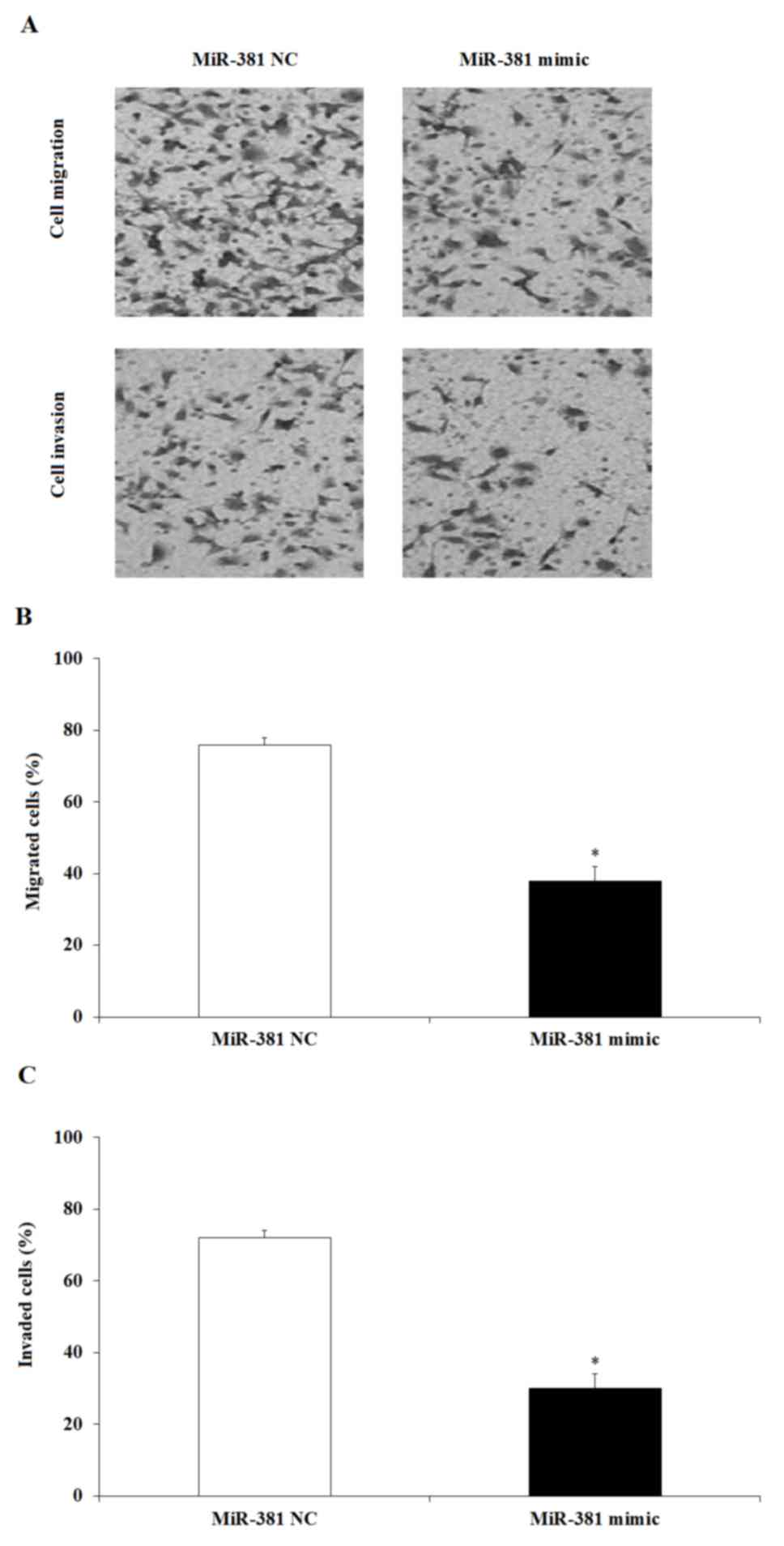
The results indicated that upregulation of miR-381 affected cell
migration and invasion in a Transwell chamber (Fig. 4A): Upregulation of miR-381
inhibited the migration and invasion capacity of SGC-7901 cells.
The cells that successfully migrated in the miR-381 upregulation
group corresponded to ~1/3 of the number of cells in the miR-381 NC
group (Fig. 4B). Therefore, the
data suggested that miR-381 may regulate GC growth through the
inhibition of cell proliferation, migration and invasion.
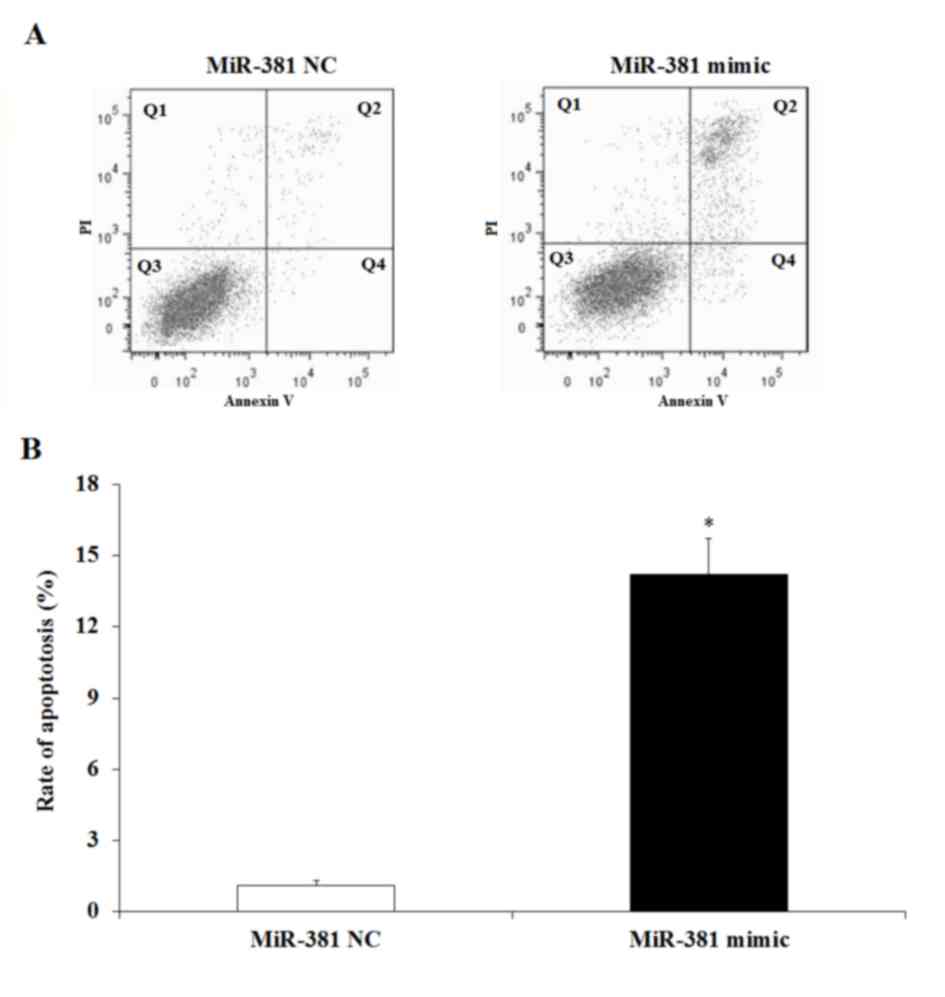
miR-381 induces GC cell apoptosis
TWIST1 was verified to be a direct target of
miR-381, and a previous study indicated that TWIST1 regulation was
associated with cell apoptosis (26). Therefore the effect of miR-381 GC
cell apoptosis was evaluated. SGC-7901 cells transfected with
miR-381 mimic or miR-381 NC were cultured for 48 h, harvested and
stained with PI and Annexin V for an FCM assay. The results
indicated that the percentage of apoptotic cells in the miR-381
mimic group was increased compared with the miR-381 NC group
(P<0.05; Fig. 5), which
suggests that expression of miR-381 may have enhanced GC cell
apoptosis.
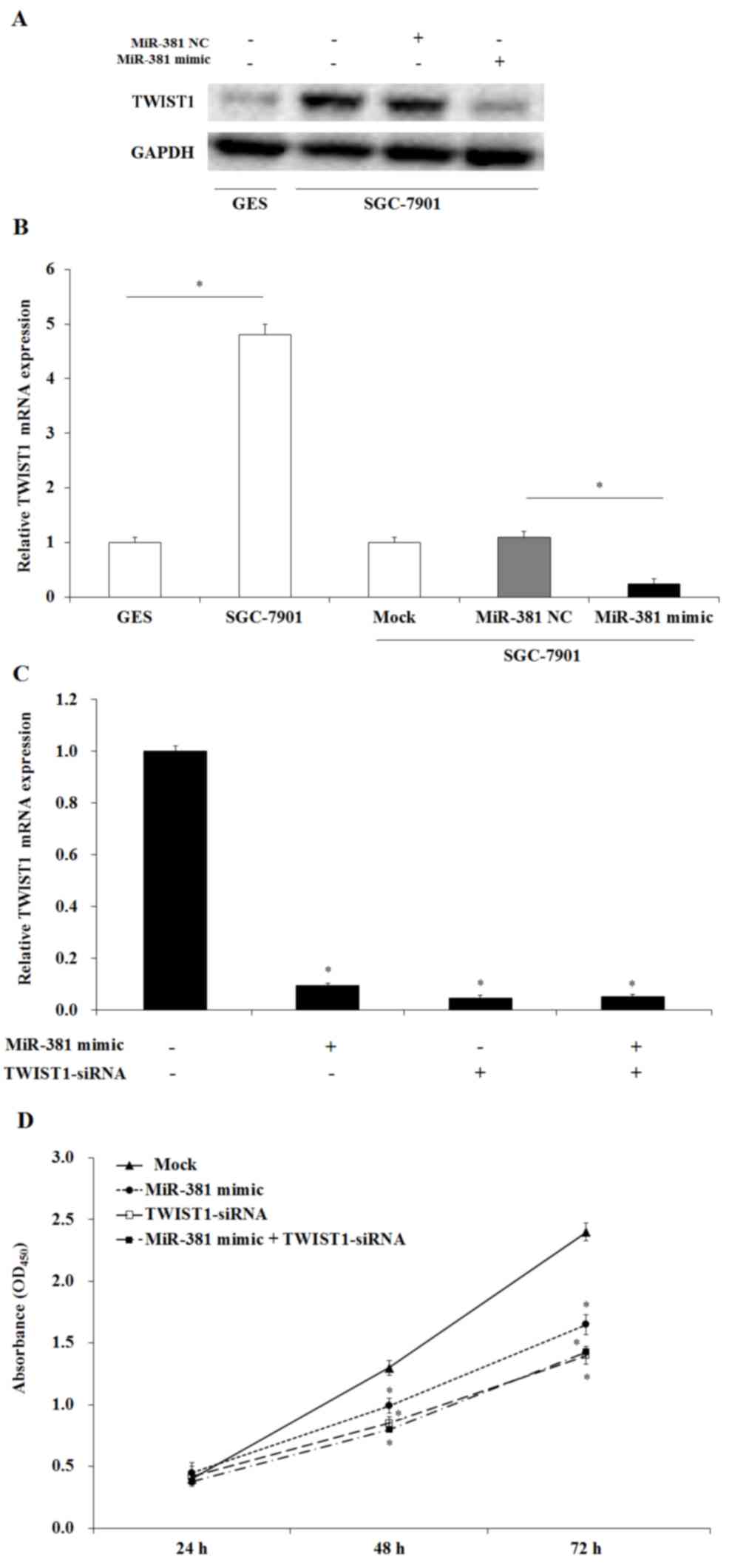
miR-381 decreases the expression of
TWIST1 in GC cells
To investigate whether the effect of miR-381 on cell
proliferation and apoptosis was caused by the regulation of TWIST1,
western blotting and RT-qPCR were performed. The results indicated
that the expression of TWIST1 was increased in GC cells compared
with the immortalized human gastric epithelial GES cells (Fig. 6A and B). Transfection of miR-381
mimic significantly decreased the expression of TWIST1 in GC cells
compared with the miR-381 NC group (P<0.05; Fig. 6A and B). RT-qPCR results also
indicated that transfection of TWIST1 siRNA decreased the
expression of TWIST1 (Fig. 6C). As
expected, TWIST1 knockout via co-transfection of miR-381 mimics and
TWIST1-siRNA also inhibited GC cell growth (Fig. 6D). Therefore, these results
demonstrated that upregulation of miR-381 may inhibit SGC-7901 GC
cell growth by targeting TWIST1.
Discussion
GC is one of the most common types of cancer in the
world (27). Approximately
one-half of patients with GC survive >5 years (1). The high mortality rate of GC has been
attributed to diagnosis at advanced stages and poor understanding
of the pathology (28). Therefore,
better assays for early-stage diagnosis and more efficient
therapeutic strategies are required for GC prevention and
treatment. Researchers have reported that miRs are associated with
cancer progression, tumor invasion and metastasis (29,30).
Various studies have also reported that miR-381 is downregulated in
a number of tumors, including cervical carcinoma, lung cancer,
prostate cancer and GC (17–20).
However, the biological role of miR-381 in GC
development/progression remained poorly understood. The present
study reported that the expression of miR-381 in GC cell lines was
significantly decreased compared with GES cells. To selectively
control the miR-381 expression in GC cells, miR-381 mimic
transfection assays were performed. Consequently, miR-381 induction
was observed to inhibit GC cell viability, invasion and migration.
Moreover, overexpression of miR-381 significantly induced GC cell
apoptosis, meaning that the observed GC cell decrease in viability
may be attributed to an increase in apoptosis.
TWIST1, a basic helix-loop-helix transcription
factor, is a known oncogene in humans (31). Previous studies indicated that
TWIST1 is a target of a number of miRs, and may be involved in the
proliferation, apoptosis and metastasis of various types of cancer.
Zhu et al (26) reported
that miR-186 regulates the cell cycle and induces cell apoptosis in
ovarian cancer by targeting Twist1. Li and Wu (32) reported that miR-32 inhibits
non-small-cell lung cancer cell proliferation,
epithelial-mesenchymal transition and metastasis by targeting
TWIST1. Bing et al (33)
reported that miR-543 inhibits tumor cell proliferation, migration
and invasion by targeting TWIST1. Additionally, Li et al
(34) demonstrated that miR-720
inhibits breast cancer cell invasion and migration through TWIST1.
TWIST1 has also been demonstrated to be expressed in human GC cell
lines, including: MKN7, MKN45, NUGC4, HSC60, AGS and SGC-7901
(35,36). In the present study, western
blotting and RT-qPCR demonstrated that the expression of TWIST1 in
SGC-7901 cells was increased compared with GES cells, while miR
target prediction programs and luciferase assays indicated that
TWIST1 is a target of miR-381. Moreover, the induction of miR-381
significantly decreased TWIST1 expression. Therefore miR-381 may
regulate GC cell viability through TWIST1.
In conclusion, the present study demonstrated that
miR-381 expression was decreased in GC cell lines compared with
normal GES cells, and low expression of miR-381 was associated with
the overexpression of TWIST1 in these cells. Moreover, forced
expression of miR-381 suppressed GC cell viability and invasion,
and induced cell apoptosis, possibly by directly targeting TWIST1.
These results provide novel insights into the regulation of gastric
tumorigenesis and progression by miRs. Upregulation of miR-381 may
constitute a novel clinical treatment against GC in the future.
However, to fully understand the effect of miR-381 in GC, further
research, including the effect of miR-381 on cell cycle
progression, is required.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YY and FZ designed the study. YY, XL and ZG
performed the experiments, analyzed the data and prepared the
manuscript. FZ reviewed the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Karpińska-Kaczmarczyk K, Lewandowska M,
Białek A, Ławniczak M and Urasińska E: Gastric hyperplastic polyps
coexisting with early gastric cancers, adenoma and neuroendocrine
cell hyperplasia. Pol J Pathol. 67:33–38. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ushijima T and Sasako M: Focus on gastric
cancer. Cancer Cell. 5:121–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smith MG, Hold GL, Tahara E and El-Omar
EM: Cellular and molecular aspects of gastric cancer. World J
Gastroenterol. 12:2979–2990. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cunningham D, Allum WH, Stenning SP,
Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ,
Falk SJ, Iveson TJ, et al: Perioperative chemotherapy versus
surgery alone for resectable gastroesophageal cancer. N Engl J Med.
355:11–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Vilaseca Cabo A, Musquera Felip M, Ribal
Caparros MJ and Alcaraz Asensio A: Metastasis of gastric carcinoma
simulating a urothelial tumor. Case report and review of the
literature. Arch Esp Urol. 66:885–889. 2013.(In Spanish).
PubMed/NCBI
|
|
7
|
Compare D, Rocco A and Nardone G: Risk
factors in gastric cancer. Eur Rev Med Pharmacol Sci. 14:302–308.
2010.PubMed/NCBI
|
|
8
|
Huang H, Han Y, Zhang C, Wu J, Feng J, Qu
L and Shou C: HNRNPC as a candidate biomarker for chemoresistance
in gastric cancer. Tumour Biol. 37:3527–3534. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Y, Gong J, Zhang Q, Lu Z, Gao J, Li Y,
Cao Y and Shen L: Dynamic monitoring of circulating tumour cells to
evaluate therapeutic efficacy in advanced gastric cancer. Br J
Cancer. 114:138–145. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oue N, Aung PP, Mitani Y, Kuniyasu H,
Nakayama H and Yasui W: Genes involved in invasion and metastasis
of gastric cancer identified by array-based hybridization and
serial analysis of gene expression. Oncology. 69 (Suppl 1):S17–S22.
2005. View Article : Google Scholar
|
|
11
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Glud M, Rossing M, Hother C, Holst L,
Hastrup N, Nielsen FC, Gniadecki R and Drzewiecki KT:
Downregulation of miR-125b in metastatic cutaneous malignant
melanoma. Melanoma Res. 20:479–484. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Croce C: Introduction to the role of
microRNAs in cancer diagnosis, prognosis, and treatment. Cancer J.
18:213–214. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Holley CL and Topkara VK: An introduction
to small non-coding RNAs: miRNA and snoRNA. Cardiovasc Drugs Ther.
25:151–159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen B, Duan L, Yin G, Tan J and Jiang X:
miR-381, a novel intrinsic WEE1 inhibitor, sensitizes renal cancer
cells to 5-FU by up-regulation of Cdc2 activities in 786-O. J
Chemother. 25:229–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tang H, Wang Z, Liu Q, Liu X, Wu M and Li
G: Disturbing miR-182 and −381 inhibits BRD7 transcription and
glioma growth by directly targeting LRRC4. PLoS One. 9:e841462014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rothschild SI, Tschan MP, Jaggi R, Fey MF,
Gugger M and Gautschi O: MicroRNA-381 represses ID1 and is
deregulated in lung adenocarcinoma. J Thorac Oncol. 7:1069–1077.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Skalsky RL and Cullen BR: Reduced
expression of brain-enriched microRNAs in glioblastomas permits
targeted regulation of a cell death gene. PLoS One. 6:e242482011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Martinez I, Gardiner AS, Board KF, Monzon
FA, Edwards RP and Khan SA: Human papillomavirus type 16 reduces
the expression of microRNA-381 in cervical carcinoma cells.
Oncogene. 27:2575–2582. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao C, Wang J, Chen W, He D and Wang H:
miR-381 regulates pituitary adenoma cell growth and migration by
targeting TWIST1. Int J Clin Exp Pathol. 10:242–249. 2017.
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sung CO, Lee KW, Han S and Kim SH: Twist1
is up-regulated in gastric cancer-associated fibroblasts with poor
clinical outcomes. Am J Pathol. 179:1827–1838. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qian J, Luo Y, Gu X, Zhan W and Wang X:
Twist1 promotes gastric cancer cell proliferation through
up-regulation of FoxM1. PLoS One. 8:e776252013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhu X, Shen H, Yin X, Long L, Xie C, Liu
Y, Hui L, Lin X, Fang Y, Cao Y, et al: miR-186 regulation of Twist1
and ovarian can-cer sensitivity to cisplatin. Oncogene. 35:323–332.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshikawa T, Aoyama T, Tanabe K, Nishikawa
K, Ito Y, Hayashi T, Cho H, Miyashita Y, Tsuburaya A and Sakamoto
J: Feasibility and safety of transhiatal approach and D2 total
gastrectomy after neoadjuvant chemotherapy for adenocarcinoma of
the Esophago-gastric junction: A subset analysis of the COMPASS
trial. Dig Surg. 33:424–430. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Burkitt MD, Varro A and Pritchard DM:
Importance of gastrin in the pathogenesis and treatment of gastric
tumors. World J Gastroenterol. 15:1–16. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nielsen CB, Shomron N, Sandberg R,
Hornstein E, Kitzman J and Burge CB: Determinants of targeting by
endogenous and exogenous microRNAs and siRNAs. RNA. 13:1894–1910.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jazdzewski K, Boguslawska J, Jendrzejewski
J, Liyanarachchi S, Pachucki J, Wardyn KA, Nauman A and de la
Chapelle A: Thyroid hormone receptor beta (THRB) is a major target
gene for microRNAs deregulated in papillary thyroid carcinoma
(PTC). J Clin Endocrinol Metab. 96:E546–E553. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bourgeois P, Stoetzel C, Bolcato-Bellemin
AL, Mattei MG and Perrin-Schmitt F: The human H-twist gene is
located at 7p21 and encodes a B-HLH protein that is 96% similar to
its murine M-twist counterpart. Mamm Genome. 7:915–917. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li L and Wu D: miR-32 inhibits
proliferation, epithelial-mesenchymal transition, and metastasis by
targeting TWIST1 in non-small-cell lung cancer cells. Onco Targets
Ther. 9:1489–1498. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bing L, Hong C, Li-Xin S and Wei G:
MicroRNA-543 suppresses endometrial cancer oncogenicity via
targeting FAK and TWIST1 expression. Arch Gynecol Obstet.
290:533–541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li LZ, Zhang CZ, Liu LL, Yi C, Lu SX, Zhou
X, Zhang ZJ, Peng YH, Yang YZ and Yun JP: miR-720 inhibits tumor
invasion and migration in breast cancer by targeting TWIST1.
Carcinogenesis. 35:469–478. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sakamoto A, Akiyama Y, Shimada S, Zhu WG,
Yuasa Y and Tanaka S: DNA methylation in the Exon 1 region and
complex regulation of Twist1 expression in gastric cancer cells.
PLoS One. 10:e01456302015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yuan W, Li T, Mo X, Wang X, Liu B, Wang W,
Su Y, Xu L and Han W: Knockdown of CMTM3 promotes metastasis of
gastric cancer via the STAT3/Twist1/EMT signaling pathway.
Oncotarget. 7:29507–29519. 2016. View Article : Google Scholar : PubMed/NCBI
|