Introduction
A mature ovarian follicle consists of several basic
elements that together form one functional unit. Theca externa is
located the most externally, followed by theca interna, while
granulosa cells (GCs) and oocytes are located inside the ovarian
follicle. Several types of follicular GCs are distinguished in the
ovarian follicle: Cells lining the inner part of the ovarian
follicle, adjacent to the basal lamina are called mural GCs,
followed by the layer forming the cumulus oophorus and cells
directly surrounding the oocyte-corona radiata. The basic functions
of GCs in the individual phases of follicle growth are the
production of hormones in response to follicle stimulating hormone
(FSH), induction of ovarian follicle atresia through specific
molecular markers, as well as the production of nexus cellular
connections for communication with the oocyte (1–5). The
well-known physiological properties of GCs have influenced the
intensive development of fields related to assisted reproduction
techniques in humans and animals (6–9).
In recent years, an increase of interest and intense
development of domains related to stem cells has been observed. The
term ‘stem cell’ describes cells that have the ability to
self-renew and differentiate into more targeted cell types. There
is a hierarchy of stem cells from those that give rise to all types
of cells to those that only differentiate into tissue and
organ-specific products. We, therefore, distinguish: Totipotent
stem cells-giving rise to embryo cells and extraembryonic tissue;
pluripotent stem cells-giving rise to all three germ layers;
multipotent stem cells-giving rise to cells from two or one germ
layer and unipotent stem cells-tissue-directed, giving rise only to
specific cell lines (10). The
research suggests that the stem cell reservoir is located in every
mature tissue, because cells undergoing apoptosis are continuously
replaced by new cells. It is suggested that the biggest stem cell
pools are located in the liver, lungs and pancreas (11–15).
In terms of clinical approaches, one of the most promising types of
such cells are mesenchymal stem cells (MSCs). MSCs are multipotent
progenitor cells that have the ability to differentiate towards
bone, cartilage and adipose cells. These cells are obtained
primarily from fetal tissues (placenta, umbilical cord blood,
umbilical cord), also being found in the body of an adult organism
(16–19). The main source of adult MSCs in
recent years has been the bone marrow and adipose tissue (20). Recent studies indicate that the
ovary can also be a source of stem cells. Kossowska-Tomaszczuk
et al (21) indicates
surprising stem-like properties of GCs. According to her research,
GCs in vitro have the properties similar to those of MSCs.
Other authors also point out that GCs have stem cell properties,
but not as broad as those of MSCs and pluripotent stem cells
(22,23). The presented research suggests that
GCs, routinely disposed of during the in vitro fertilization
procedure, may become a valuable source of cells used to obtain
osteoblast populations. Such osteoblasts could be used in the
treatment of diseases related to skeletal system pathologies.
Materials and methods
Part of the material and methods section is based on
other publications of the same research team, presenting results
from the same cycle of studies related to human ovarian GCs
(24,25).
Granulosa cell collection
The study group consisted of 8 patients, aged 18–40
years, enrolled in in vitro fertilization (IVF) procedure in
the Division of Infertility and Reproductive Endocrinology, Poznan
University of Medical Sciences, Poland. Follicular fluid containing
the GCs from patients undergoing in vitro fertilization
(IVF) procedures was collected.
The IVF procedure was based on a controlled ovarian
hyperstimulation protocol adapted to the patient's initial cause of
infertility, as well as predicted and current ovarian response.
Stimulation with human recombinant FSH (Gonal-F; Merck-Serono;
Merck KGaA) and highly purified human menopausal gonadotropin
(hMG-HP; Menopur; Ferring) has been performed according to
protocol. Gonadotropin-releasing hormone (GnRH)
antagonist-cetrorelix acetate (Cetrotide; Merck-Serono; Merck
KGaA), injection has been given at the appropriate dose to suppress
the function of the pituitary gland. Induction of ovulation was
based on the subcutaneous injection of 6.500 h of human chorionic
gonadotropin (hCG; Ovitrelle; Merck-Serono; Merck KGaA). The doses
of gonadotropins and GnRH antagonist have been precisely controlled
and recorded for every patient. The follicular fluid has been
collected during transvaginal ultrasound-guided oocyte pick-up, 36
h after administration of human chorionic gonadotropin. GCs have
been taken from follicles with a diameter of over 16 mm. Directly
after ovarian puncture, the complete content of the ovarian
follicle (follicular fluid containing GCs and oocytes) was passed
on to a qualified embryologist that extracted all of contained
oocytes that were subsequently used in further stages of the IVF
procedure (conducted at Division of Infertility and Reproductive
Endocrinology, Department of Gynecology, Obstetrics and
Gynecological Oncology, Poznan University of Medical Sciences,
Poznan, Poland). Meanwhile, the remaining granulosa cell containing
follicular fluid, usually discarded after this step, was passed on
to the employees of Department of Anatomy, Poznan University of
Medical Sciences, in which the further research was conducted.
Patients with a potential risk of inadequate ovarian
stimulation-according to Bologna criteria of poor ovarian
responders, published by European Society of Human Reproduction and
Embryology (ESHRE) in 2011 (26)
have been excluded, accepting serum antimullerian hormone (AMH) 0,7
ng/ml as a cut-off value. Moreover, patients with serum level of
FSH above 15 mU/ml on the 2nd-3rd day of the cycle, as well as
patients with polycystic ovary syndrome and endometriosis, have
also been excluded from the study. Only ovarian GCs, usually a part
of the discarded remnant material of the IVF procedure, were used
in the research. This study has been approved with resolution
558/17 by Poznan University of Medical Sciences Bioethical
Committee. All participants gave their written informed consent for
use of their material in research.
Primary cell culture
The GCs, suspended in follicular fluid, were washed
twice by centrifugation at 200 × g for 10 min at RT. Medium
consisted of Dulbecco's Modified Eagle's Medium (DMEM,
Sigma-Aldrich Co.; Merck KGaA), 2% fetal bovine serum FBS (FBS;
Sigma-Aldrich; Merck KGaA), 4 mM L-glutamine (Invitrogen; Thermo
Fisher Scientific, Inc.), 10 mg/ml gentamycin (Invitrogen; Thermo
Fisher Scientific, Inc.), 10,000 U/ml penicillin, and 10,000 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific, Inc.). Cells
were cultivated at 37°C under aerobic conditions (5%
CO2) in 25 cm3 culture flasks (Corning Inc.,
Corning, NY, USA). The cells were passaged upon reaching 90%
confluence; they were detached with 0.05% trypsin-EDTA (Invitrogen;
Thermo Fisher Scientific, Inc.) for 1–2 min and counted using an
ADAM Cell Counter and Viability Analyzer (Bulldog Bio). After
counting, the cells were seeded onto a number of flasks appropriate
for total cell number (2–3×106 cells per 25
cm3 flask). GCs were then cultivated for 30 days, the
morphology was checked daily and photographed using an Olympus
inverted microscope (Olympus). The medium was changed twice a week.
Finally, total RNA was isolated from GCs after 1, 7, 15 and 30
days. The viability of each collected sample was tested using the
ADAM CCVA, with only samples containing 95% or more viable cells
used for subsequent molecular analyses.
Total RNA isolation
RNA was isolated at 4 time periods, after 1, 7, 15,
and 30 days cultivation. The Chomczyński-Sacchi method was used to
isolate the total RNA (27). The
GCs were suspended in 1 ml mixture of guanidine thiocyanate and
phenol in monophase solution (TRI Reagent®;
Sigma-Aldrich; Merck KGaA). In the next step, the chloroform was
added and centrifuged to separate 3 phases. RNA has been located in
an aqueous phase. The resulting RNA was intact with no
contaminating DNA and protein. In the last step, the RNA has been
precipitated with 2-propanol (Sigma-Aldrich, cat. no. I9516), in
amount accurate per 1 ml of TRI-reagent and has been washed with
75% ethanol. Resulting RNA has been used for further analysis. The
total mRNA was determined from the optical density at 260 nm and
the RNA purity was estimated using the 260/280 nm absorption ratio
(NanoDrop spectrophotometer, Thermo Scientific). Samples with
absorbance ratio 260/280 greater than 1.8 have been used to the
presented study.
Microarray expression analysis
The microarray procedure was conducted according to
protocols used in previous studies of our team (5,28–32).
Total RNA (100 ng) from each pooled sample was subjected to two
rounds of sense cDNA amplification (Ambion® WT
Expression Kit). The obtained cDNA was used for biotin labelling
and fragmentation using Affymetrix GeneChip® WT Terminal
Labeling and Hybridization (Affymetrix). Biotin-labelled fragments
of cDNA (5.5 µg) were hybridized to the Affymetrix®
Human Genome U219 Array (48°C /20 h). Microarrays were then washed
and stained according to the technical protocol using the
Affymetrix GeneAtlas Fluidics Station. The array strips were
scanned employing Imaging Station of the GeneAtlas System.
Preliminary analysis of the scanned chips was performed using
Affymetrix GeneAtlas™ Operating Software. The quality of gene
expression data was confirmed according to the quality control
criteria provided by the software. The obtained CEL files were
imported into downstream data analysis software.
Reverse transcription-quantitative PCR
(RT-qPCR)
The RT-qPCR method was performed to confirm the
results obtained in the analysis of expression microarrays. Three
genes were selected from each heatmap: The ones showing highest,
lowest, and most intermediate-level of expression. Changes in the
level of expression of those genes were then examined. In each
group, three independent samples were analyzed, each coming from a
different patient (referred to as a biological repeat). Each test
was performed in 3 replicates. Reverse transcription was based on
the protocols and reagents of SABiosciences (RT2 First
Stand Kit-330401), using a Veritimer 96 well Thermal Cycler. 1 µg
of each gene's RNA transcript was used for reverse transcription.
qPCR was performed using the Light Cycler® 96 (Roche
Diagnostic GmbH, Germany), RT2 SYBR® Green
ROX™ qPCR Master Mix (Qiagen Sciences, Inc.,) and sequence-specific
primers (Table I).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin
(ACTB) and hypoxanthine phosphoribosyltransferase 1
(HRPT1) were used as reference genes. Gene expression was
analyzed using the 2−ΔΔCq method (33). The qPCR starters were designed
using the Primer3Plus software (http://primer3plus.com/cgi-bin/dev/primer3plus.cgi).
 | Table I.Oligonucleotide sequences of primers
used for RT-qPCR analysis. |
Table I.
Oligonucleotide sequences of primers
used for RT-qPCR analysis.
| Gene name | Primer sequence
(5′-3′) | Product size
(bp) |
|---|
| SYNCRIP | F:
TGTGGGAAAGATCCCAAGAG | 231 |
|
| R:
TTGGCAACTGAGATGCAGAC |
|
| MRC2 | F:
CACCAAACTCCGGTATTGCT | 189 |
|
| R:
TGGATCTCGGGTTCTGATTC |
|
| RRAS2 | F:
AGCACGGCAGCTTAAGGTAA | 165 |
|
| R:
TGGCAGCCTTTCTTGTCTTT |
|
| TPM4 | F:
TTGAGGAGGAGTTGGACAGG | 159 |
|
| R:
GCTGCATCTCCTGAATCTCC |
|
| SPP1 | F:
GCCGAGGTGATAGTGTGGTT | 242 |
|
| R:
GTGGGTTTCAGCACTCTGGT |
|
| FHL2 | F:
CTCATCCAAGTGCCAGGAAT | 175 |
|
| R:
CTCATAGCAGGGCACACAGA |
|
| SKI | F:
CAGCAGAAGGTTGTGAGCAG | 165 |
|
| R:
CGAGTCCTTGTCCTCCTCTG |
|
| EPHA2 | F:
GAGGGCGTCATCTCCAAATA | 236 |
|
| R:
TCAGACACCTTGCAGACCAG |
|
| CYR61 | F:
CTCCCTGTTTTTGGAATGGA | 241 |
|
| R:
TGGTCTTGCTGCATTTCTTG |
|
| CDK6 | F:
TGCACAGTGTCACGAACAGA | 150 |
|
| R:
ACCTCGGAGAAGCTGAAACA |
|
| SFRP1 | F:
CGAGTTTGCACTGAGGATGA | 190 |
|
| R:
GAAGTGGTGGCTGAGGTTGT |
|
| CTHRC1 | F:
GCTCACTTCGGCTAAAATGC | 165 |
|
| R:
CCACAGAAGAAGTGCGATGA |
|
| CREB3L1 | F:
AGGTGGAGACCCTGGAGAAT | 223 |
|
| R:
AGGGGGTCTTCCTTCACAGT |
|
| GLI2 | F:
CACCAACCAGAACAAGCAGA | 246 |
|
| R:
ACCTCAGCCTCCTGCTTACA |
|
| IGFBP5 | F:
GAGCTGAAGGCTGAAGCAGT | 237 |
|
| R:
GAATCCTTTGCGGTCACAAT |
|
| TWIST | F:
GTCCGCAGTCTTACGAGGAG | 159 |
|
| R:
CCAGCTTGAGGGTCTGAATC |
|
| BMP4 | F:
CTGGTCCACCACAATGTGAC | 162 |
|
| R:
CGATCGGCTAATCCTGACAT |
|
| COL1A1 | F:
GTGCTAAAGGTGCCAATGGT | 228 |
|
| R:
CTCCTCGCTTTCCTTCCTCT |
|
| LRRC17 | F:
CAACCCCTGGCACTGTACTT | 225 |
|
| R:
ACCTCAGGCTTGATGACTGG |
|
| GAPDH | F:
TCAGCCGCATCTTCTTTTGC | 90 |
|
| R:
ACGACCAAATCCGTTGACTC |
|
| ACTB | F:
AAAGACCTGTACGCCAACAC | 132 |
|
| R:
CTCAGGAGGAGCAATGATCTTG |
|
| HPRT | F:
TGGCGTCGTGATTAGTGATG | 141 |
|
| R:
ACATCTCGAGCAAGACGTTC |
|
Statistical analysis
All of the presented analyses and graphs were
performed using Bioconductor and R programming languages. Each CEL
file was merged with a description file. In order to correct
background, normalize, and summarize results, we used the Robust
Multiarray Averaging (RMA) algorithm. To determine the statistical
significance of the analyzed genes, moderated t-statistics from the
empirical Bayes method were performed. The obtained P-value was
corrected for multiple comparisons using Benjamini and Hochberg's
false discovery rate. The selection of significantly altered genes
was based on a P-value beneath 0.05 and expression higher than
two-fold. The differentially expressed gene list (separated for up-
and down-regulated genes) was uploaded to the DAVID software
(Database for Annotation, Visualization and Integrated Discovery)
(34).
Subsequently, sets of differentially expressed genes
from selected GO BP terms were applied to STRING software (Search
Tool for the Retrieval of Interacting Genes/Proteins) for
interaction prediction. STRING is a huge database containing
information about protein/gene interactions, including experimental
data, computational prediction methods and public text
collections.
Finally, the functional interactions between genes
that belong to the chosen GO BP terms were investigated by REACTOME
FIViz application to the Cytoscape 3.6.0 software. The
ReactomeFIViz app is designed to find pathways and network patterns
related to cancer and other types of diseases. This app accesses
the pathways stored in the Reactome database, allowing to do
pathway enrichment analysis for a set of genes, visualize hit
pathways using manually laid-out pathway diagrams directly in
Cytoscape, and investigate functional relationships among genes in
hit pathways. The app can also access the Reactome Functional
Interaction (FI) network, a highly reliable, manually curated
pathway-based protein functional interaction network covering over
60% of human proteins.
Additionally, a statistical analysis of the RT-qPCR
results (dependent sample Student's t-test corrected for multiple
comparisons using Benjamini and Hochberg's false discovery rate)
was conducted for every analyzed sample mean. Samples were only
considered further if P<0.05. This analysis employed the Real
Statistics Resource Pack add-on for MS Excel 2016 (Microsoft
Corporation).
Results
Whole transcriptome profiling by Affymetrix
microarray allowed us to analyze the expression changes in GCs,
after 1, 7, 15 and 30 days of culture. By Affymetrix®
Human HgU 219 Array, we examined the expression of 22480
transcripts. Genes with a fold change higher then abs (2) and with a corrected P-value lower than
0.05 were considered as differentially expressed. This set of genes
consisted of 2278 different transcripts.
DAVID (Database for Annotation, Visualization and
Integrated Discovery) software was used for extraction of the gene
ontology biological process terms (GO BP). Up and down-regulated
gene sets were subjected to the DAVID search separately and only
gene sets of adj. P-values <0.05 were selected. The DAVID
software analysis showed that differentially expressed genes
belonged to 582 Gene Ontology Biological Process (GO BP) terms and
45 KEGG pathways. In this report, we focused on ‘osteoblast
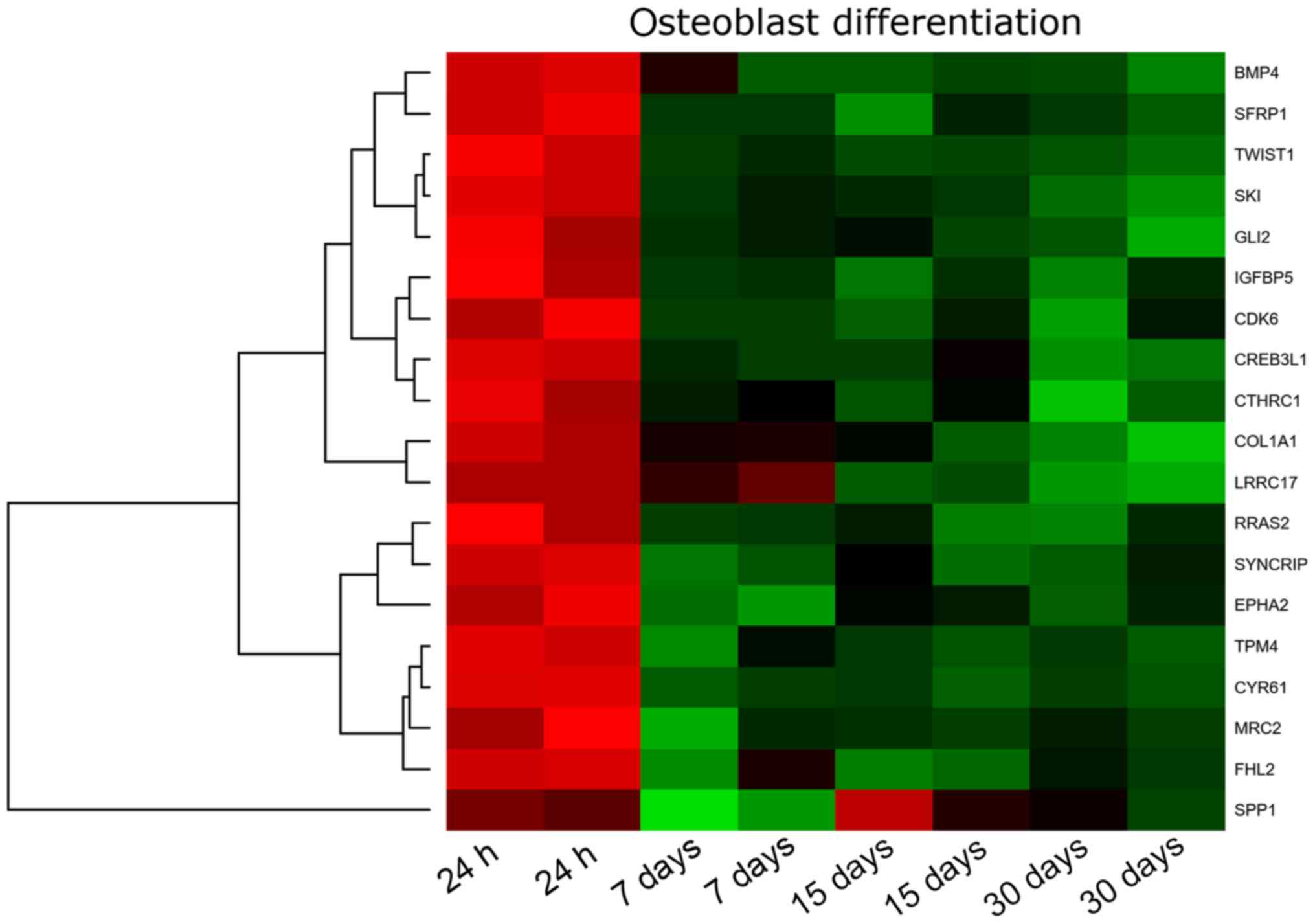
differentiation’ GO BP term. This set of genes was subjected to
hierarchical clusterization procedure and presented as a heatmap
(Fig. 1). The gene symbols, fold
changes in expression, Entrez gene IDs and corrected P-values of
these genes were shown in Table
II.
 | Table II.Gene symbols, fold changes in
expression, Entrez gene IDs and corrected P-values of studied
genes. |
Table II.
Gene symbols, fold changes in
expression, Entrez gene IDs and corrected P-values of studied
genes.
| Symbol | Entrez Gene ID | Fold change
D7/D1 | Fold change
D15/D1 | Fold change
D30/D1 | Adj. P-value
D7/D1 | Adj. P-value
D15/D1 | Adj. P-value
D30/D1 |
|---|
| SYNCRIP | 10492 | 2.060 | 1.867 | 1.885 | 0.013 | 0.018 | 0.016 |
| MRC2 | 9902 | 2.337 | 2.031 | 1.974 | 0.018 | 0.028 | 0.028 |
| RRAS2 | 22800 | 2.242 | 2.358 | 2.427 | 0.025 | 0.019 | 0.015 |
| TPM4 | 7171 | 2.538 | 2.478 | 2.493 | 0.013 | 0.012 | 0.011 |
| SPP1 | 6696 | 9.788 | 0.953 | 2.840 | 0.028 | 0.965 | 0.188 |
| FHL2 | 2274 | 3.073 | 3.898 | 2.849 | 0.028 | 0.014 | 0.028 |
| SKI | 6497 | 2.463 | 2.503 | 3.342 | 0.003 | 0.002 | 0.001 |
| EPHA2 | 1969 | 4.820 | 2.854 | 3.519 | 0.003 | 0.008 | 0.004 |
| CYR61 | 3491 | 4.318 | 4.249 | 4.154 | 0.001 | 0.001 | 0.001 |
| CDK6 | 1021 | 3.725 | 3.762 | 4.232 | 0.024 | 0.022 | 0.014 |
| SFRP1 | 6422 | 5.257 | 6.419 | 5.765 | 0.009 | 0.006 | 0.006 |
| CTHRC1 | 115908 | 3.349 | 4.023 | 6.926 | 0.040 | 0.023 | 0.007 |
| CREB3L1 | 90993 | 6.916 | 5.727 | 12.541 | 0.004 | 0.005 | 0.001 |
| GLI2 | 2736 | 6.935 | 7.104 | 14.113 | 0.019 | 0.016 | 0.006 |
| IGFBP5 | 3488 | 12.733 | 16.566 | 17.230 | 0.014 | 0.009 | 0.008 |
| TWIST1 | 7291 | 13.411 | 16.061 | 20.741 | 0.001 | 0.001 | 0.001 |
| BMP4 | 652 | 10.489 | 17.221 | 21.243 | 0.020 | 0.010 | 0.007 |
| COL1A1 | 1277 | 8.195 | 19.443 | 85.847 | 0.028 | 0.009 | 0.002 |
| LRRC17 | 10234 | 3.985 | 34.428 | 103.392 | 0.016 | 0.001 | 0.000 |
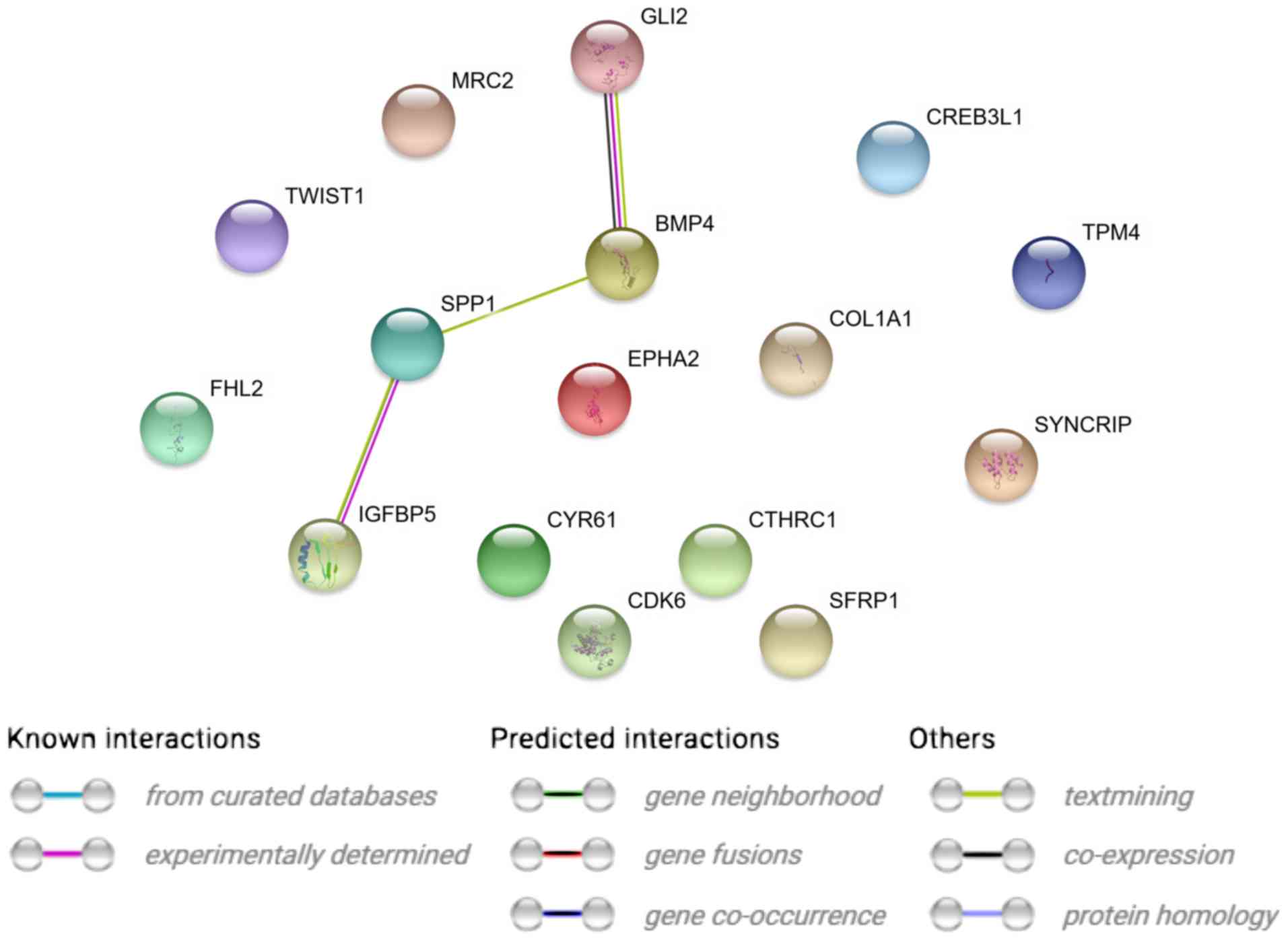
STRING interaction network was generated among
differentially expressed genes belonging to each of selected GO BP
terms. Using such a prediction method provided us with a molecular
interaction network formed between protein products of studied
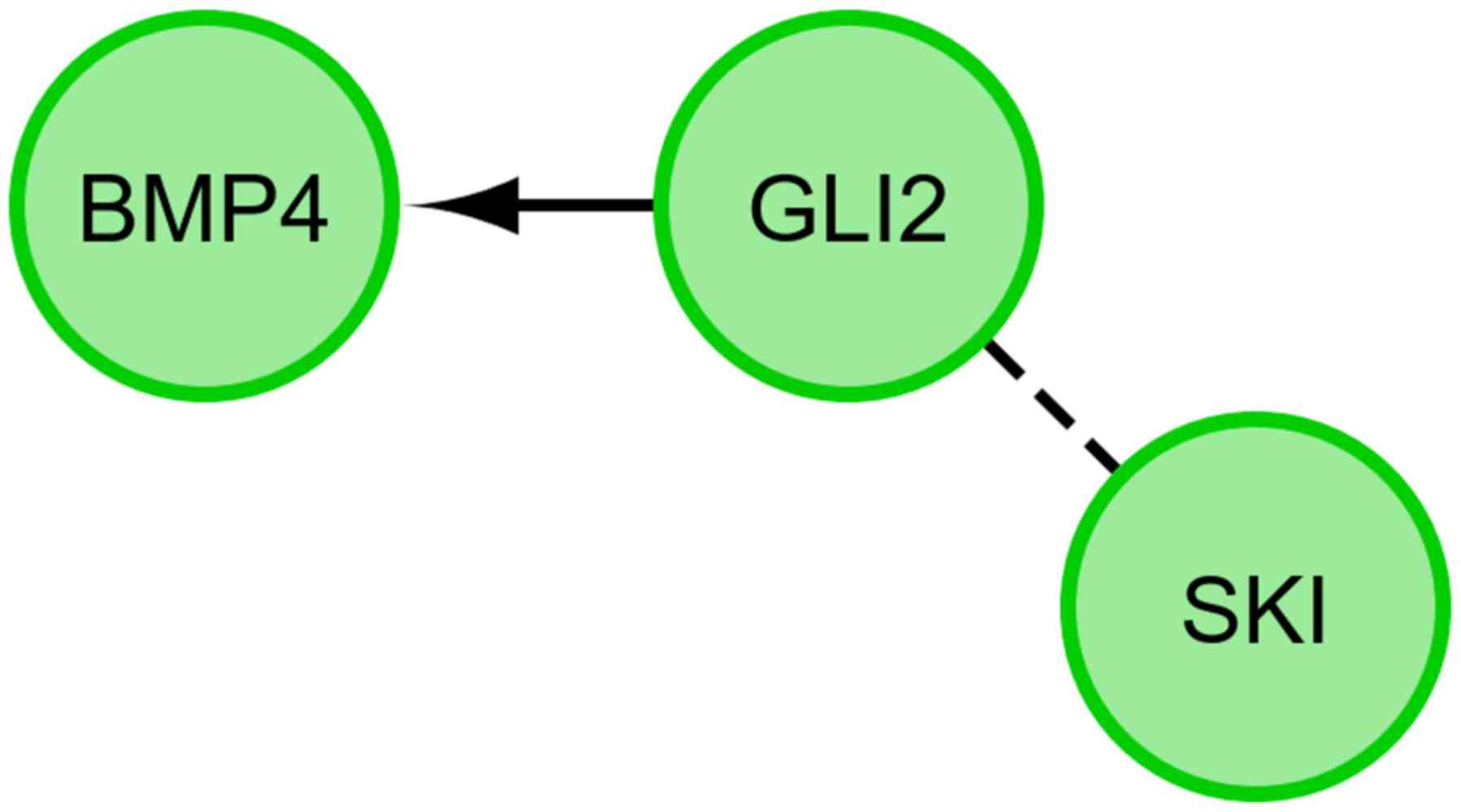
genes (Fig. 2). Finally, we
investigated the functional interactions between chosen genes with
REACTOME FIViz app to Cytoscape 3.6.0 software. The results were
shown in Fig. 3.
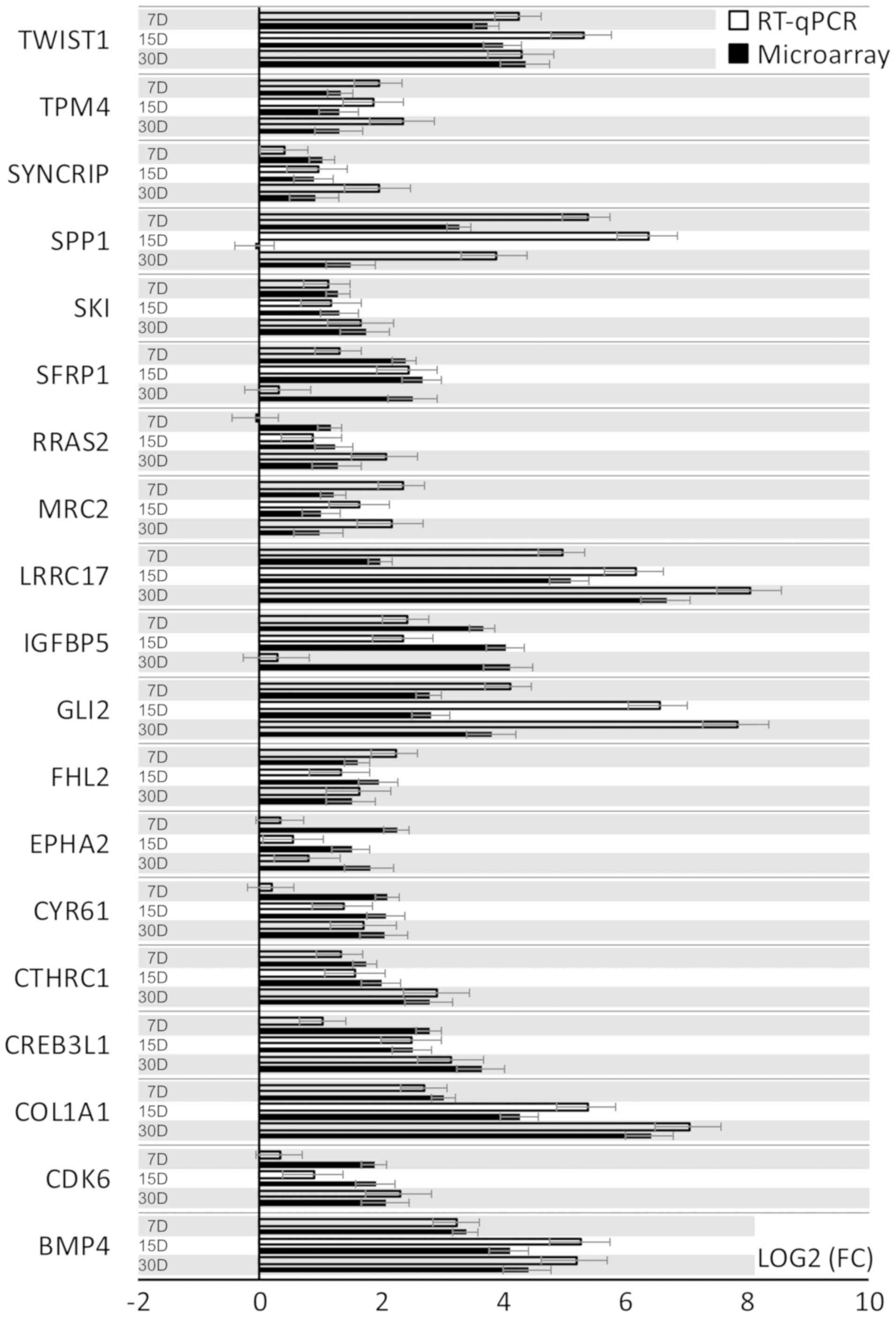
RT-qPCR was conducted to validate the results
obtained during microarray analysis. The outcomes were presented
and compared in a form of a bar graph (Fig. 4).
As can be seen, the direction of changes in
expression was confirmed in most examples. Nevertheless, the
microarray approach, used to analyze the full transcriptome of the
cells, is largely qualitative, which can be observed as validation
of the results with quantitative RT-qPCR in two examples gives
variable results. For SPP1 transcript expression level, microarray
results indicate downregulation in 15 days of cell culture, while
RT-qPCR results give a different direction of changes. A similar
situation may be observed for RRAS1 mRNA levels. This might be due
to the fact that the microarrays account for multiple available
exons forming many variants of the expressed gene, which is not
usually the case with RT-qPCR, as it probes for a specific gene
sequence. Likewise, the scale of differences in transcript levels
varied between both of the methods analysed.
Discussion
The process of MSC differentiation towards
osteoblasts is regulated by a number of transcription factors, the
list of which is still incomplete and systematically supplemented
and updated (35,36). During the development of the
skeleton, MSCs may give rise to two cell lines: i) Osteoblasts and
ii) chondroblasts. The dual development potential of MSC is made
possible due to expression of two basic bone formation regulators:
Runx-2 (CBFA-core binding factor Ralpha/osteoblast-specific factor
2-OSF-2) and SOX-9. The further way of differentiation (towards
osteoblasts or chondroblasts) is decided by the inclusion of
further transcription factors. Osterix (OSX), and Runx-2 are some
of the factors that facilitate the osteogenesis process (37,38).
The increase in expression of OSX factor influences the increase of
expression and secretion of osteoblast-specific proteins-collagen
type I, osteopontin, sialoprotein and alkaline phosphatase
(39). Other important factors
regulating osteoblastogenesis are involved in the Wnt/β-catenin
signalling pathway. Activation of Wnt pathway proteins stimulates
osteoblast proliferation (36,38).
Bone morphogenetic proteins (BMPs) are yet another important group
of factors regulating the process of osteoblastogenesis. These
proteins have a stimulating effect on osteoblast activity through
BMP serine-threonine kinase receptors. Many other factors also
contribute to the stimulation of osteoblast activity such as
parathormone (PTH)-acting via insulin-like growth factor (IGF),
dexamethasone, lectin etc. (40–43).
As mentioned above, the literature provides a number
of factors that influence the differentiation of MSCs towards
osteoblasts in vitro. Many authors indicate that the source
of these stem cells is also the ovary. Kossowska-Tomaszczuk et
al (21) were the first to
suggest that GCs have the potential of stem cells (mesenchymal stem
cells), due to the expression of markers characteristic for this
type of cells. It was the first to prove that GCs can differentiate
into osteoblasts in long-term in vitro culture under the
influence of a suitable differentiating medium. GCs thus shed new
properties and could be successfully used as a starting material
for obtaining stable populations of osteoblasts used in
regenerative medicine of skeletal-related disorders (21,44,45).
The results of the presented studies confirm the possibility of
differentiating GCs towards osteoblasts. We can observe that GCs is
subject to such differentiation without differentiating factors
constituting the supplement of the culture medium. We can,
therefore, conclude that GCs undergoes a number of changes in gene
expression during long-term in vitro culture, the effect of
which is their entry into the pathway of osteoblast
differentiation.
The presented studies also indicate that GCs may
differentiate towards osteoblasts under long-term in vitro
culture conditions. Moreover, it was shown that GCs express genes
characteristic for this process, which can be considered genetic
markers of differentiation of GCs towards osteoblasts. GCs in the
presented studies were cultivated without the addition of
supplements considered necessary for the process of cell
differentiation towards osteoblasts. The basal medium did not
contain any supplements such as dexamethasone, BMP-2, vitamin D3,
ascorbic acid, β-glycerophosphate, valproic acid, which are
considered to be key factors osteoblast differentiation (15,19,46).
The results of the presented research on the potential of GCs
differentiation towards osteoblasts are confirmed in the literature
of recent years (21,23,44).
The ‘osteoblast differentiation’ ontological group
defines a group of genes responsible for the biological process
during which the differentiation of less specialized cells towards
osteoblasts occurs. The included heat map presents a set of genes
that are characteristic of the process of GCs differentiation
towards osteoblasts.
As can be seen in the attached table, during the
30-day in vitro culture, the expression of 19 genes is
changed, with SYNCRIP, MRC2, RRAS2 and TPM4
exhibiting the lowest expression, and LRRC17, COL1A1, BMP4,
TWIST1, IGFBP5, GLI2, CTHRC1 showing the highest expression.
This report is focused on the genes of the highest expression and
their mutual relation.
Analyzing the relationships and interactions between
the 19 genes of interest, we can only observe some between two
pairs of genes (BMP4 and GLI2 and SPP1 and
IGFBP5).
The highest expression from all genes has been
demonstrated by LRRC17 (Leucine-rich repeat containing
17). It is not only one of the genes that regulate the
osteoblastogenesis process, but also a gene that is expressed by
the ovary. The above result, on one hand, can confirm that the
obtained cells are cells derived from the ovary, but on the other
hand can be a factor regulating the process of differentiation of
GCs towards osteoblasts (47,48).
It is a gene that is highly expressed in osteoblasts under
physiological conditions (47).
The high expression change of this gene during long-term in
vitro GC culture indicates that these cells can spontaneously
gain osteoblast properties.
Another very important gene demonstrating the
differentiation of a given population towards osteoblasts is the
expression of the collagen type I gene and protein. The
expression of collagen type I is significantly increased after 30
days of GC in vitro culture. We can, therefore, suppose that
the process of GCs differentiation towards osteoblasts is also
regulated by typical factors of MSC osteoblastogenesis. COL1A1
(Collagen type I alpha 1 chain) is a gene providing inductions
for the synthesis of a large molecular molecule called collagen
type I. Collagens are a family of proteins that are part of most
organs in the human body: They build cartilage, tendons, skin,
sclera, and above all bones. Collagen molecules form long fibrils,
connected by transverse bonds between them in intercellular spaces.
Such structure and interactions between collagen fibres are called
cross-linking, which results in the formation of very strong type 1
collagen fibres (49). The
expression of the COL1A1 gene indicates that GCs have the
potential to differentiate towards the bone tissue. Under
physiological conditions, the COL1A1 gene is not expressed
in GCs. Only the presence of collagen type I in the theca cells of
the ovarian follicle has been proven (50,51).
However, these are not the subject of the research presented. Only
GCs building an internal layer of ovarian follicle were used in the
study.
Osteogenesis is the process leading to the formation
of bone tissue. It begins during the formation of the embryo and
continues throughout the entire life of the organism by maintaining
a balance between bone formation and resorption (52). Bone morphogenetic proteins belong
to the superfamily of TGF-βs (Transforming growth factors
β). The family of these proteins is responsible for the formation
of bone and cartilage in vivo but also fulfils important
roles in the female reproductive system (53,54).
One of the strongest inductors in bone formation through osteoblast
differentiation stimulation is BMP-4. Under physiological
conditions, BMP-4 transduced signals through heterodimer formation
of Type II and Type I cognate complex (BMPRIA and BMPRIB) through
serine/threonine receptors. This leads to phosphorylation of Sma
and Mad proteins playing an important role in the differentiation
of cells derived from the mesenchymal line (55–57).
The group of BMP proteins also plays a role in the regulation of
follicular development and has an effect on GCs proliferation and
steroidogenesis. Tanwar and Mcfarlane (58) indicate, in their mice studies, that
BMP-4 is expressed in the ovary, uterus and oviduct epithelium. It
has also been shown that BMP-4 acts as a paracrine/autocrine
modulator of steroidogenesis of GCs cells. BMP-4 together with
BMP-6, BMP-7 stimulates Smad-1 accumulation and release of
estradiol (E2) stimulated by IGF (59,60).
Other studies have shown that another types of BMP, above all
BMP-15, are necessary for the proper functioning of the
reproductive system of the female. It is BMP-15 that affects the
proliferation and differentiation of GCs into individual layers
within the follicle (61). The
presented studies do not clearly indicate whether the expression of
BMP-4 is associated with the the process of steroidogenesis
occurring in vitro culture or whether this expression is
associated with osteogenesis. In a broad sense, we can conclude
that the presence of other genes involved in the BMP-4 osteogenesis
process could confirm its occurrence in the culture. This
conclusion is supported by the expression of the previously
mentioned SPP1 gene (secreted phosphoprotein; osteopontin)
(62). SPP1 is a gene responsible
for the proper bone mineralization in the process of osteogenesis.
Kim et al (63) suggested
that SPP1 could be a marker of ovarian cancer. Under physiological
conditions, the expression of SPP1 increases in the antral
follicles (64) suggesting that
SPP1 does not indicate a differentiation of GCs towards
osteoblasts. Kulterer et al (65) proved that during the
differentiation of MSCs towards osteoblasts, SPP1 and COL1A1 are
expressed (66). Our research
confirms this scheme; however, it should be emphasized that in the
presented results P-values were <0.05 only after 7 days of
culture, bringing their actual significance into question. In
addition, another gene supporting the GCs' tendency for osteogenic
differentiation in long-term in vitro is the GLI2
(GLI-Kruppel Family Member 2). This gene belongs to the
family of zinc finger proteins. Under physiological conditions, the
GLI family proteins play an important role in embryonic
development. Abnormal operation of these genes causes development
defects, eg: Mutations in the GLI2 gene cause defects in the
development of the skeleton. In addition, GLI2 has been shown to
play a large role in regulating BMP-2 protein expression during
osteoblast differentiation (67).
The presented research results confirm the relationship between the
expression of GLI2 and BMP-4. During the differentiation of GCs
towards osteoblasts in long-term in vitro culture, GLI2 may
interact with BMP-4 in a positive manner (Figs. 2 and 3).
IGFBP-5 (Insulin-like growth factor-binding
protein 5) is another factor involved in the process of
osteoblast formation. This substance is detected during
osteoblastogenesis. It is most intensively released during the
first days of in vitro culture before mature osteoblasts
arise. It is known that the amount of this factor decreases during
long-term in vitro culture (68). In addition, IGFBP-5 produced by
osteoblasts stimulates osteoclastogenesis and thus acts as an
osteoblast-osteoclast coupling agent (69). Liu and Ling (70) proved that the rat ovary produces 5
types of IGFBP, including IGFBP-5 in GCs of atretic preantral
follicles. In addition, the presented study results suggest that
IGFBP5 interacts with the SPP1 gene described above (Fig. 2).
According to the results presented, growth factors
such as BMP-4 or IGFBP5 interact with genes that, in addition to
their role in the ovary, also play key roles in osteogenesis.
Therefore, these factors may be potential regulators of the
expression of the above genes, as well as the process of GC
differentiation towards osteoblasts. As already mentioned, these
genes play a key role in the process of osteoblast differentiation
from mesenchymal stem cells. The results of the presented studies
therefore suggest one of the probable ways of osteo-differentiation
of GCs.
Another gene that is expressed by GCs after 30 days
of in vitro culture is TWIST (Transcription Factor
TWIST). Under physiological conditions, this gene is expressed
in cells that express RUNX2 expression. As mentioned earlier, the
RUNX2 gene is involved in the process of osteoblast formation. The
basic function of TWIST is the inhibition of the RUNX2 function
during skeletogenesis (71). The
TWIST expression can, therefore, explain the lack of RUNX2
expression in the presented GCs after 30 days of culture.
Osteoblast differentiation from stem cells is
possible thanks to CTHRC1. Wang et al (72) indicated that this gene, under
physiological conditions, is involved in bone remodelling, and also
shows the presence in osteocytes, bone matrix and periodontal
ligament cells in rat. In presented research CTHRC1 has been
activated during long-term in vitro culture of GCs.
In addition to gene expression changes observed
during the course of the long-term in vitro culture, we have
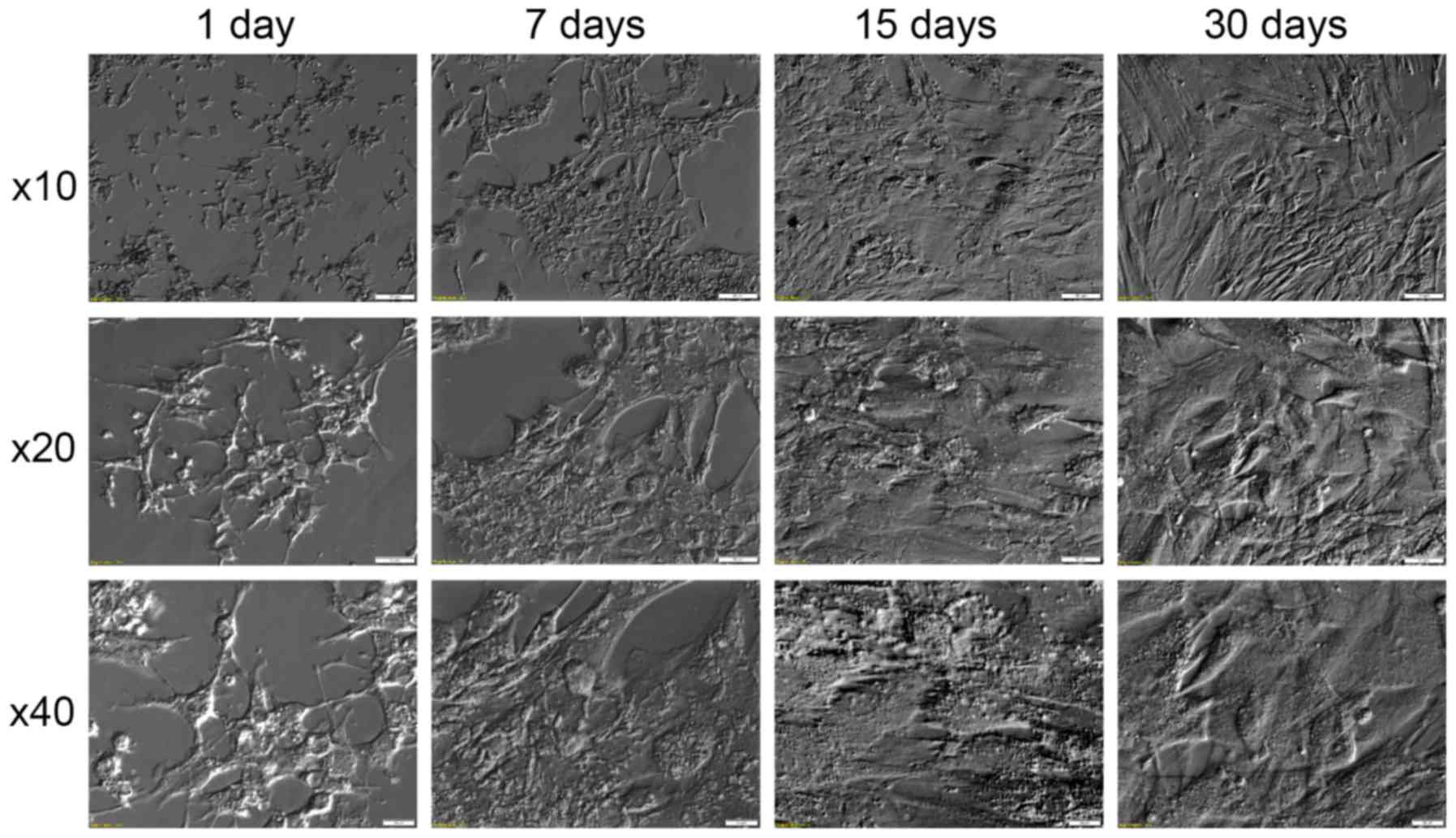
also observed significant changes of morphology. These changes,
documented in Fig. 5, show that in
the first days of culture the cells assume star-like shape,
followed by their elongation and transition into fibroblast like
shape in the later stages of culture. Similar results can be found
in literature (22,44). This fact can be associated with the
assumed loss of granulosa specific gene expression, caused by the
absence of the physiological extracellular environment, and
assumption of new, culture-specific phenotype. This process is also
accompanied by upregulation of expression of non-granulosa specific
growth factors (such as BMP4 and IGFBP5), which may further explain
the assumption of new morphology.
Summing up the conducted research, it can be
concluded that there is a lot of evidence for the possibility of GC
differentiation towards osteoblasts. Obtaining stable cultures of
differentiated osteoblasts derived from GCs may find wide
application in the treatment of skeletal disorders. However, it
needs to be noted that this is an entry level transcriptomic study,
which only accounts for the gene changes observed in the granulosa
cell culture and refers them to the available literature. Despite
that, the presented research is an excellent “signpost” for
further, more detailed studies, possibly including comparisons to
osteoblasts cultured in similar conditions, or based on proteomic
approaches (which are much better translatable to in vivo
knowledge). Detailed analysis of pathways involved in the
differentiation of GCs towards osteoblasts is required.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from
Poznan University of Medical Sciences (grant no.
502-14-02227367-10694) and Polish National Science Centre (grant
no. 2018/31/B/NZ5/02475).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MBrą provided resources (provision of study
materials and patients), designed the experiments and methodology,
and wrote the original draft of the manuscript. WK performed the
investigation (conducted the research and investigation process,
performed the experiments, or data/evidence collection), developed
the methodology, and wrote the original draft. PC developed the
software, created and presented the published work, wrote the
initial draft, and performed the formal analysis and visualization.
KO conducted the experiments, acquired data and wrote the
manuscript. JBT conducted the experiments, acquired data and wrote
the manuscript. MJ designed the methodology and created the models.
LP designed the study and revised the medical methodology. MBru
contributed to medical procedure design and approved the final
draft of the manuscript. MN supervised the study, designed the
experiments and provided editorial supervision. MZ revised the
methodology, analyzed data and approved the final draft of the
manuscript. BK conceived the study, contributed to project
administration, acted as the senior author and provided major
assistance during the study. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
This study has been approved with resolution 558/17
by Poznan University of Medical Sciences Bioethical Committee. All
participants gave their written informed consent for use of their
material in research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang H, Vollmer M, De Geyter M,
Litzistorf Y, Ladewig A, Dürrenberger M, Guggenheim R, Miny P,
Holzgreve W and De Geyter C: Characterization of an immortalized
human granulosa cell line (COV434). Mol Hum Reprod. 6:146–153.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brůcková L, Soukup T, Moos J, Moosová M,
Pavelková J, Rezábek K, Vísek B and Mokrý J: The cultivation of
human granulosa cells. Acta Medica (Hradec Kralove). 51:165–172.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rybska M, Knap S, Jankowski M, Jeseta M,
Bukowska D, Antosik P, Nowicki M, Zabel M, Kempisty B and Jaśkowski
JM: Characteristic of factors influencing the proper course of
folliculogenesis in mammals. Med J Cell Biol. 6:33–38. 2018.
View Article : Google Scholar
|
|
4
|
Kranc W, Jankowski M, Budna J, Celichowski
P, Khozmi R, Bryja A, Borys S, Dyszkiewicz-Konwińska M, Jeseta M,
Magas M, et al: Amino acids metabolism and degradation is regulated
during porcine oviductal epithelial cells (OECs) primary culture in
vitro-signaling pathway activation approach. Med J Cell Biol.
6:18–26. 2018. View Article : Google Scholar
|
|
5
|
Rybska M, Knap S, Jankowski M, Jeseta M,
Bukowska D, Antosik P, Nowicki M, Zabel M, Kempisty B and Jaśkowski
JM: Cytoplasmic and nuclear maturation of oocytes in mammals-living
in the shadow of cells developmental capability. Med J Cell Biol.
1:13–17. 2018. View Article : Google Scholar
|
|
6
|
Kempisty B, Ziółkowska A, Piotrowska H,
Ciesiółka S, Antosik P, Bukowska D, Zawierucha P, Woźna M,
Jaśkowski JM, Brüssow KP, et al: Short-term cultivation of porcine
cumulus cells influences the cyclin-dependent kinase 4 (Cdk4) and
connexin 43 (Cx43) protein expression-a real-time cell
proliferation approach. J Reprod Dev. 59:339–345. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ciesiółka S, Budna J, Jopek K, Bryja A,
Kranc W, Chachuła A, Borys S, Dyszkiewicz Konwińska M, Ziółkowska
A, Antosik P, et al: Influence of estradiol-17beta on progesterone
and estrogen receptor mRNA expression in porcine follicular
granulosa cells during short-term, in vitro real-time cell
proliferation. Biomed Res Int. 2016:84310182016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Spanel-Borowski K and Sterzik K:
Ultrastructure of human preovulatory granulosa cells in follicular
fluid aspirates. Arch Gynecol. 240:137–146. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Neubourg DD, Robins A, Fishel S and Gibbon
L: Flow cytometric analysis of granulosa cells from follicular
fluid after follicular stimulation. Hum Reprod. 11:2211–2214. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ratajczak MZ and Suszyńska M: Quo vadis
regenerative medicine? Acta Haematol Pol. 44:161–170. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Weiss DJ: Concise review: Current status
of stem cells and regenerative medicine in lung biology and
diseases. Stem Cells. 32:16–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Desai TJ, Brownfield DG and Krasnow MA:
Alveolar progenitor and stem cells in lung development, renewal and
cancer. Nature. 507:190–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Matthews VB and Yeoh GC: Liver stem cells.
IUBMB Life. 57:549–553. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murtaugh LC and Kopinke D: Pancreatic Stem
CellsStemBook [Internet]. Harvard Stem Cell Institute; Cambridge,
MA: 2008, View Article : Google Scholar
|
|
15
|
Lee RH, Kim B, Choi I, Kim H, Choi HS, Suh
K, Bae YC and Jung JS: Characterization and expression analysis of
mesenchymal stem cells from human bone marrow and adipose tissue.
Cell Physiol Biochem. 14:311–324. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pereira LO, Rubini MR, Silva JR, Oliveira
DM, Silva ICR, Poças-Fonseca MJ and Azevedo RB: Comparison of stem
cell properties of cells isolated from normal and inflamed dental
pulps. Int Endod J. 45:1080–1090. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karahuseyinoglu S, Kocaefe C, Balci D,
Erdemli E and Can A: Functional structure of adipocytes
differentiated from human umbilical cord stroma-derived stem cells.
Stem Cells. 26:682–691. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Malekshah AK, Moghaddam AE and Daraka SM:
Comparison of conditioned medium and direct co-culture of human
granulosa cells on mouse embryo development. Indian J Exp Biol.
44:189–192. 2006.PubMed/NCBI
|
|
19
|
Kern S, Eichler H, Stoeve J, Klüter H and
Bieback K: Comparative analysis of mesenchymal stem cells from bone
marrow, umbilical cord blood, or adipose tissue. Stem Cells.
24:1294–1301. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rojas M, Xu J, Woods CR, Mora AL, Spears
W, Roman J and Brigham KL: Bone marrow-derived mesenchymal stem
cells in repair of the injured lung. Am J Respir Cell Mol Biol.
33:145–152. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kossowska-Tomaszczuk K, De Geyter C, De
Geyter M, Martin I, Holzgreve W, Scherberich A and Zhang H: The
multipotency of luteinizing granulosa cells collected from mature
ovarian follicles. Stem Cells. 27:210–219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brevini TA, Pennarossa G, Rahman MM,
Paffoni A, Antonini S, Ragni G, deEguileor M, Tettamanti G and
Gandolfi F: Morphological and molecular changes of human granulosa
cells exposed to 5-azacytidine and addressed toward muscular
differentiation. Stem Cell Rev Rev. 10:633–642. 2014. View Article : Google Scholar
|
|
23
|
Aghadavod E, Zarghami N, Farzadi L, Zare
M, Barzegari A, Movassaghpour AA and Nouri M: Isolation of
granulosa cells from follicular fluid; applications in biomedical
and molecular biology experiments. Adv Biomed Res. 4:2502015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kranc W, Brązert M, Budna J, Celichowski
P, Bryja A, Nawrocki MJ, Ożegowska K, Jankowski M Chermuła B,
Dyszkiewicz-Konwińska M, et al: Genes responsible for
proliferation, differentiation, and junction adhesion are
significantly up-regulated in human ovarian granulosa cells during
a long-term primary in vitro culture. Histochem Cell Biol.
151:125–143. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kranc W, Brązert M, Ożegowska K, Nawrocki
MJ, Budna J, Celichowski P, Dyszkiewicz-Konwińska M, Jankowski M,
Jeseta M, Pawelczyk L, et al: Expression profile of genes
regulating steroid biosynthesis and metabolism in human ovarian
granulosa cells-A primary culture approach. Int J Mol Sci. 18(pii):
E26732017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ferraretti AP, La Marca A, Fauser BC,
Tarlatzis B, Nargund G and Gianaroli L; ESHRE working group on Poor
Ovarian Response Definition, : ESHRE consensus on the definition of
‘poor response’ to ovarian stimulation for in vitro fertilization:
The Bologna criteria. Hum Reprod. 26:1616–1624. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kranc W, Brązert M, Ożegowska K,
Budna-Tukan J, Celichowski P, Jankowski M, Bryja A, Nawrocki MJ,
Popis M, Jeseta M and Pawelczyk L: Response to abiotic and organic
substances stimulation belongs to ontologic groups significantly
up-regulated in porcine immature oocytes. Med J Cell Biol.
6:91–100. 2018. View Article : Google Scholar
|
|
29
|
Bryja A, Dyszkiewicz-Konwińska M,
Jankowski M, Celichowski P, Stefańska K, Chamier-Gliszczyńska A,
Popis M, Mehr K, Bukowska D, Antosik P, et al: Ion homeostasis and
transport are regulated by genes differentially expressed in
porcine buccal pouch mucosal cells during long-term culture in
vitro-a microarray approach. Med J Cell Biol. 6:75–82. 2018.
View Article : Google Scholar
|
|
30
|
Chamier-Gliszczyńska A, Brązert M,
Sujka-Kordowska P, Popis M, Ożegowska K, Stefańska K, Kocherova I,
Celichowski P, Kulus M, Bukowska D, et al: Genes involved in
angiogenesis and circulatory system development are differentially
expressed in porcine epithelial oviductal cells during long-term
primary in vitro culture-a transcriptomic study. Med J Cell Biol.
6:163–173. 2018. View Article : Google Scholar
|
|
31
|
Stefańska K, Chamier-Gliszczyńska A,
Jankowski M, Celichowski P, Kulus M, Rojewska M, Antosik P,
Bukowska D, Bruska M, Nowicki M, et al: Epithelium morphogenesis
and oviduct development are regulated by significant increase of
expression of genes after long-term in vitro primary culture - a
microarray assays. Med J Cell Biol. 6:195–204. 2018. View Article : Google Scholar
|
|
32
|
Nawrocki MJ, Celichowski P, Jankowski M,
Kranc W, Bryja A, Borys-Wójcik S, Jeseta M, Antosik P, Bukowska D,
Brusk M, et al: Ontology groups representing angiogenesis and blood
vessels development are highly up-regulated during porcine
oviductal epithelial cells long-term real-time proliferation-a
primary cell culture approach. Med J Cell Biol. 6:186–194. 2018.
View Article : Google Scholar
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang DW, Sherman BT, Tan Q, Kir J, Liu D,
Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC and Lempicki RA:
DAVID Bioinformatics resources: Expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35((Web Server Issue)): W169–W175. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rahman MS, Akhtar N, Jamil HM, Banik RS
and Asaduzzaman SM: TGF-b/BMP signaling and other molecular events:
Regulation of osteoblastogenesis and bone formation. Bone Res.
3:150052015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zuo C, Huang Y, Bajis R, Sahih M, Li YP,
Dai K and Zhang X: Osteoblastogenesis regulation signals in bone
remodeling. Osteoporos Int. 23:1653–1663. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Akiyama H, Kim JE, Nakashima K, Balmes G,
Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T and
de Crombrugghe B: Osteo-chondroprogenitor cells are derived from
Sox9 expressing precursors. Proc Natl Acad Sci USA.
102:14665–14670. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bennett CN, Longo KA, Wright WS, Suva LJ,
Lane TF, Hankenson KD and MacDougald OA: Regulation of
osteoblastogenesis and bone mass by Wnt10b. Proc Natl Acad Sci USA.
102:3324–3329. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao Y, Zhou Z, de Crombrugghe B, Nakashima
K, Guan H, Duan X, Jia SF and Kleinerman ES: Osterix, a
transcription factor for osteoblast differentiation, mediates
antitumor activity in murine osteosarcoma. Cancer Res.
65:1124–1128. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sánchez-Duffhues G, Hiepen C, Knaus P and
ten Dijke P: Bone morphogenetic protein signaling in bone
homeostasis. Bone. 80:43–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yuasa M, Yamada T, Taniyama T, Masaoka T,
Xuetao W, Yoshii T, Horie M, Yasuda H, Uemura T, Okawa A and Sotome
S: Dexamethasone enhances osteogenic differentiation of bone
marrow- and muscle-derived stromal cells and augments ectopic bone
formation induced by bone morphogenetic protein-2. PLoS One.
10:e01164622015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Włodarski KH, Galus R, Brodzikowska A and
Włodarski PK: Sclerostin, an osteocytes-derived bone-forming
inhibitor. Pol Orthop Traumatol. 78:151–154. 2013.PubMed/NCBI
|
|
43
|
Włodarski K and Włodarski P: Leptin as a
modulator of osteogenesis. Ortop Traumatol Rehabil. 11:1–6.
2009.PubMed/NCBI
|
|
44
|
Kossowska-Tomaszczuk K and De Geyter C:
Cells with stem cell characteristics in somatic compartments of the
ovary. Biomed Res Int. 2013:3108592013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kossowska-Tomaszczuk K, Pelczar P, Güven
S, Kowalski J, Volpi E, De Geyter C and Scherberich A: A novel
three-dimensional culture system allows prolonged culture of
functional human granulosa cells and mimics the ovarian
environment. Tissue Eng Part A. 16:2063–2073. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chamberlain G, Fox J, Ashton B and
Middleton J: Concise Review: Mesenchymal stem cells: Their
phenotype, differentiation capacity, immunological features, and
potential for homing. Stem Cells. 25:2739–2749. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kim T, Kim K, Lee SH, So HS, Lee J, Kim N
and Choi Y: Identification of LRRc17 as a negative regulator of
receptor activator of NF-kappaB ligand (RANKL)-induced osteoclast
differentiation. J Biol Chem. 284:15308–15316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Coveney C, Boocock DJ, Rees RC, Deen S and
Ball GR: Data mining of gene arrays for biomarkers of survival in
ovarian cancer. Microarrays (Basel). 4:324–338. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Saito M and Marumo K: Collagen cross-links
as a determinant of bone quality: A possible explanation for bone
fragility in aging, osteoporosis, and diabetes mellitus. Osteoporos
Int. 21:195–214. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hatzirodos N, Hummitzsch K, Irving-Rodgers
HF, Harland ML, Morris SE and Rodgers RJ: Transcriptome profiling
of granulosa cells from bovine ovarian follicles during atresia.
BMC Genomics. 15:402014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhao Y and Luck MR: Gene expression and
protein distribution of collagen, fibronectin and laminin in bovine
follicles and corpora lutea. J Reprod Fertil. 104:115–123. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Luyten FP, Cunningham NS, Ma S,
Muthukumaran N, Hammonds RG, Nevins WB, Woods WI and Reddi AH:
Purification and partial amino acid sequence of osteogenin, a
protein initiating bone differentiation. J Biol Chem.
264:13377–13380. 1989.PubMed/NCBI
|
|
53
|
Bandyopadhyay A, Tsuji K, Cox K, Harfe BD,
Rosen V and Tabin CJ: Genetic analysis of the roles of BMP2, BMP4,
and BMP7 in limb patterning and skeletogenesis. PLoS Genet.
2:e2162006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bayne RA, Donnachie DJ, Kinnell HL, Childs
AJ and Anderson RA: BMP signalling in human fetal ovary somatic
cells is modulated in a gene-specific fashion by GREM1 and GREM2.
Mol Hum Reprod. 22:622–633. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chang SF, Chang TK, Peng HH, Yeh YT, Lee
DY, Yeh CR, Zhou J, Cheng CK, Chang CA and Chiu JJ: BMP-4 induction
of arrest and differentiation of osteoblast-like cells via p21 CIP1
and p27 KIP1 regulation. Mol Endocrinol. 23:1827–1838. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fujii M, Takeda K, Imamura T, Aoki H,
Sampath TK, Enomoto S, Kawabata M, Kato M, Ichijo H and Miyazono K:
Roles of bone morphogenetic protein type I receptors and Smad
proteins in osteoblast and chondroblast differentiation. Mol Biol
Cell. 10:3801–3813. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yamaguchi A, Komori T and Suda T:
Regulation of osteoblast differentiation mediated by bone
morphogenetic proteins, hedgehogs, and Cbfa1. Endocr Rev.
21:393–411. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tanwar PS and Mcfarlane JR: Dynamic
expression of bone morphogenetic protein 4 in reproductive organs
of female mice. Reproduction. 142:573–579. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Glister C, Kemp CF and Knight PG: Bone
morphogenetic protein (BMP) ligands and receptors in bovine ovarian
follicle cells: Actions of BMP-4, −6 and −7 on granulosa cells and
differential modulation of Smad-1 phosphorylation by follistatin.
Reproduction. 127:239–254. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Dooley CA, Attia GR, Rainey WE, Moore DR
and Carr BR: Bone morphogenetic protein inhibits ovarian androgen
production. J Clin Endocrinol Metab. 85:3331–3337. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Moore RK, Otsuka F and Shimasaki S:
Molecular basis of bone morphogenetic protein-15 signaling in
granulosa cells. J Biol Chem. 278:304–310. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Harvey NT, Hughes JN, Lonic A, Yap C, Long
C, Rathjen PD and Rathjen J: Response to BMP4 signalling during ES
cell differentiation defines intermediates of the ectoderm lineage.
J Cell Sci. 123:1796–1804. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kim JH, Skates SJ, Uede T, Wong KK,
Schorge JO, Feltmate CM, Berkowitz RS, Cramer DW and Mok SC:
Osteopontin as a potential diagnostic biomarker for ovarian cancer.
JAMA. 287:1671–1679. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Skinner MK, Schmidt M, Savenkova MI,
Sadler-Riggleman I and Nilsson EE: Regulation of granulosa and
theca cell transcriptomes during ovarian antral follicle
development. Mol Reprod Dev. 75:1457–1472. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kulterer B, Friedl G, Jandrositz A,
Sanchez-Cabo F, Prokesch A, Paar C, Scheideler M, Windhager R,
Preisegger KH and Trajanoski Z: Gene expression profiling of human
mesenchymal stem cells derived from bone marrow during expansion
and osteoblast differentiation. BMC Genomics. 8:702007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Bilousova G, Jun DH, King KB, De Langhe S,
Chick WS, Torchia EC, Chow KS, Klemm DJ, Roop DR and Majka SM:
Osteoblasts derived from induced pluripotent stem cells form
calcified structures in scaffolds both in vitro and in vivo. Stem
Cells. 29:206–216. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhao M, Qiao M, Harris SE, Chen D, Oyajobi
BO and Mundy GR: The zinc finger transcription factor Gli2 mediates
bone morphogenetic protein 2 expression in osteoblasts in response
to hedgehog signaling. Mol Cell Biol. 26:6197–6208. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Thrailkill KM, Quarles LD, Nagase H,
Suzuki K, Serra DM and Fowlkes JL: Characterization of insulin-like
growth factor-binding protein 5-degrading proteases produced
throughout murine osteoblast differentiation. Endocrinology.
136:3527–3533. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Peruzzi B, Cappariello A, Del Fattore A,
Rucci N, De Benedetti F and Teti A: c-Src and IL-6 inhibit
osteoblast differentiation and integrate IGFBP5 signalling. Nat
Commun. 3:6302012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu XJ and Ling N: Regulation of IGFBP-4
and −5 expression in rat granulosa cells. Adv Exp Med Biol.
343:367–376. 1994. View Article : Google Scholar
|
|
71
|
Bialek P, Kern B, Yang X, Schrock M, Sosic
D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, et al: A twist code
determines the onset of osteoblast differentiation. Dev Cell.
6:423–435. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang C, Gu W, Sun B, Zhang Y, Ji Y, Xu X
and Wen Y: CTHRC1 promotes osteogenic differentiation of
periodontal ligament stem cells by regulating TAZ. J Mol Histol.
48:311–319. 2017. View Article : Google Scholar : PubMed/NCBI
|