Introduction
Lower back pain (LBP) and other clinical symptoms
occur as a result of degenerative spinal disorders, which includes
disc degeneration, facet joint degeneration, and adjacent segment
disease (1). Intervertebral disc
degeneration (IDD) and ligamentum flavum hypertrophy (LFH) are most
common degenerative spinal disorders (2,3).
Intervertebral discs (IVDs) are elastic joint tissues between
vertebral bodies that bear the load of daily activities and are
more susceptible to degeneration because of the upright posture of
humans (4). IDD is a major cause
of LBP and sciatica, with the severity of LBP depending on the
pathological grading of IDD because the pathology depresses the
normal function of IVDs (5,6). LFH
and IDD are exacerbated by increased cellular apoptosis and
senescence, and the upregulation of pro-inflammatory cytokines and
proteins, which results from the turnover of the matrix in IVDs and
the ligamentum flavum (LF) (7,8).
Furthermore, aging (especially over the age of 50) (2) and several environmental factors (such
as oxygen, mechanical stress and osmotic pressure) have been
reported to trigger the onset and progression of IDD (9). Nevertheless, the pathophysiological
mechanisms of IDD and LFH remain poorly understood.
A number of high-throughput proteomic analysis
techniques, such as isobaric tags for relative and absolute
quantification (iTRAQ), have been used to profile the proteomic map
of the annulus fibrosus (AF) and nucleus pulposus (NP; Table I). NP is a gel-like tissue which is
surrounded by AF, a layered cartilaginous structure. The present
review summarizes the results of quantitative proteomic studies of
the LF, AF, and NP, as well as body fluids, including cerebrospinal
fluid (CSF) and serum from patients with degenerative spinal
disorders. The aim of this review was to identify the crucial
proteins mediating the onset and progression of IDD and LFH, which
may facilitate the development of novel potential therapies for
these disorders.
 | Table I.Significantly differentially
expressed proteins in degenerative disc disease. |
Table I.
Significantly differentially
expressed proteins in degenerative disc disease.
| Author, year | Sample source | Increased protein
expression | Decreased protein
expression | (Refs.) |
|---|
| Kamita et
al, 2015 | Ligamentum
flavum | Chondroadherin,
cartilage intermediate layer protein, lysophosphatidic acid
receptor 1, SLRPs, prolargin, FN1, HTRA serine peptidase 1,
tenascin | Asporin | (11) |
| Johnson et
al, 2006 | Annulus
fibrosus |
Ubiquitin-associated domain-containing
protein 1, potassium voltage-gated channel subfamily D member
3 | NA | (39) |
| Battié et
al, 2008 |
| FN1, clusterin,
aggrecan, decorin, prolargin, | COL2, type XI
collagen, COL1, COL6 | (9) |
| Yee et al,
2016 |
| Guanine
nucleotide-binding protein G(i) subunit α-2, transmembrane protein
51, adenosine receptor A3, FAR1, lipid phosphate
phosphatase-related protein type 2 | Heat shock cognate
71-kDA protein, glucose-6-phosphate dehydrogenase,
protocadherin-23 | (10) |
| Battié et
al, 2008 | Nucleus
pulposus | Prolargin, FN1,
COMP, clusterin, SLRPs | COL1 | (9) |
| Honsho et
al, 2010 | Notochordal cell
conditioned medium | Laminin B1,
glycosaminoglycan, SOX9, COL2, transforming growth factor β3 | Type III
collagen | (56) |
| Elliott and Setton,
2001 | Murine
intervertebral discs | FN1, prolargin,
COMP, COL6, type XII collagen, type XV collagen, SOX9, COL2 | NA | (61) |
| Markolf and Morris,
1974 | Cerebrospinal
fluid | APO A-IV, vitamin
D-binding protein, neurofilament triplet L protein, tetranectin,
hemoglobin, immunoglobulin G | ProSAAS,
prostaglandin D2 synthase, creatine kinase B, superoxide dismutase
1, peroxiredoxin 2 | (63) |
| Yang et al,
2009 | Serum | APO L1 | apolipoprotein M,
tetranectin | (66) |
Kyoto Encyclopedia of Genes and Genomes
(KEGG) analysis
Methods
To conduct KEGG pathway analysis, 54 differentially
expressed proteins (DEPs) from LF tissue, 15 DEPs from the AF
(soluble, the supernatant of samples), 10 DEPs from AF (insoluble,
the lyophilized pellet of samples), 21 DEPs from NP (soluble), and
7 DEPs from NP (insoluble) were selected based on previous
proteomic studies (Table SI)
(10,11); patient information is provided in
Table SII. Briefly, for the LF,
the DEPs were selected according to the protein expression ratio
between lumbar spinal stenosis (LSS) and the control (individuals
with disc herniation; protein expression ratio=LSS/control, ≥2 or
≤0.5) (11). For the AF and NP,
genes which encoded DEPs that were significantly increased or
decreased in the degenerative samples compared with control samples
were selected (P<0.05) (10).
The KEGG pathway analysis database (http://www.genome.jp/kegg) was used to identify
signaling pathways enriched by genes which encoded DEPs and the
KEGG Orthology-Based Annotation System (KOBAS) version 3.0
(http://kobas.cbi.pku.edu.cn) was used to
investigate gene/protein functional annotation and gene set
enrichment (12–14). The module of KOBAS called ‘Gene
List Enrichment’, based on the first gene set enrichment method,
overrepresentation analysis, was used for KEGG pathway analysis.
Statistical analysis was performed using a hypergeometric test and
Fisher's exact test. P<0.05 was considered to indicate a
statistically significant difference.
Results
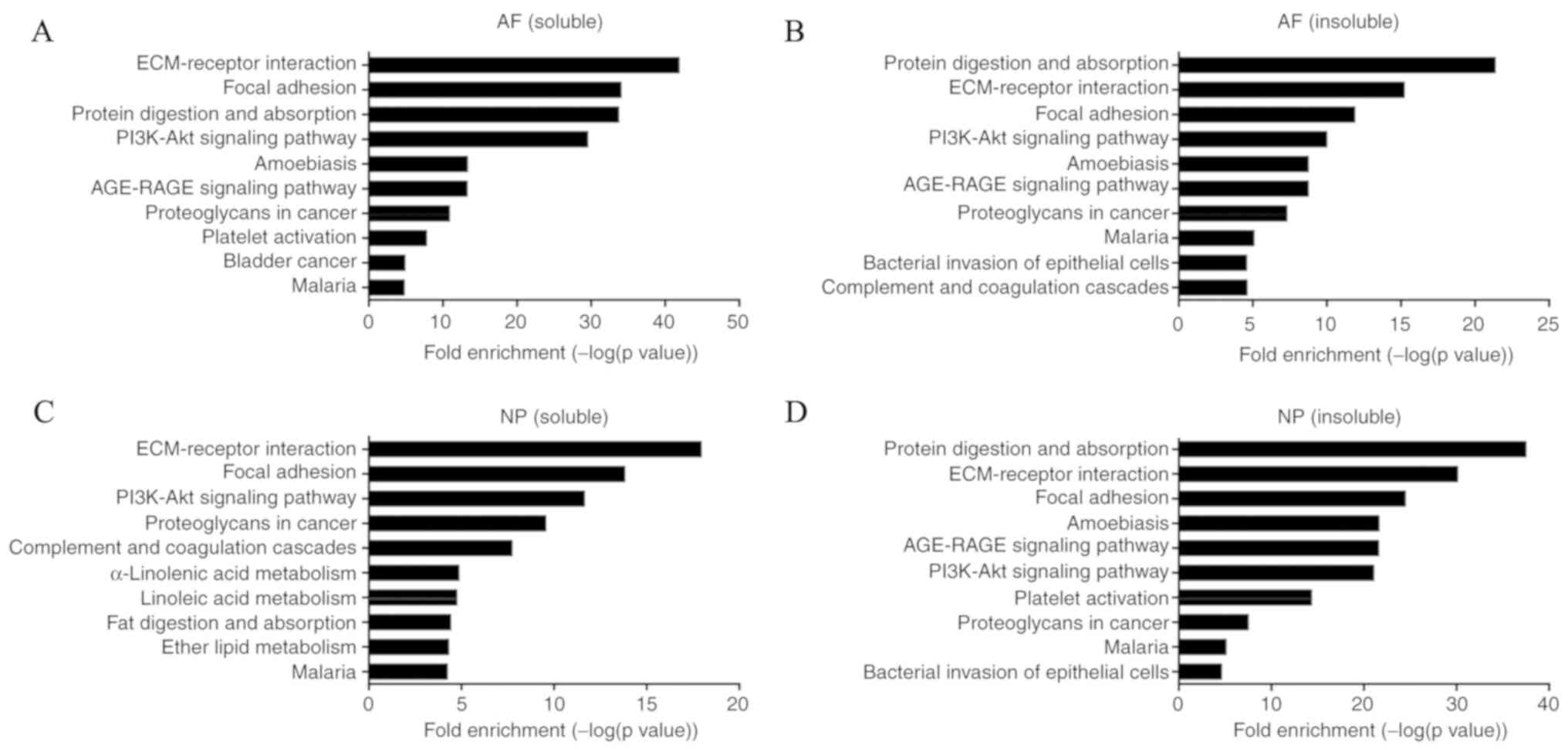
KEGG pathway analysis indicated that multiple
pathways were involved in LFH and the degeneration of AF and NP.
Some were the same in LF, AF and NP, such as ECM-receptor
interaction and focal adhesion. The total enrichment pathways are
presented in Table SIII. The top
10 KEGG pathways enriched with DEPs, including protein digestion
and absorption, ECM-receptor interaction and focal adhesion, are
presented in Figs. 1B and 2. The p53 signaling pathway, advanced
glycation endproduct-receptor for advanced glycation endproducts
signaling pathway (AGE-RAGE), PI3K/AKT signaling pathway, and
transforming growth factor (TGF)-β signaling pathway were enriched
by these DEPs (Table II). These
four signaling pathways were not in the top 10 pathways in LF, but
they were still enriched by DEPs (Table SIII). This review combines past
proteomic analyses with present analysis to provide a deeper
understanding of the molecular mechanisms of degenerative diseases
of the spine.
 | Table II.Differentially expressed proteins
enriched in the PI3K/AKT, AGE-RAGE, p53 and TGF-β signaling
pathways. |
Table II.
Differentially expressed proteins
enriched in the PI3K/AKT, AGE-RAGE, p53 and TGF-β signaling
pathways.
| Tissue | Enriched signaling
pathway | Increased protein
expression | Decrease protein
expression |
|---|
| LF | PI3K/AKT | Type VI collagen,
FN1, COMP, chondroadherin | COL1A1, COL1A2 |
|
| AGE-RAGE | FN1 | COL1A1, COL1A2 |
|
| p53 | Insulin-like growth
factor-binding protein | NA |
| AF (soluble) | PI3K-AKT | THBS1, FN1 | COL1A2, COL2A1,
COL1A1, α-2 type VI collagen, α-3 type VI collagen |
|
| AGE-RAGE | FN1 | COL1A2, COL1A1 |
|
| p53 | THBS1 | NA |
|
| TGF-β | THBS1 | NA |
| AF (insoluble) | PI3K/AKT | COMP, FN1 | COL2A1 |
|
| AGE-RAGE | Α-1 type III
collagen, FN1 | NA |
|
| TGF-β | DCN | NA |
| NP (soluble) | PI3K/AKT | COMP, FN1 | Chondroadherin,
COL2A1 |
|
| AGE-RAGE | FN1 | NA |
|
| TGF-β | DCN | NA |
| NP (insoluble) | PI3K/AKT | COL1A2, FN1, COMP,
COL1A1 | COL2A1 |
|
| AGE-RAGE | COL1A2, FN1,
COL1A1 | COL2A1 |
Proteomic analysis of the human LF
Structural proteomic analysis of the
LF
Hypertrophy and ossification of the LF are major
causes of LSS (15,16). However, the proteins involved in
hypertrophy of the LF remain unknown. To clarify the molecular
events during LSS disease progression and to identify targets for
treatment, LF proteomic analysis was employed using 2-dimensional
image converted analysis of liquid chromatography (2DICAL)-based
label-free proteomics. A set of small leucine-rich proteoglycans
(SLRPs), including asporin, decorin, and fibromodulin were
identified, in addition to the large proteoglycans (PGs), versican
and aggrecan (11). These protein
components suggest that the LF structure shares common features
with other elastic tissues and that within normal physiology the LF
is more elastic compared to other ligaments and tendons (17). This is demonstrated through the
increased presence of fibulins (FBLN 1/2/3/5), elastin, and
microfibril-associated protein 4 (10). Redox proteins present in the LF,
including lysyl oxidase homolog 1 and superoxide dismutase (SOD) 3,
are considered to be involved in the formation and regulation of
these elastic fibers (18,19). The proteins identified in the human
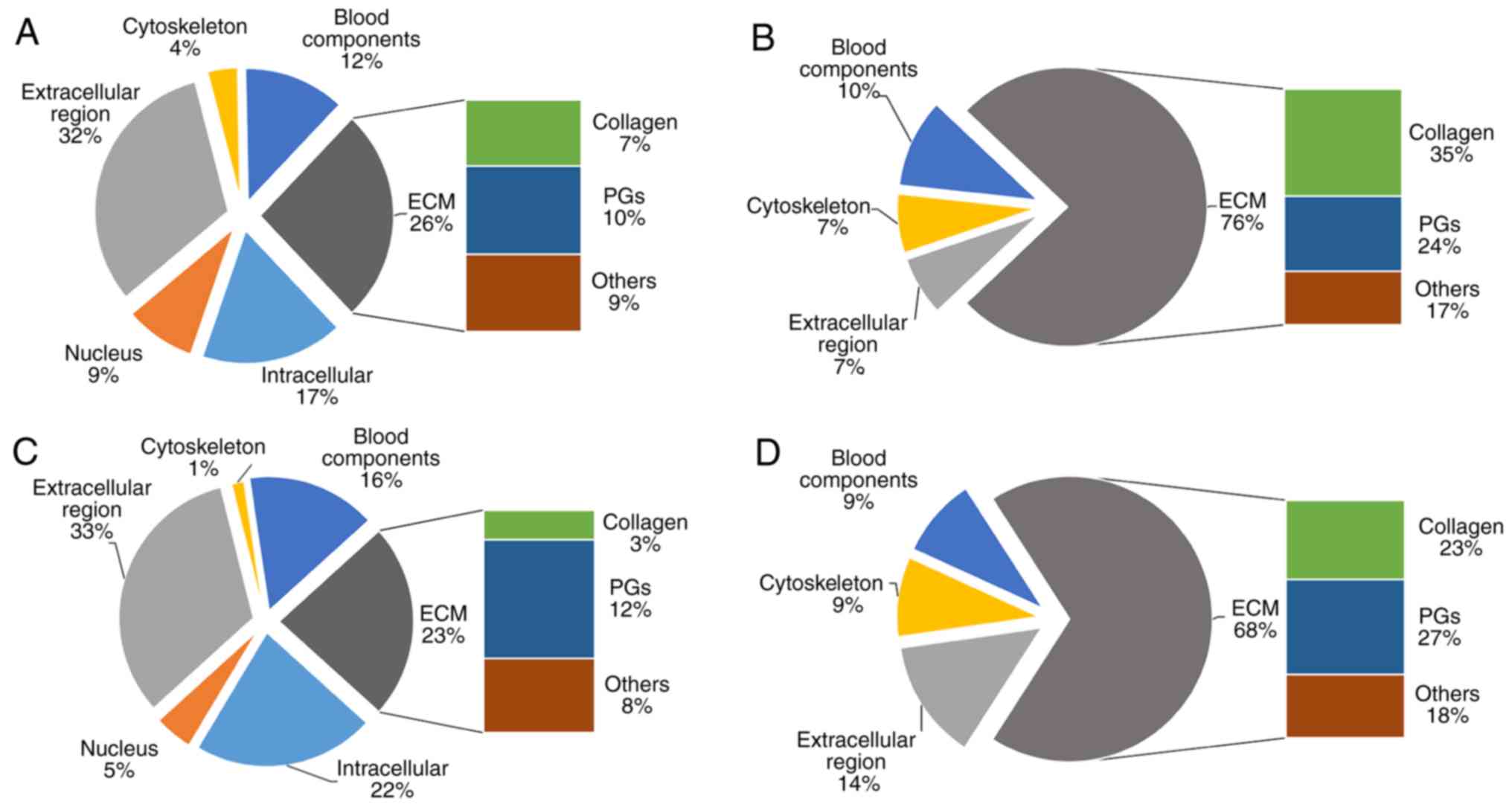
LF were classified into 24 cellular components by gene ontology
(GO) term enrichment analysis (Fig.
1A) (11).
Comparative proteomic analysis of the
LF
A number of proteins were identified through
Selective Reaction Monitoring/Multi Reaction Monitoring in LFH
samples (20) (Table I). Notably, vasculature is thought
to be implicated in LFH owing to the significantly upregulated
expression of plasma proteins, including fibrinogen,
apolipoproteins (APOs), and transthyretin. Chondrometaplasia is
also observed in degenerated LF (20); with multiple proteins involved in
chondrometaplasia consistently upregulated in LFH, including
chondroadherin (CHAD), prolargin, cartilage intermediate layer
proteins, and aggrecan (21).
These proteins have been reported to be associated with the
ossification of the LF (21).
CHAD is a leucine-rich repeat (LRR) protein that is
highly expressed in cartilaginous tissues (22). LRR proteins are involved in
promoting interactions with other extracellular matrix (ECM)
molecules and collagen fibrillogenesis (23), and are often regulated by TGF-β
(24). In addition, prolargin and
SLRPs isolated from mice lacking decorin are reportedly involved in
collagen fiber assembly and fibril abnormalities (25). Thus, the interactions between
molecules in the ECM and collagen fibrillogenesis may directly
influence the structure of the LF, and may be involved in the
process of degeneration.
In vitro studies have demonstrated that
cartilage intermediate layer protein (CILP) modulates TGF-β
signaling (26), and TGF-β has
been detected in the early stages of degenerative hypertrophy of
the LF (27). Furthermore, the
expression of lysophosphatidic acid (LPA), and its receptor, LPA
receptor 1 (LPAR1), are significantly upregulated in samples
isolated from LFH specimens (11).
Previous studies from Japan and Finland have reported that LPA is
closely related to the process of IDD (26,28),
and that upon LPA interacting with LPAR1, the protein can promote
LF cell proliferation and further induce LFH, through the LPAR1/AKT
signaling pathway (29).
The expression levels of fibronectin 1 (FN1),
tenascin, and serine protease HTRA1 (HTRA1) are positively
correlated with LFH, whereas asporin expression is negatively
correlated in LFH (11). The level
of peptides derived from FN1 is influenced by HTRA1, and the HTRA1
mutation causes diseases such as cerebral autosomal recessive
arteriopathy and leukoencephalopathy (30). In addition, HTRA1 upregulation is
observed in many degenerative disorders, including age-related
macular degeneration, osteoarthritis (OA), and lumbar disc
degeneration (31–33), with a previous study reporting that
FNS is regulated by HTRA1 in joints affected by OA (34).
Proteomic analysis of the human AF
Structural proteomic analysis of the
AF
The spine resists multidirectional loading from the
radial, axial, and circumferential directions, and the upright
posture of humans imposes greater mechanical loading and
accelerates the process of IDD (35). IDD pathological features are
accompanied by NP fibrosis, AF fissuring, and protein structure
disorganization (36). Type II
collagen (COL2), chondroitin sulfate, and PGs are produced by AF
cells (37), and in healthy IVDs,
the AF contains 65–70% water; with the dry weight composed of 20%
PGs, ~60% collagen, and 2% elastin (38). Type I collagen (COL1) extracted
from AF cells is also significantly upregulated compared to COL1
extracted from NP cells, but the level of chondroitin-6-sulfated
PGs demonstrates the opposite pattern (39). In IDD, changes in the level of PGs
can be detected. In the early stages, AF cells proliferate with
increasing biosynthetic processes, whereas in degenerated IVDs, the
level of aggrecan is decreased, and the levels of decorin,
biglycan, and fibromodulin (which are small PGs) are upregulated in
AF cells (40). Tenomodulin levels
were also reported to be increased in degenerated AF cells
(41). Other genes that are
correlated with AF cells in degenerated IVDs have been identified,
including the gene encoding pleiotrophin, which increases in the AF
with age (42); increased
vascularization in the degenerated AF tissues may also be present
(43). The proteome of a normal
IVD from a 35-year old patient (male) has been established
(10), and the cellular components
from the AF (soluble and insoluble) determined by GO term
enrichment analysis are presented in Fig. 3A and B. Compared with the AF
(soluble), ECM proteins are present at a higher proportion in the
AF (insoluble), and nuclear proteins only exist in the AF
(soluble).
Comparative proteomic analysis of the
AF
A previous study has identified a total of 759
proteins in non-degenerative AF tissue (44). DEPs of importance in the
degenerated AF include the ubiquitin-associated domain-containing
protein 1 (UBAC1), aspartyl transfer RNA synthetase, potassium
voltage-gated channel subfamily D member 3 (KCND3), structural
proteins, and signaling factors such as Indian hedgehog protein
(44) (Table I). UBAC1 serves a prominent role in
lysosomal and proteasomal degeneration (45,46),
and KCND3, a voltage-activated A-type potassium ion channel, is
involved in degenerative diseases such as spinocerebellar ataxia
(47).
Semi-quantitative analysis of silver-stained 2D
electrophoresis gels of AF cells isolated from normal and
degenerated IVDs has demonstrated that the expression levels of
glucose-6-phosphate 1-dehydrogenase (G6PD), heat shock cognate
71-kDa protein (HSPA8), and protocadherin-23 are decreased, whereas
SOD, transmembrane protein 51 (TMEM51), guanine nucleotide-binding
protein G(i) subunit α-2 (GNAI2), 26S protease regulatory subunit
8, adenosine A3 receptor (ADORA3), fatty acyl-CoA reductase 1
(FAR1), and lipid phosphate phosphatase-related protein type 2
(LPPR2) expression levels are increased (48).
HSPA8 is a heat shock cognate protein that represses
pre-mRNA splicing and forms an essential part of the spliceosome,
where it is thought to be involved in spliceosome assembly
(49). Thus, the low expression of
HSAP8 suggests that the reduced repression of pre-mRNA splicing is
a potential regulatory mechanism in the degenerated AF. G6PD is a
member of the dehydrogenase family that provides pentose phosphates
and serves as a reductant in fatty acid and nucleic acid synthesis
(50). A significant decrease in
G6PD expression in the AF suggests an important role for oxidative
stress in the process of degeneration. These findings suggest that
oxidative stress may be a major contributor to the process of
degeneration (51). The main
function of protocadherin-23 is to provide adhesion and it is
involved in morphogenesis during development (52). However, the correlation between
protocadherin-23 and IDD remains to be elucidated. GNAI2 may serve
as a signal transducer or modulator in multiple types of
transmembrane signaling systems, as the α-subunit GTP activating
protein, GNAI2 is a regulator for the effector interaction
(53). ADORA3 is an adenosine
receptor and serve a role in duplication (54). Both GNAI2 and ADORA3 are associated
with G-proteins (55), thus,
G-proteins of the transmembrane signaling systems may be involved
in the process of AF degeneration. FAR1 reduces saturated fatty
acids into fatty alcohols (56).
In addition, fatty alcohols accumulated in specific cell lines
defective in plasmalogen biosynthesis (56). Thus, the upregulation of FAR1
suggests that plasmalogen biosynthesis may be involved in the AF
degenerative process. The function of LPPR2 is similar to that of
other lipid phosphate phosphatase superfamily members, which serve
roles in signal transduction and extracellular concentrations of
lipid phosphate esters (57);
TMEM51 is a member of the multipass membrane proteins (58). However, the roles of LPPR2 and
TMEM51 in AF cells are unknown, but the data suggest that these
proteins may influence the process of AF cell degeneration.
Proteomic analysis of the human NP
Structural proteomic analysis of the
NP
NP cells are commonly described as
‘chondrocyte-like’ or ‘stem cell-like’ owing to their morphology
and the cell markers that they synthesize; the main constituents of
the NP are PGs, collagen, elastin and water (38,59).
The PGs absorb the water in the NP, and account for 35–65% of the
dry weight (60), whereas COL2
fibrils form an incompact frame structure that holds the NP tissues
together. A previous study demonstrated that NP water levels
decrease with age, and a similar decrease may occur in PGs
(38), which would decrease the
size of the NP by ~50%. Although the morphology of the NP is closer
to a solid form than a fluid structure due to the dehydration
(61), the proteome of the NP
(soluble and insoluble) is similar to the AF (Fig. 3C and D). Nevertheless, PGs are more
abundant in the NP (soluble and insoluble) compared with the AF
(10).
Comparative proteomic analysis of the
NP
Similar to the LF, cartilage oligomeric matrix
protein (COMP), prolargin, FN1, and clusterin expression were
upregulated in both soluble and insoluble fractions of the NP from
IDD specimens (10) (Table I). Furthermore, the expression of
SLRPs, such as biglycan and decorin, and extracellular SOD, were
increased in the soluble fraction (10). However, Erwin et al reported
that SLRPs were intact in both chondrodystrophic canines that
developed early disc degeneration and non-chondrodystrophic animals
(62). As previously discussed,
COL2 and CHAD are closely associated with collagen fibrillogenesis
and are observed to be downregulated with age (11), thus indicating that substantial
matrix remodeling is involved in NP degeneration. The majority of
abnormal changes in proteins are similar to those observed in the
LF, except for COL1. COL1, the major component of the insoluble
fraction of the degenerative NP, was present in increased amounts
(10). It has been reported that
COL1 is capable of achieving cross-linking, mediated by enzymatic
or non-enzymatic processes (10),
which is consistent with the increasing trends in the insoluble
fraction of the degenerative NP. These data suggested that COL1 may
be a major contributor to reducing protein solubility in
degenerative discs.
The function of local or migratory cells in IVD is
not completely understood. Nevertheless, self-repair induced by
local or migratory cells has been observed in dogs with IDD induced
by enzymatic digestion (63). A
number of studies have transplanted bone marrow-derived mesenchymal
stromal cells (BM-MSCs) and stem-like or progenitor-like cells in
IDD models (64,65). The transplantation of these cells
activates a set of native, uncharacterized cells, which express
both α-1 COL2 (COL2A1) and SRY-related protein 9 (SOX9) (66), which postponed the onset of IDD in
humans and sheep (64,65). However, this evidence is not
sufficiently robust to support cellular transplantation as a
clinical therapy. In fact, another study reported that there was no
clinically significant difference between MSC and sham treatment
for IDD, regardless of allogeneic or autologous transplantation
methods (67). Therefore, the
development of progenitor cell therapies and the identification of
specific biomarkers will require a deeper understanding of
progenitor cells.
Notochordal cells (NCs), which are potential
progenitor cells, can induce the differentiation of MSCs to NP
cells by synthesizing PGs and resisting the expression and
hypertrophy of collagen fiber; this is noted through the increased
production of glycosaminoglycans (GAG), laminin B1, and type III
collagen (COL3) observed in human MSCs cultured in porcine
notochordal conditioned medium (68). Furthermore, following MSC
transplantation in animals, NCs in the native tissue promoted the
upregulation expression of SOX9, COL2, and transforming growth
factor β3 (TGF-β3), which are also detected in healthy NP (69–71).
These data suggest that laminin B1, GAG, COL3, SOX9, and TGF-β3 may
serve vital roles in the transformation of MSCs into NP cells
(72).
Comparative proteomic analysis of IDD model
mice
Previous studies have established SM/J and LG/J
mouse models of IDD. The former display cellular and matrix changes
in IVDs similar to those in degenerative human IVDs, whereas the
latter maintain abundant vacuolated NC-like cells in the NP
(72). FN1, Prolargin, and COMP
upregulated in SM/J mice, which is consistent with observations in
degenerative human IVDs (73). In
addition, the upregulation of collagen, such as α-1 type VI and
type V collagen expression, is observed in SM/J mice (73). These changes indicate ECM
enrichment in SM/J mice, with processes such as chondrogenic
differentiation and fibrillogenesis likely to be taking place.
Notably, chondrocyte markers such as SOX9 and COL2A1 are detected
at the edge of the NP region, close to the AF, which has been
observed in other mouse strains (74). Thus, chondrocyte markers may serve
an important role in the process of IVD degeneration in mice.
Clinical proteomic analysis of body fluids
from patients with degenerative spinal disorders
The majority of lumbar disk herniation (LDH) cases
are caused by IDD (75); however,
the pathophysiological mechanism of disc herniation is not fully
understood. Owing to the compression of the NP on the nerve root,
many of the DEPs in the CSF of patients with LDH are associated
with neurons and pain; for example, Lin et al reported that
a total of nine proteins were detected at high levels in the CSF of
LDH patients, including cystatin C and APO A-IV, whereas five
proteins were found to be decreased, including creatine kinase
B-type and SOD1 (Table I)
(76). In addition, APOL1, which
exists in endothelial cells and is closely related to
atherosclerotic iliac arteries, is regulated by tumor necrosis
factor-α (TNF-α), and upregulated in the serum of people suffering
from LDH (77,78). This higher expression of APOL1 may
be responsible for the degeneration, because TNF-α is involved in
the inflammation induced by LDH (79). Similarly, APOM which was
downregulated in serum of LDH patients is an immunity-associated
gene adjacent to the TNF-α and lymphotoxin genes (80). However, the explanation behind the
downregulation of APOM in the serum of LDH patients requires
further study.
Differentially regulated proteins and their
signaling pathways in degenerative spinal disorders
α-2 COL1 forms one of the chains for COL1 and the
structural disturbance of this protein is involved in bone
development (81). There is not
enough evidence to prove the link between bone development and IDD
except for the downregulation of COLA2. COL2 is a major component
of cartilage, and mutations in this protein have been reported to
contribute to type II collagenopathies (82). The expression levels of FN1, one of
the major components of the ECM that binds a large number of
molecules related to signal transduction and cell adhesion, is
reportedly increased in degenerating discs, which is related to the
change in the organizational structure (83,84).
Furthermore, COMP, which is found in the ECM as an integral part of
ligaments and tendons, is believed to serve a role in cellular
proliferation and apoptosis, in addition to regulating cell
movement and attachment (85). A
previous study demonstrated that the abnormal expression of COMP
and SOX9 is associated with cartilage degeneration in OA (86), and another study reported that
alongside CILP and HTRA1, COMP serves as a biomarker that may be
involved in the process of IDD (10).
A previous study indicated that downregulated
microRNAs (including hsa-miR-125b-1-3p and hsa-miR-1184) and
upregulated genes (including AP2 associated kinase 1 and hemoglobin
subunit β) between normal and degenerative discs are involved in
the PI3K/AKT signaling pathway (87). The activation of the PI3K/AKT
pathway serves an important role in protecting cells from harmful
physiological processes, such as cellular apoptosis, oxidative
damage, and a hypoxic microenvironment, and has been found to
protect against IDD (88).
Furthermore, it was recently demonstrated that bone morphogenetic
protein 2 can inhibit cellular apoptosis and suppress the synthesis
of matrix proteins via the PI3K/AKT signaling pathway, which
further alleviates IDD (89).
Activation of the AGE-RAGE signaling pathway in
diabetic polyneuropathy, a complication of diabetes, is commonly
observed, and increased AGE-RAGE signaling exacerbates degenerative
disorders of peripheral neurons (90). In fact, a recent study reported no
significant difference in the GAG content or histological features
between discs from non-obese diabetic mice and euglycemic
littermates, but noted increases in cellular apoptosis and matrix
aggrecan fragmentation (91). To
determine the link between diabetes and IDD, the role of the
AGE-RAGE signaling pathway in IDD requires further research.
The p53 signaling pathway is an important indicator
of cellular apoptosis, and decreased expression of wild-type
p53-induced phosphatase 1 is closely related to p53 activation and
neuronal apoptosis (92). During
periods of replicative senescence, the p53/p21/retinoblastoma
pathway is activated to alleviate telomere erosion and DNA damage
response (93,94). Furthermore, the effects of small
ubiquitin-related modifier 2 on the proliferation and senescence of
NP cells has been investigated through the mediation of the p53
signaling pathway in rat models of IDD (95) which indicated that SUMOS was a
potential target for IDD treatment.
TGF-β signaling is an extensive pathway involved in
developmental programs of cells, including proliferation,
differentiation, homeostasis, and regeneration (96). In the IVD, TGF-β is a major
regulatory cytokine that maintains cellular differentiation and
homeostasis (97). Previous
research has reported that the upregulation of TGF-β causes a
decrease in NF-κB (98). The
inhibition of NF-κB may play an important role in IDD (99). Furthermore, TGF-β-dependent AF cell
proliferation and progressive vertebral fusions due to the loss of
filamin B was involved in IDD (100). Moreover, the increased expression
of COL2 and aggrecan regulated by TGF-β1 alleviates the
degeneration of IVDs (101).
Conclusions
Main components of the matrix such as prolargin,
FN1, CILP, COMP, COL1 and COL2 are significantly changed in the
degenerative LF, AF, and NP. COMP is involved in cartilage
degeneration in OA (102), but it
has not been fully studied in IDD. Moreover, The role of AGE-RAGE
signaling pathway in IDD requires further research. Despite the
limitations of GO and KEGG pathway analysis, proteomic analysis
still provides novel targets that aid in understanding IDD
pathophysiology.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant nos. 81672215, 81572186,
81472076, 81271982 and 81401801).
Availability of data and materials
All data generated or analyzed during the present
study are included in the article.
Authors' contributions
CL conceived and designed the experiment, analyzed
the data and wrote the manuscript. MY and LL collected the data and
analyzed the data. YaZ, QZ, CH and HW performed data analysis and
provided interpretation. YQZ and HL provided technical support and
analyzed and interpreted the data. CQL and BH critically revised
the article and interpreted the data. CF and YZ were involved in
designing the experiment, analyzing the data, revising the
manuscript for important intellectual content and final approval of
the version to be published.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ADORA3
|
adenosine receptor A3
|
|
AF
|
annulus fibrosus
|
|
AGE-RAGE
|
advanced glycation
endproducts-receptor for advanced glycation endproducts
|
|
APO
|
apolipoprotein
|
|
CHAD
|
chondroadherin
|
|
COL1
|
type I collagen
|
|
COL2
|
type II collagen
|
|
COL3
|
type III collagen
|
|
COL2A1
|
α-1 type II collagen
|
|
COMP
|
cartilage oligomeric matrix
protein
|
|
CSF
|
cerebrospinal fluid
|
|
DEP
|
differentially expressed protein
|
|
FAR1
|
fatty acyl-CoA reductase 1
|
|
FN1
|
fibronectin 1
|
|
G6PD
|
glucose-6-phosphate
1-dehydrogenase
|
|
GAG
|
glycosaminoglycan
|
|
GNAI2
|
guanine nucleotide-binding protein
G(i) subunit α-2
|
|
HSPA8
|
heat shock cognate 71-kDa protein
|
|
HTRA1
|
serine protease HTRA1
|
|
IDD
|
intervertebral disc degeneration
|
|
IVD
|
intervertebral disc
|
|
KCND3
|
potassium voltage-gated channel
subfamily D member 3
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
LBP
|
lower back pain
|
|
LDH
|
lumbar disk herniation
|
|
LF
|
ligamentum flavum
|
|
LFH
|
ligamentum flavum hypertrophy
|
|
LPA
|
lysophosphatidic acid
|
|
LPPR2
|
lipid phosphate phosphatase-related
protein 2
|
|
LSS
|
lumbar spinal stenosis
|
|
MSC
|
mesenchymal stem cell
|
|
NC
|
notochordal cell
|
|
NP
|
nucleus pulposus
|
|
OA
|
osteoarthritis
|
|
PG
|
proteoglycan
|
|
SLRP
|
small leucine-rich proteoglycan
|
|
SOD
|
superoxide dismutase
|
|
SOX9
|
SRY-related protein 9
|
|
TGF-β
|
transforming growth factor-β
|
|
TMEM51
|
transmembrane protein 51
|
|
TNF-α
|
tumor necrosis factor-α
|
|
UBAC1
|
ubiquitin-associated
domain-containing protein 1
|
References
|
1
|
Iorio JA, Jakoi AM and Singla A:
Biomechanics of degenerative spinal disorders. Asian Spine J.
10:377–384. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cheung KM, Karppinen J, Chan D, Ho DW,
Song YQ, Sham P, Cheah KS, Leong JC and Luk KD: Prevalence and
pattern of lumbar magnetic resonance imaging changes in a
population study of one thousand forty-three individuals. Spine
(Phila Pa 1976). 34:934–940. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Szpalski M and Gunzburg R: Lumbar spinal
stenosis in the elderly: An overview. Eur Spine J. 12 (Suppl
2):S170–S175. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alini M, Eisenstein SM, Ito K, Little C,
Kettler AA, Masuda K, Melrose J, Ralphs J, Stokes I and Wilke HJ:
Are animal models useful for studying human disc
disorders/degeneration? Eur Spine J. 17:2–19. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Adams MA and Roughley PJ: What is
intervertebral disc degeneration, and what causes it? Spine (Phila
Pa 1976). 31:2151–2161. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takatalo J, Karppinen J, Niinimäki J,
Taimela S, Näyhä S, Mutanen P, Sequeiros RB, Kyllönen E and
Tervonen O: Does lumbar disc degeneration on magnetic resonance
imaging associate with low back symptom severity in young Finnish
adults? Spine (Phila Pa 1976). 36:2180–2189. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Risbud MV and Shapiro IM: Role of
cytokines in intervertebral disc degeneration: Pain and disc
content. Nat Rev Rheumatol. 10:44–56. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang C, Chen Z, Meng X, Li M, Zhang L and
Huang A: The involvement and possible mechanism of pro-inflammatory
tumor necrosis factor alpha (TNF-α) in thoracic ossification of the
ligamentum flavum. PLoS One. 12:e01789862017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Battié MC, Videman T, Levälahti E, Gill K
and Kaprio J: Genetic and environmental effects on disc
degeneration by phenotype and spinal level: A multivariate twin
study. Spine (Phila Pa 1976). 33:2801–2808. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yee A, Lam MP, Tam V, Chan WC, Chu IK,
Cheah KS, Cheung KM and Chan D: Fibrotic-like changes in degenerate
human intervertebral discs revealed by quantitative proteomic
analysis. Osteoarthritis Cartilage. 24:503–513. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kamita M, Mori T, Sakai Y, Ito S, Gomi M,
Miyamoto Y, Harada A, Niida S, Yamada T, Watanabe K and Ono M:
Proteomic analysis of ligamentum flavum from patients with lumbar
spinal stenosis. Proteomics. 15:1622–1630. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ai C and Kong CL: GPS: A machine
learning-based approach integrating multiple gene set analysis
tools for better prioritization of biologically relevant pathways.
J Genet Genomics. 45:489–504. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu J, Mao X, Cai T, Luo J and Wei L: KOBAS
server: A web-based platform for automated annotation and pathway
identification. Nucleic Acids Res. 34:W720–W724. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong
S, Kong L, Gao G, Li CY and Wei L: KOBAS 2.0: A web server for
annotation and identification of enriched pathways and diseases.
Nucleic Acids Res. 39:W316–W322. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kudo S, Ono M and Russell WJ: Ossification
of thoracic ligamenta flava. AJR Am J Roentgenol. 141:117–121.
1983. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hur JW, Kim BJ, Park JH, Kim JH, Park YK,
Kwon TH and Moon HJ: The mechanism of ligamentum flavum
hypertrophy: Introducing angiogenesis as a critical link that
couples mechanical stress and hypertrophy. Neurosurgery.
77:274–282. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Misawa H, Ohtsuka K, Nakata K and
Kinoshita H: Embryological study of the spinal ligaments in human
fetuses. J Spinal Disord. 7:495–498. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nguyen AD, Itoh S, Jeney V, Yanagisawa H,
Fujimoto M, Ushio-Fukai M and Fukai T: Fibulin-5 is a novel binding
protein for extracellular superoxide dismutase. Circ Res.
95:1067–1074. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Zhao Y, Gao J, Pawlyk B, Starcher
B, Spencer JA, Yanagisawa H, Zuo J and Li T: Elastic fiber
homeostasis requires lysyl oxidase-like 1 protein. Nat Genet.
36:178–182. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Postacchini F, Gumina S, Cinotti G,
Perugia D and DeMartino C: Ligamenta flava in lumbar disc
herniation and spinal stenosis. Light and electron microscopic
morphology. Spine (Phila Pa 1976). 19:917–922. 1976. View Article : Google Scholar
|
|
21
|
Wang B, Chen Z, Meng X, Li M, Yang X and
Zhang C: iTRAQ quantitative proteomic study in patients with
thoracic ossification of the ligamentum flavum. Biochem Biophys Res
Commun. 487:834–839. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Batista MA, Nia HT, Önnerfjord P, Cox KA,
Ortiz C, Grodzinsky AJ, Heinegård D and Han L: Nanomechanical
phenotype of chondroadherin-null murine articular cartilage. Matrix
Biol. 38:84–90. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
McEwan PA, Scott PG, Bishop PN and Bella
J: Structural correlations in the family of small leucine-rich
repeat proteins and proteoglycans. J Struct Biol. 155:294–305.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hildebrand A, Romarís M, Rasmussen LM,
Heinegård D, Twardzik DR, Border WA and Ruoslahti E: Interaction of
the small interstitial proteoglycans biglycan, decorin and
fibromodulin with transforming growth factor beta. Biochem J.
302:527–534. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang G, Ezura Y, Chervoneva I, Robinson
PS, Beason DP, Carine ET, Soslowsky LJ, Iozzo RV and Birk DE:
Decorin regulates assembly of collagen fibrils and acquisition of
biomechanical properties during tendon development. J Cell Biochem.
98:1436–1449. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Seki S, Kawaguchi Y, Chiba K, Mikami Y,
Kizawa H, Oya T, Mio F, Mori M, Miyamoto Y, Masuda I, et al: A
functional SNP in CILP, encoding cartilage intermediate layer
protein, is associated with susceptibility to lumbar disc disease.
Nat Genet. 37:607–612. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sairyo K, Biyani A, Goel V, Leaman D,
Booth R Jr, Thomas J, Gehling D, Vishnubhotla L, Long R and
Ebraheim N: Pathomechanism of ligamentum flavum hypertrophy: A
multidisciplinary investigation based on clinical, biomechanical,
histologic, and biologic assessments. Spine (Phila Pa 1976).
30:2649–2656. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kelempisioti A, Eskola PJ, Okuloff A,
Karjalainen U, Takatalo J, Daavittila I, Niinimäki J, Sequeiros RB,
Tervonen O, Solovieva S, et al: Genetic susceptibility of
intervertebral disc degeneration among young Finnish adults. BMC
Med Genet. 12:1532011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou T, Du L, Chen C, Han C, Li X, Qin A,
Zhao C, Zhang K and Zhao J: Lysophosphatidic acid induces
ligamentum flavum hypertrophy through the LPAR1/Akt pathway. Cell
Physiol Biochem. 45:1472–1486. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hara K, Shiga A, Fukutake T, Nozaki H,
Miyashita A, Yokoseki A, Kawata H, Koyama A, Arima K, Takahashi T,
et al: Association of HTRA1 mutations and familial ischemic
cerebral small-vessel disease. N Engl J Med. 360:1729–1739. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tiaden AN and Richards PJ: The emerging
roles of HTRA1 in musculoskeletal disease. Am J Pathol.
182:1482–1488. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dewan A, Liu M, Hartman S, Zhang SS, Liu
DT, Zhao C, Tam PO, Chan WM, Lam DS, Snyder M, et al: HTRA1
promoter polymorphism in wet age-related macular degeneration.
Science. 314:989–992. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsuchiya A, Yano M, Tocharus J, Kojima H,
Fukumoto M, Kawaichi M and Oka C: Expression of mouse HtrA1 serine
protease in normal bone and cartilage and its upregulation in joint
cartilage damaged by experimental arthritis. Bone. 37:323–336.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grau S, Richards PJ, Kerr B, Hughes C,
Caterson B, Williams AS, Junker U, Jones SA, Clausen T and Ehrmann
M: The role of human HtrA1 in arthritic disease. J Biol Chem.
281:6124–6129. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bogduk N: Functional anatomy of the spine.
Handb Clin Neurol. 136:675–688. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Feng C, Liu H, Yang M, Zhang Y, Huang B
and Zhou Y: Disc cell senescence in intervertebral disc
degeneration: Causes and molecular pathways. Cell Cycle.
15:1674–1684. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gruber HE, Hoelscher G, Ingram JA and
Hanley EN Jr: Culture of human anulus fibrosus cells on polyamide
nanofibers: Extracellular matrix production. Spine (Phila Pa 1976).
34:4–9. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Antoniou J, Steffen T, Nelson F,
Winterbottom N, Hollander AP, Poole RA, Aebi M and Alini M: The
human lumbar intervertebral disc: Evidence for changes in the
biosynthesis and denaturation of the extracellular matrix with
growth, maturation, ageing, and degeneration. J Clin Invest.
98:996–1003. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Johnson WE, Wootton A, El Haj A,
Eisenstein SM, Curtis AS and Roberts S: Topographical guidance of
intervertebral disc cell growth in vitro: Towards the development
of tissue repair strategies for the anulus fibrosus. Eur Spine J.
15 (Suppl 3):S389–S396. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Singh K, Masuda K, Thonar EJ, An HS and
Cs-Szabo G: Age-related changes in the extracellular matrix of
nucleus pulposus and anulus fibrosus of human intervertebral disc.
Spine (Phila Pa 1976). 34:10–6. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Minogue BM, Richardson SM, Zeef LA,
Freemont AJ and Hoyland JA: Transcriptional profiling of bovine
intervertebral disc cells: Implications for identification of
normal and degenerate human intervertebral disc cell phenotypes.
Arthritis Res Ther. 12:R222010. View
Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rutges J, Creemers LB, Dhert W, Milz S,
Sakai D, Mochida J, Alini M and Grad S: Variations in gene and
protein expression in human nucleus pulposus in comparison with
annulus fibrosus and cartilage cells: Potential associations with
aging and degeneration. Osteoarthritis Cartilage. 18:416–423. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Johnson WE, Patterson AM, Eisenstein SM
and Roberts S: The presence of pleiotrophin in the human
intervertebral disc is associated with increased vascularization:
An immunohistologic study. Spine (Phila Pa 1976). 32:1295–1302.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sarath Babu N, Krishnan S, Brahmendra
Swamy CV, Venkata Subbaiah GP, Gurava Reddy AV and Idris MM:
Quantitative proteomic analysis of normal and degenerated human
intervertebral disc. Spine J. 16:989–1000. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hofmann K and Falquet L: A
ubiquitin-interacting motif conserved in components of the
proteasomal and lysosomal protein degradation systems. Trends
Biochem Sci. 26:347–350. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Feng P, Scott CW, Cho NH, Nakamura H,
Chung YH, Monteiro MJ and Jung JU: Kaposi's sarcoma-associated
herpesvirus K7 protein targets a
ubiquitin-like/ubiquitin-associated domain-containing protein to
promote protein degradation. Mol Cell Biol. 24:3938–3948. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Duarri A, Nibbeling E, Fokkens MR, Meijer
M, Boddeke E, Lagrange E, Stevanin G, Brice A, Durr A and Verbeek
DS: Erratum to: The L450F [Corrected] mutation in KCND3 brings
spinocerebellar ataxia and Brugada syndrome closer together.
Neurogenetics. 16:2432015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ye D, Liang W, Dai L, Zhou L, Yao Y, Zhong
X, Chen H and Xu J: Comparative and quantitative proteomic analysis
of normal and degenerated human annulus fibrosus cells. Clin Exp
Pharmacol Physiol. 42:530–536. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Tsukahara F, Yoshioka T and Muraki T:
Molecular and functional characterization of HSC54, a novel variant
of human heat-shock cognate protein 70. Mol Pharmacol.
58:1257–1263. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Persico MG, Viglietto G, Martini G,
Toniolo D, Paonessa G, Moscatelli C, Dono R, Vulliamy T, Luzzatto L
and D'Urso M: Isolation of human glucose-6-phosphate dehydrogenase
(G6PD) cDNA clones: Primary structure of the protein and unusual
5′non-coding region. Nucleic Acids Res. 14:2511–2522. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Feng C, Zhang Y, Yang M, Lan M, Liu H,
Huang B and Zhou Y: Oxygen-sensing Nox4 generates genotoxic ROS to
induce premature senescence of nucleus pulposus cells through MAPK
and NF-κB pathways. Oxid Med Cell Longev. 2017:74264582017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kamboh MI, Barmada MM, Demirci FY, Minster
RL, Carrasquillo MM, Pankratz VS, Younkin SG, Saykin AJ;
Alzheimer's Disease Neuroimaging Initiative, ; Sweet RA, et al:
Genome-wide association analysis of age-at-onset in Alzheimer's
disease. Mol Psychiatry. 17:1340–1346. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Brown DA and Sihra TS: Presynaptic
signaling by heterotrimeric G-proteins. Handb Exp Pharmacol.
207–60. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sajjadi FG and Firestein GS: cDNA cloning
and sequence analysis of the human A3 adenosine receptor. Biochim
Biophys Acta. 1179:105–107. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Salvatore CA, Jacobson MA, Taylor HE,
Linden J and Johnson RG: Molecular cloning and characterization of
the human A3 adenosine receptor. Proc Natl Acad Sci USA.
90:10365–10369. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Honsho M, Asaoku S and Fujiki Y:
Posttranslational regulation of fatty acyl-CoA reductase 1, Far1,
controls ether glycerophospholipid synthesis. J Biol Chem.
285:8537–8542. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Samland AK and Sprenger GA: Transaldolase:
From biochemistry to human disease. Int J Biochem Cell Biol.
41:1482–1494. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gauci S, Helbig AO, Slijper M, Krijgsveld
J, Heck AJ and Mohammed S: Lys-N and trypsin cover complementary
parts of the phosphoproteome in a refined SCX-based approach. Anal
Chem. 81:4493–4501. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Dickson IR, Happey F, Pearson CH, Naylor A
and Turner RL: Variations in the protein components of human
intervertebral disk with age. Nature. 215:52–53. 1967. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sivan SS, Hayes AJ, Wachtel E, Caterson B,
Merkher Y, Maroudas A, Brown S and Roberts S: Biochemical
composition and turnover of the extracellular matrix of the normal
and degenerate intervertebral disc. Eur Spine J. 23 (Suppl
3):S344–S353. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Elliott DM and Setton LA: Anisotropic and
inhomogeneous tensile behavior of the human anulus fibrosus:
Experimental measurement and material model predictions. J Biomech
Eng. 123:256–263. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Erwin WM, DeSouza L, Funabashi M, Kawchuk
G, Karim MZ, Kim S, Mӓdler S, Matta A, Wang X and Mehrkens KA: The
biological basis of degenerative disc disease: Proteomic and
biomechanical analysis of the canine intervertebral disc. Arthritis
Res Ther. 17:2402015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Markolf KL and Morris JM: The structural
components of the intervertebral disc. A study of their
contributions to the ability of the disc to withstand compressive
forces. J Bone Joint Surg Am. 56:675–687. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Pettine KA, Murphy MB, Suzuki RK and Sand
TT: Percutaneous injection of autologous bone marrow concentrate
cells significantly reduces lumbar discogenic pain through 12
months. Stem Cells. 33:146–156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shu CC, Smith MM, Smith SM, Dart AJ,
Little CB and Melrose J: A histopathological scheme for the
quantitative scoring of intervertebral disc degeneration and the
therapeutic utility of adult mesenchymal stem cells for
intervertebral disc regeneration. Int J Mol Sci. 18(pii):
E10492017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Yang F, Leung VY, Luk KD, Chan D and
Cheung KM: Mesenchymal stem cells arrest intervertebral disc
degeneration through chondrocytic differentiation and stimulation
of endogenous cells. Mol Ther. 17:1959–1966. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Meisel HJ, Agarwal N, Hsieh PC, Skelly A,
Park JB, Brodke D, Wang JC, Yoon ST and Buser Z: Cell therapy for
treatment of intervertebral disc degeneration: A systematic review.
Global Spine J. 9 (1 Suppl):39S–52S. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Korecki CL, Taboas JM, Tuan RS and
Iatridis JC: Notochordal cell conditioned medium stimulates
mesenchymal stem cell differentiation toward a young nucleus
pulposus phenotype. Stem Cell Res Ther. 1:182010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Steck E, Bertram H, Abel R, Chen B, Winter
A and Richter W: Induction of intervertebral disc-like cells from
adult mesenchymal stem cells. Stem Cells. 23:403–411. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sive JI, Baird P, Jeziorsk M, Watkins A,
Hoyland JA and Freemont AJ: Expression of chondrocyte markers by
cells of normal and degenerate intervertebral discs. Mol Pathol.
55:91–97. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Risbud MV, Di Martino A, Guttapalli A,
Seghatoleslami R, Denaro V, Vaccaro AR, Albert TJ and Shapiro IM:
Toward an optimum system for intervertebral disc organ culture:
TGF-beta 3 enhances nucleus pulposus and anulus fibrosus survival
and function through modulation of TGF-beta-R expression and ERK
signaling. Spine (Phila Pa 1976). 31:884–890. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Purmessur D, Schek RM, Abbott RD, Ballif
BA, Godburn KE and Iatridis JC: Notochordal conditioned media from
tissue increases proteoglycan accumulation and promotes a healthy
nucleus pulposus phenotype in human mesenchymal stem cells.
Arthritis Res Ther. 13:R812011. View
Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhang Y, Xiong C, Kudelko M, Li Y, Wang C,
Wong YL, Tam V, Rai MF, Cheverud J, Lawson HA, et al: Early onset
disc degeneration in SM/J mice is associated with ion transport
systems and fibrotic changes. Matrix Biol. 70:123–139. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Tam V, Chan WCW, Leung VYL, Cheah KSE,
Cheung KMC, Sakai D, McCann MR, Bedore J, Séguin CA and Chan D:
Histological and reference system for the analysis of mouse
intervertebral disc. J Orthop Res. 36:233–243. 2018.PubMed/NCBI
|
|
75
|
Donnally IC, Hanna A and Varacallo M:
Lumbar degenerative disk disease, in StatPearls. StatPearls
Publishing. StatPearls Publishing LLC.; Treasure Island (FL):
2019
|
|
76
|
Liu XD, Zeng BF, Xu JG, Zhu HB and Xia QC:
Proteomic analysis of the cerebrospinal fluid of patients with
lumbar disk herniation. Proteomics. 6:1019–1028. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Xie P, Liu B, Chen R, Yang B, Dong J and
Rong L: Comparative analysis of serum proteomes: Identification of
proteins associated with sciatica due to lumbar intervertebral disc
herniation. Biomed Rep. 2:693–698. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Horrevoets AJ, Fontijn RD, van Zonneveld
AJ, de Vries CJ, ten Cate JW and Pannekoek H: Vascular endothelial
genes that are responsive to tumor necrosis factor-alpha in vitro
are expressed in atherosclerotic lesions, including inhibitor of
apoptosis protein-1, stannin, and two novel genes. Blood.
93:3418–3431. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Murata Y, Nannmark U, Rydevik B, Takahashi
K and Olmarker K: The role of tumor necrosis factor-alpha in
apoptosis of dorsal root ganglion cells induced by herniated
nucleus pulposus in rats. Spine (Phila Pa 1976). 33:155–162. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Luo G, Zhang X, Nilsson-Ehle P and Xu N:
Apolipoprotein M. Lipids Health Dis. 3:212004. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Palomo T, Vilaca T and Lazaretti-Castro M:
Osteogenesis imperfecta: Diagnosis and treatment. Curr Opin
Endocrinol Diabetes Obes. 24:381–388. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Deng H, Huang X and Yuan L: Molecular
genetics of the COL2A1-related disorders. Mutat Res Rev Mutat Res.
768:1–13. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Zollinger AJ and Smith ML: Fibronectin,
the extracellular glue. Matrix Biol. 60-61:27–37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Oegema TR Jr, Johnson SL, Aguiar DJ and
Ogilvie JW: Fibronectin and its fragments increase with
degeneration in the human intervertebral disc. Spine (Phila Pa
1976). 25:2742–2747. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kook SH, Lim SS, Cho ES, Lee YH, Han SK,
Lee KY, Kwon J, Hwang JW, Bae CH, Seo YK and Lee JC:
COMP-angiopoietin 1 increases proliferation, differentiation, and
migration of stem-like cells through Tie-2-mediated activation of
p38 MAPK and PI3K/Akt signal transduction pathways. Biochem Biophys
Res Commun. 455:371–377. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang Q, Ji Q, Wang X, Kang L, Fu Y, Yin
Y, Li Z, Liu Y, Xu X and Wang Y: SOX9 is a regulator of
ADAMTSs-induced cartilage degeneration at the early stage of human
osteoarthritis. Osteoarthritis Cartilage. 23:2259–2268. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Mo S, Liu C, Chen L, Ma Y, Liang T, Xue J,
Zeng H and Zhan X: KEGG-expressed genes and pathways in
intervertebral disc degeneration: Protocol for a systematic review
and data mining. Medicine (Baltimore). 98:e157962019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Ouyang ZH, Wang WJ, Yan YG, Wang B and Lv
GH: The PI3K/Akt pathway: A critical player in intervertebral disc
degeneration. Oncotarget. 8:57870–57881. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tan Y, Yao X, Dai Z, Wang Y and Lv G: Bone
morphogenetic protein 2 alleviated intervertebral disc degeneration
through mediating the degradation of ECM and apoptosis of nucleus
pulposus cells via the PI3K/Akt pathway. Int J Mol Med. 43:583–592.
2019.PubMed/NCBI
|
|
90
|
Zochodne DW: Mechanisms of diabetic neuron
damage: Molecular pathways. Handb Clin Neurol. 126:379–399. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Russo F, Ambrosio L, Ngo K, Vadalà G,
Denaro V, Fan Y, Sowa G, Kang JD and Vo N: The role of type I
diabetes in intervertebral disc degeneration. Spine (Phila Pa
1976). 44:1177–1185. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ma X, Han J, Wu Q, Liu H, Shi S, Wang C,
Wang Y, Xiao J, Zhao J, Jiang J and Wan C: Involvement of
dysregulated Wip1 in manganese-induced p53 signaling and neuronal
apoptosis. Toxicol Lett. 235:17–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ben-Porath I and Weinberg RA: The signals
and pathways activating cellular senescence. Int J Biochem Cell
Biol. 37:961–976. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Muller M: Cellular senescence: Molecular
mechanisms, in vivo significance, and redox considerations.
Antioxid Redox Signal. 11:59–98. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Jin LZ, Lu JS and Gao JW: Silencing SUMO2
promotes protection against degradation and apoptosis of nucleus
pulposus cells through p53 signaling pathway in intervertebral disc
degeneration. Biosci Rep. 38(pii): BSR201715232018. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Massagué J: TGFβ signalling in context.
Nat Rev Mol Cell Biol. 13:616–630. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chen G, Deng C and Li YP: TGF-β and BMP
signaling in osteoblast differentiation and bone formation. Int J
Biol Sci. 8:272–288. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Cao C, Zou J, Liu X, Shapiro A, Moral M,
Luo Z, Shi Q, Liu J, Yang H and Ebraheim N: Bone marrow mesenchymal
stem cells slow intervertebral disc degeneration through the NF-κB
pathway. Spine J. 15:530–538. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lu L, Hu J, Wu Q, An Y, Cui W, Wang J and
Ye Z: Berberine prevents human nucleus pulposus cells from
IL1betainduced extracellular matrix degradation and apoptosis by
inhibiting the NFkappaB pathway. Int J Mol Med. 43:1679–1686.
2019.PubMed/NCBI
|
|
100
|
Zieba J, Forlenza KN, Khatra JS,
Sarukhanov A, Duran I, Rigueur D, Lyons KM, Cohn DH, Merrill AE and
Krakow D: TGFβ and BMP dependent cell fate changes due to loss of
filamin b produces disc degeneration and progressive vertebral
fusions. PLoS Genet. 12:e10059362016. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Yang H, Cao C, Wu C, Yuan C, Gu Q, Shi Q
and Zou J: TGF-βl suppresses inflammation in cell therapy for
intervertebral disc degeneration. Sci Rep. 5:132542015. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Posey KL, Coustry F and Hecht JT:
Cartilage oligomeric matrix protein: COMPopathies and beyond.
Matrix Biol. 71-72:161–173. 2018. View Article : Google Scholar : PubMed/NCBI
|