Introduction
Angiogenesis is a process in which blood vessels are
formed from pre-existing vessels, and is vital for vascularization
during embryonic development and vessel formation in various organs
and tissues (1,2). Vascular endothelial growth factor
(VEGF) has been studied as a fundamental growth factor involved in
blood vessel formation (3).
Moreover, tight regulation of VEGF expression is required during
angiogenesis, but this regulation may be altered by certain
physiological signals, particularly in hypoxia (3,4).
Hypoxia-inducible factor 1 (HIF-1) exerts its effects on every
stage of angiogenesis via activation of various angiogenic factors,
such as VEGF, or regulation of proangiogenic chemokines (5–7).
VEGF also increases the expression of other proangiogenic factors,
including fibroblast growth factor (FGF) (5). FGF1 and FGF2 bind to FGF receptors
and serve crucial roles in the angiogenic process (8). Hypoxia is primarily caused by
ischemia in vivo, which can result in severe diseases, such
as coronary artery diseases (9).
Coronary artery diseases are one of the leading causes of mortality
worldwide (10). Furthermore,
ischemic heart disease results in >1 million mortalities
worldwide each year, and the morbidity and mortality rates have
remained high, particularly in developing countries (10,11).
It has been demonstrated that brief continual myocardial ischemia
enhances blood vessel formation in the adult heart, resulting in
improvement to blood supply and improvement in ischemic areas,
which may prevent extended ischemic disorders (12). However, the detailed mechanisms of
ischemia/hypoxia-induced angiogenesis are not fully understood.
Long non-coding RNAs (lncRNAs), which are non-coding
sequences >200 nucleotides in length, are involved in various
physiological and pathological processes, and diseases (13,14).
In the heart, a conserved lncRNA mechanism, known as the
Mhrt-related circuit, participates in chromatin remodelling and
prevent myocardial hypertrophy (15). It has been revealed that cardiac
apoptosis-related lncRNA affects abnormal mitochondrial fission and
apoptosis, which may improve the treatment of myocardial infarction
(16). Furthermore, lncRNA
non-coding RNA activated by DNA damage (NORAD), which is involved
in the maintenance of genomic stability, has been identified
(17). As its activation is
strongly associated with a poor prognosis and survival in breast
cancer and pancreatic cancer, and high expression of NORAD is
observed in gastric cancer cells and tissues, NORAD is considered
to be a potential oncogene (18–20).
Additionally, NORAD may serve a role in the development of lung
carcinoma by regulating the expression of transforming growth
factor-β (TGF-β) and its associated phenotype (21). Furthermore, among the family of
microRNAs (miRNAs/miRs), which are small non-coding RNAs ~22
nucleotides in length, hsa-miR-590-5p has been shown to
downregulate the TGF-β signalling pathway, resulting in a decreased
generation of cardiac cells (22,23).
Previous studies have also revealed that hypoxia modulates the
activities of abundantly expressed miRNAs by regulating proteins
involved in post-transcriptional processes of miRNAs (24).
It has been shown that lncRNAs serve an important
role in the regulation of angiogenesis to further affect cancer
development or diseases (25).
However, the function and mechanisms of the majority of lncRNAs
have not been previously determined. It was hypothesized that NORAD
may regulate endothelial cell properties via miR-590-3p, and
modulated angiogenesis under hypoxic conditions. In the present
study, the effects of NORAD and miR-590-3p on angiogenesis in human
umbilical vein endothelial cells (HUVECs), and whether miR-590-3p
affected NORAD-mediated regulation of endothelial cell activities
were investigated.
Materials and methods
Cell culture and grouping
treatment
HUVECs were purchased from the Shanghai Zhong Qiao
Xin Zhou Biotechnology Company and cultured in M199 complete medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 20% FBS
(Sigma-Aldrich; Merck KGaA) at 37°C with 5% CO2. When
cells reached ~70% confluency within 3 passages, cells were divided
into two groups (normoxia and hypoxia) for hypoxia treatment in
subsequent experiments. Normoxia cells were cultured in 5%
CO2 and 21% O2 at 37°C for 24 h, and cells
that underwent hypoxia were incubated in a tri-gas incubator
(Shanghai Lishen Scientific Equipment Co., Ltd.) with 5%
CO2, 1% O2 and 94% N2 at 37°C for
24 h. All subsequent experiments were performed 24 h after
culturing. 293T cells were also obtained from the Shanghai Zhong
Qiao Xin Zhou Biotechnology Company, and cultured in DMEM (HyClone;
GE Healthcare Life Sciences) with 10% FBS at 37°C with 5%
CO2 for 24 h.
Transfection
Specific short hairpin (sh)RNA against lncRNA NORAD
(shNORAD) and control shRNA (shNC1) were obtained from Wanleibio
Co., Ltd. miR-590-3p mimics (5′-UAAUUUUAUGUAUAAGCUAGU-3′),
miR-590-3p inhibitor (5′-ACUAGCUUAUACAUAAAAUUA-3′) and scrambled
control (mimics-NC and inhibitor-NC) were purchased from JTS
Scientific Co., Ltd. The overall length of NORAD was 5,378 bp, and
the overexpression plasmid pcDNA3.1+ (Clontech Laboratories, Inc.)
containing the first 2,000 bp of sequence NORAD
(NORAD1-2000-OE) was established. This sequence was used
to ensure the potential binding sequence with miR-590-3p was
included, and to avoid the binding site of shNORAD. HUVECs were
transfected with 100 pmol of shNORAD (shNC1), miR-590-3p mimics
(mimics-NC), NORAD1-2000-OE plasmid (empty
plasmid/vector) or shNORAD combined with miR-590-3p inhibitor
(inhibitor-NC) using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) according to the manufacturer
protocol. Then, 48 h after transfection, cells were underwent
hypoxia treatment as described above.
Cell Counting Kit-8 (CCK-8) assay
A total of 3×103 cells/well HUVECs or
transfected HUVECs were plated in 96-well plates with 5 replicates
per condition. Following incubation at 37°C with 5% CO2
for 24 h, cells underwent normoxia or hypoxia treatment and were
cultured for 12 or 24 h, respectively. Subsequently, 10 µl CCK-8
reagent (Sigma-Aldrich; Merck KGaA) was added and incubated at 37°C
for 1 h according to the manufacturer's instructions. Absorbance
was measured at 450 nm using a microplate reader (BioTek
Instruments, Inc.).
Transwell migration assay
Transwell chambers with 8.0 µm pore polycarbonate
membrane inserts (Corning, Inc.) were placed in 24-well-plates. A
total of 4×103 normal or transfected HUVECs were
suspended in serum-free M199 medium and seeded into the upper
chamber, and 800 µl medium supplemented with 20% FBS was added to
the lower chamber. Cells were cultured under normoxic or hypoxic
conditions at 37°C for 24 h and washed twice with PBS. Cells that
had migrated through the filter were fixed with 4% paraformaldehyde
(Sinopharm Chemical Reagent Co., Ltd.) for 20 min and stained using
0.1% crystal violet (Amresco, LLC) for 5 min at room temperature.
The stained cells were visualized using an inverted phase contrast
microscope (magnification, ×200) (Olympus Corporation), and the
number of cells in 5 randomly chosen fields was counted.
Tube formation assay
Matrigel (BD Biosciences) was thawed overnight at
4°C, and used to coat the wells of a 96-well plate (50 µl per well)
at 37°C for 2 h. Subsequently, HUVECs and transfected HUVECs were
suspended and seeded in the coated 96-well-plates
(1×104/well), followed by culturing the cells under
normoxia or hypoxia at 37°C for 24 h. Tube formation was observed
and imaged using an inverted phase contrast microscope
(magnification, ×100) (Olympus Corporation), and relative tube
length was calculated using Image-Pro Plus version 6 (Media
Cybernetics, Inc.). Tube formation was assessed in 3 randomly
selected fields of view.
Dual luciferase reporter assay
Potential target sequences between miR-590-3p and
lncRNA NORAD, or other target genes, were predicted using the
Bielefeld Bioinformatics Server (BiBiServ)-RNAhybrid (26) tool online. Luciferase reporter
assays were performed using 293T cells. Cells (70% confluency) were
seeded in 12-well-plates with serum-free DMEM medium treatment for
1 h prior to transfection. The medium in each well was replaced
with complete medium and transfection mixture containing: 1.5 µg
reporter plasmid pmirGLO (GenScript Co., Ltd.), which included the
3′ untranslated region (UTR) of the target genes (NORAD, VEGFA,
FGF1 and FGF2) with wild-type or mutated binding sequences, and 75
pmol synthetic nucleic acid segment in 200 µl Opti-minimum
essential medium (Gibco; Thermo Fisher Scientific, Inc.) with 9 µl
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Cells were cultured at 37°C for 48 h, and
luciferase activity was measured in comparison with Renilla
luciferase activity using a dual luciferase detection assay system
(Promega Corporation) according to the manufacturer's protocol.
ELISA
ELISA kits for VEGF A (VEGFA; SEA143Hu), FGF1
(SEA032Hu) and FGF2 (CEA551Hu) were used to measure the respective
protein levels in the supernatant cell culture of in different
groups under hypoxia according to the manufacturer's protocol
(Wuhan USCN Business Co., Ltd.).
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from HUVECs and tissues
using TRIpure reagent (BioTeke Corporation), and RT into cDNA using
Super M-MLV reverse transcriptase (BioTeke Corporation), dNTPs,
RNase inhibitor (BioTeke Corporation), 5X reaction buffer, and
primers (RT primer for miRNA, Oligo(dT)15 for other
genes) at 42°C for 30 min and 70°C for 10 min (miRNA) or 42°C for
50 min and 80°C for 10 min (other genes). Subsequently, qPCR was
performed using diluted 2× Power Taq PCR MasterMix (BioTeke
Corporation) with SYBR-Green (Sigma-Aldrich; Merck KGaA). The
thermocycling conditions for miRNA were: 94°C for 4 min, 40 cycles
of 94°C for 15 sec, 60°C for 20 sec and 72°C for 15 sec; for other
genes they were: 94°C for 5 min, 40 cycles of 94°C for 15 sec, 60°C
for 25 sec and 72°C for 30 sec. The expression of hsa-miR-590-3p
was normalized to 5S, and the rest of the genes was normalized to
β-actin. Primers were purchased from GenScript and the sequences
were: hsa-miR-590-3p/mmu-miR-590-3p forward,
5′-TAATTTTATGTATAAGCTAGT-3′; hsa-miR-590-3p/mmu-miR-590-3p reverse,
5′-GCAGGGTCCGAGGTATTC-3′; 5S forward, 5′-GATCTCGGAAGCTAAGCAGG-3′;
5S reverse, 5′-TGGTGCAGGGTCCGAGGTAT-3′; mmu-5S forward,
5′-CTAAAGATTTCCGTGGAGAG-3′; mmu-5S reverse,
5′-TGGTGCAGGGTCCGAGGTAT-3′; lncRNA NORAD forward,
5′-GGAGAATCGCTTGAACT-3′; lncRNA NORAD reverse,
5′-CAAACACCCAATGAATAG-3′; mmu-lncRNA NORAD forward,
5′-GATTGCCGACGCAGGGTA-3′; mmu-lncRNA NORAD reverse,
5′-CTGAACAAACAGGGACGA-3′; HIF-1α forward,
5′-GAAACTTCTGGATGCTGGTG-3′; HIF-1α reverse,
5′-CAAACTGAGTTAATCCCATG-3′; VEGFA forward,
5′-TCACCAAGGCCAGCACATAG-3′; VEGFA reverse,
5′-GGGCACCAACGTACACGCT-3′; FGF1 forward, 5′-GAGCGACCAGCACATTCAG-3′;
FGF1 reverse, 5′-TCTCCTCCAGCCTTTCCAG-3′; FGF2 forward,
5′-AGAAGAGCGACCCTCACAT-3′; FGF2 reverse,
5′-AAAGAAACACTCATCCGTAA-3′; β-actin forward,
5′-GGCACCCAGCACAATGAA-3′; β-actin reverse,
5′-TAGAAGCATTTGCGGTGG-3′; mmu-β-actin forward,
5′-CTGTGCCCATCTACGAGGGCTAT-3′; and mmu-β-actin reverse,
5′-TTTGATGTCACGCACGATTTCC-3′. Fold changes of gene expressions were
calculated using the 2−ΔΔCq method (27).
Western blotting
Cell Lysis Buffer for Western or IP (Beyotime
Institute of Biotechnology) was used to extract the total protein.
Protein concentration was detected using a bicinchoninic acid
protein assay kit (Beyotime Institute of Biotechnology). A total of
20–40 µg was loaded on an 8–12% SDS gel and resolved using
SDS-PAGE. The resolved proteins were transferred to PVDF membranes
(EMD Millipore) and were blocked in 5% non-fat milk (Yili Group) at
room temperature for 1 h. Membranes were incubated with primary
antibodies against HIF-1α (AF1009, Affinity Biosciences) and
β-actin (sc-47778; Santa Cruz Biotechnology, Inc.) both at 1:1,000
at 4°C overnight. Incubation of membranes with horseradish
peroxidase-conjugated goat anti-rabbit (A0208, 1:5,000; Beyotime)
or anti-mouse (A0216, 1:5,000; Beyotime) secondary antibody was
performed at 37°C for 45 min. An ECL reagent (Beyotime Institute of
Biotechnology) was used to visualize the signals, and densitometry
analysis was performed using Gel-Pro-Analyzer software (version 4;
Media Cybernetics, Inc.).
Animal experiment
A total of 20 male C57 mice (8 weeks old, 22±2 g;
Beijing HFK Bioscience Co., Ltd.) were used to establish a
myocardial infarction (MI) model via left anterior descending (LAD)
ligation surgery. Prior to surgery, all mice were maintained in a
12 h light/dark cycle with constant temperature of 22±1°C and
45–55% humidity, with free access to food and water. After week,
mice were immobilized and anesthetized. Then, endotracheal
intubation and ventilation were performed following incision of the
neck trachea, followed by left thoracotomy between the third and
fourth intercostal space. Blood flow in the anterior descending
branch was blocked using a sterile 8/0 suture ligation. Mice in the
control group (n=6) underwent the same operation, but without
ligation. After 24 h following suture, the left ventricle tissues
were removed for analysis. Each group contained six mice. Animal
experiments were performed in accordance with the principles
described in the Guide for The Care and Use of Laboratory Animals
(28), and were approved by The
Ethics Committee of The First Hospital of China Medical
University.
Statistical analysis
Data were presented as the mean ± SD of 3 repeats
for the in vitro experiments, and for the animal experiments
there were 6 animals per group. GraphPad Prism version 8 (GraphPad
Prism Software, Inc.) was used to perform the statistical analysis.
Unpaired t-tests were used to compare two independent groups, and
differences between ≥3 groups were compared using the one-way ANOVA
combined with Tukey's multiple comparisons post hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
HUVEC tube-formation ability is
increased under hypoxic conditions
To determine the effects of non-coding RNAs on
hypoxia exposed HUVECs, experiments were performed to assess the
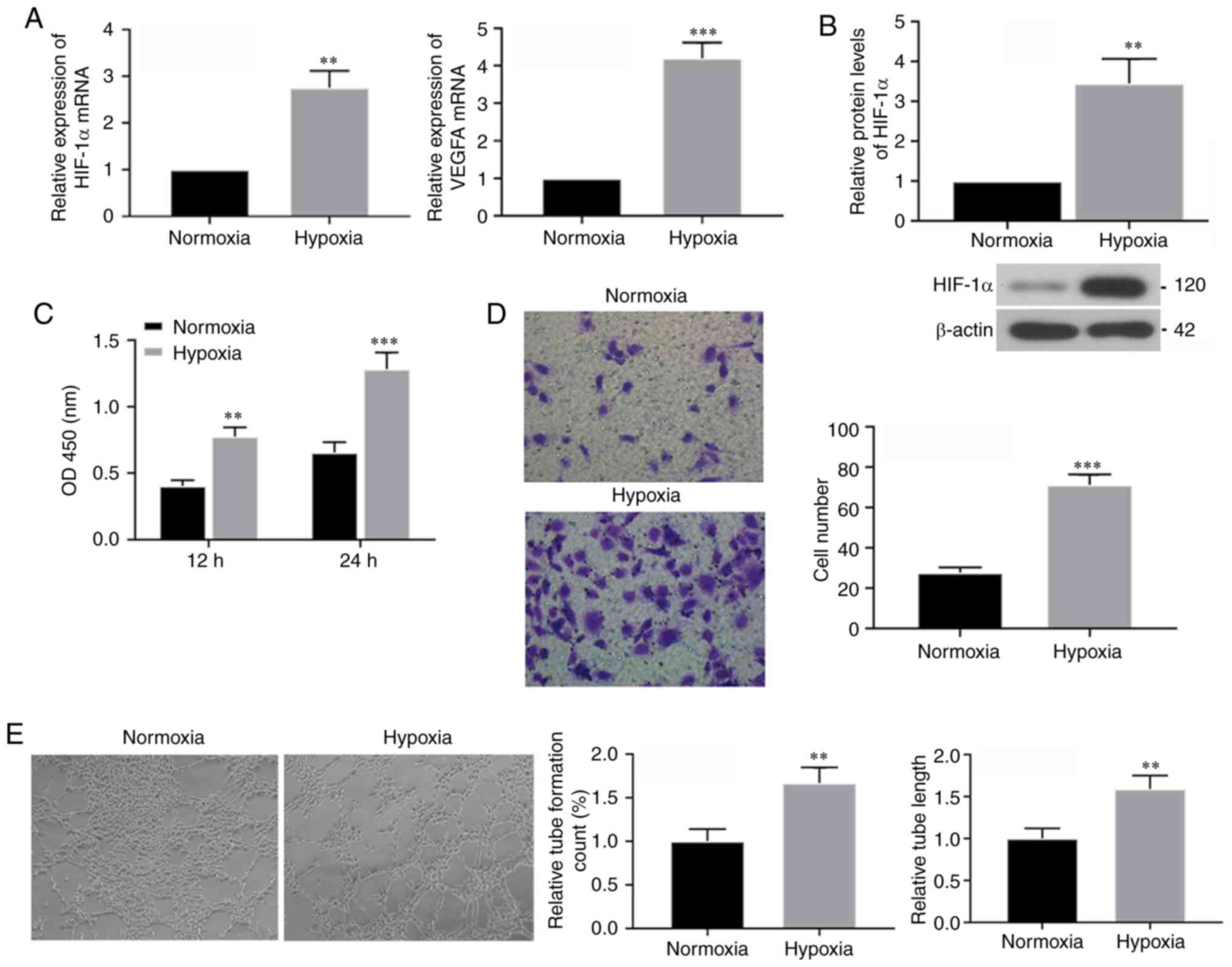
activities of HUVECs under hypoxic conditions. mRNA expression
levels of HIF-1α and its downstream target VEGFA were determined 24
h after hypoxia culturing using RT-qPCR, and it was identified that
both had increased expression levels following hypoxia treatment
(Fig. 1A). Western blotting
results also demonstrated increased protein expression of HIF-1α
under hypoxic conditions (Fig.
1B). Thus, these results suggested that hypoxia treatment was
effective. Furthermore, the CCK-8 assay results indicated that,
compared with cells in the normoxic condition, cell viability of
HUVECs was increased after 24 h of hypoxia exposure (Fig. 1C). Moreover, Transwell assays were
performed to assess the migratory ability of cells following
hypoxia for 24 h, and it was identified that hypoxia significantly
increased migration (Fig. 1D).
Furthermore, results from the tube formation assay in the
hypoxia-exposed HUVECs demonstrated that tube count and length were
increased compared with normoxic cells (Fig. 1E). Therefore, the present results
suggested that short-term hypoxia exposure caused increased
tube-formation in HUVECs.
lncRNA NORAD expression is increased
and miR-590-3p expression is decreased in hypoxic HUVECs in vitro
and MI left ventricular tissues in vivo
The relative mRNA expression levels of lncRNA NORAD
and miR-590-3p in HUVECs under hypoxia were assessed using RT-qPCR.
It was demonstrated that NORAD expression was increased and
miR-590-3p expression was decreased in HUVECs exposed to hypoxia
for 24 h (Fig. 2A). In
vivo, mice MI models were also used to assess the expression
levels of these 2 factors in the left ventricle following MI
modelling for 24 h, and similar results were observed as those
identified in vitro (Fig
2B). Thus, it was identified that hypoxia increased expression
of lncRNA NORAD, and decreased expression of miR-590-3p, in HUVECs
in vitro and in left ventricular tissues in vivo.
lncRNA NORAD knockdown inhibits
tube-formation ability of HUVECs under hypoxia
To investigate how lncRNA NORAD affected the HUVECs
under hypoxia, cells were transfected with shNORAD and shNC1.
RT-qPCR was performed to detect the relative expression of NORAD in
transfected HUVECs, and it was revealed that shNORAD effectively
knocked down the expression of lncRNA NORAD in HUVECs (Fig. 3A). Moreover, hypoxic treatment for
24 h in shNORAD transfected HUVECs also demonstrated that NORAD
expression was significantly knocked down. It was also identified
that NORAD downregulation caused an increase in miR-590-3p
expression under hypoxia (Fig.
3B). Cell viability of transfected HUVECs was assessed using a
CCK-8 assay. In contrast to hypoxia-exposed shNC1 cells, knockdown
of NORAD resulted in decreased viability of shNORAD transfected
HUVECs (Fig. 3C). Furthermore, the
Transwell migration assays showed that HUVECs transfected with
shNORAD exhibited a lower migratory capacity compared with cells
transfected with shNC1 under hypoxia (Fig. 3D). NORAD knockdown also decreased
tube formation ability of HUVECs exposed to hypoxia compared with
the NC cells (Fig. 3E). Therefore,
the present results suggested that lncRNA NORAD knockdown inhibited
the angiogenesis-associated abilities of HUVECs under hypoxia,
indicating decreased blood vessel formation.
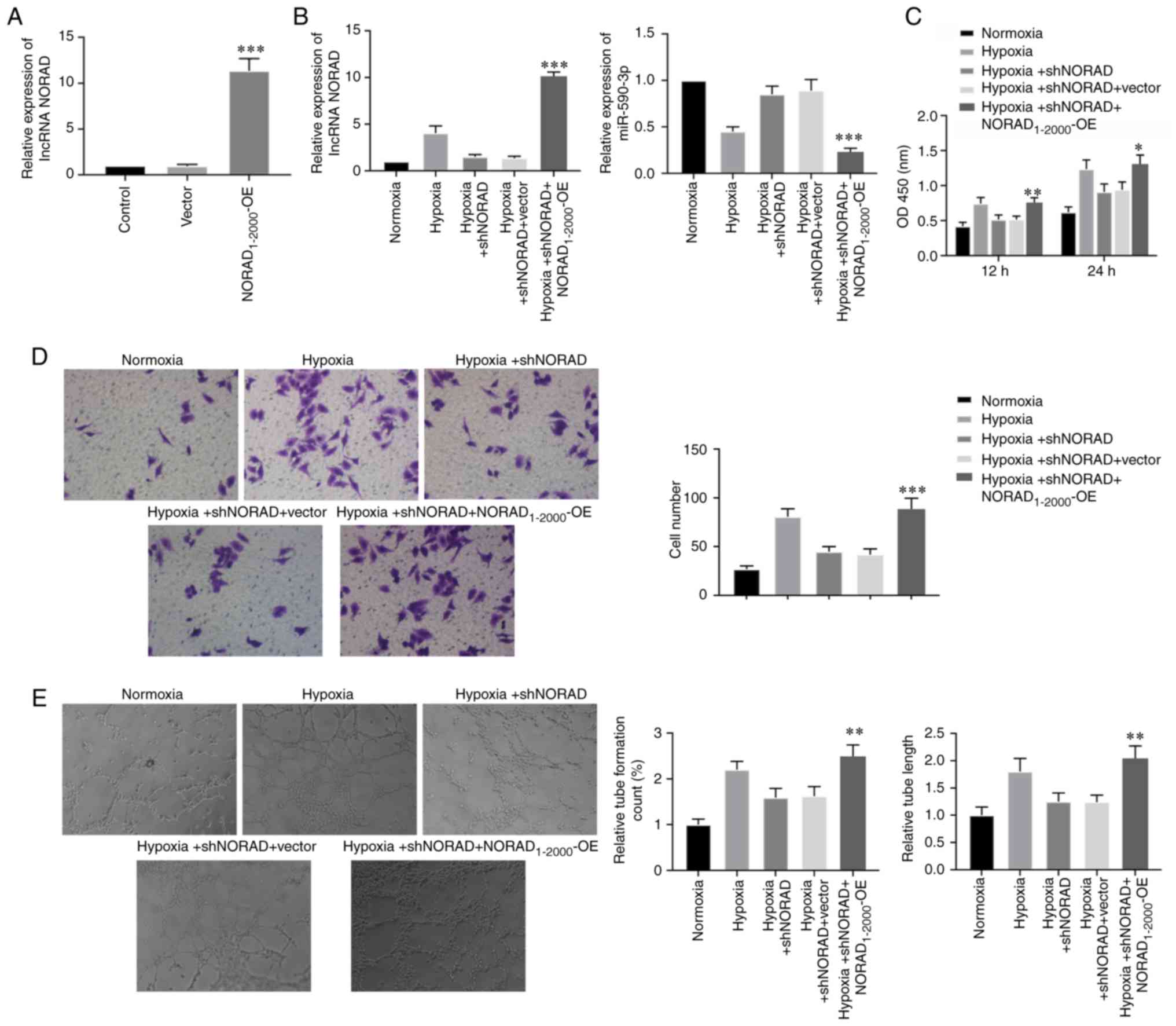
lncRNA NORAD overexpression promotes
tube-formation in HUVECs under hypoxia
Cells were transfected with the
NORAD1-2000-OE plasmid to study the effects of NORAD
overexpression on HUVECs under hypoxia. Under normoxic conditions,
the transfection of NORAD1-2000-OE plasmid in HUVECs was
highly efficient (Fig. 4A).
Moreover, the relative expression of NORAD was significantly
upregulated, and the expression of miR-590-3p was decreased in the
NORAD1-2000-OE transfected cells under hypoxia (Fig. 4B). Cell viability, migratory
capacity and tube formation, including tube count and length, were
assessed in NORAD overexpressed cells under hypoxia, and were
identified to be increased (Fig.
4C-E). Collectively, the present results indicated that NORAD
overexpression in HUVECs reversed the reduced tube-formation
ability caused by NORAD knockdown under hypoxia.
Overexpression of miR-590-3p
suppresses tube formation in HUVECs under hypoxia
To determine the role of miR-590-3p in the
vasculogenic ability of HUVECs under hypoxia, cells were
transfected with miR-590-3p mimics and mimics-NC. The RT-qPCR
results indicated that miR-590-3p was efficiently overexpressed
when cells were transfected with miR-590-3p mimics, under both
normoxic and hypoxic conditions (Fig.
5A). Furthermore, overexpression of miR-590-3p resulted in
similar effects on cell viability, cell migration and tube
formation ability as knockdown of NORAD under hypoxia (Fig. 5B-D). In the dual luciferase assay,
293T cells co-transfected with NORAD-wild-type and miR-590-3p
mimics exhibited significantly decreased luciferase activity
compared with cells transfected with mutant NORAD and miR-590-3p
mimics (Fig. 5E), which suggested
that NORAD could bind with miR-590-3p directly. Thus, it was
hypothesized that miR-590-3p also affected HUVEC tube-forming
ability under hypoxia, which may be associated with NORAD
targeting.
 | Figure 5.Overexpression of miR-590-3p
suppresses tube-formation in HUVECs under hypoxia. (A) HUVECs were
transfected with miR-590-3p mimics and mimics-NC. Relative
expression of miR-590-3p following transfection was assessed prior
and subsequent to hypoxia treatment. ***P<0.001 vs. respective
NC controls. (B) Cell viability, (C) cell migration (magnification,
×200) and (D) tube formation (magnification 100×) were examined in
cells transfected with miR-590-3p mimics under hypoxia. *P<0.05,
**P<0.01 and ***P<0.001 vs. Hypoxia + mimics-NC group. (E)
Relative luciferase activity was measured 48 h after
co-transfection of miR-590-3p mimics with NORAD-wt or NORAD-mut in
293T cells. The binding sequences of the wt and mut NORAD are
shown. **P<0.01 vs. NORAD-mut + miR-590-3p mimics group. lncRNA,
long non-coding RNA; lncRNA NORAD, lncRNA non-coding RNA activated
by DNA damage; miR, microRNA; NC, negative control; mut, mutant;
wt, wild-type; OD, optical density. |
lncRNA NORAD affects angiogenic
characteristics of HUVECs under hypoxia via miR-590-3p
To investigate whether the effects of NORAD on HUVEC
tube-formation ability were regulated by miR-590-3p under hypoxia,
HUVECs were co-transfected with shNORAD and miR-590-3p inhibitor.
The transfection efficiency of the miR-590-3p inhibitor under
normoxic conditions was assessed using RT-qPCR, which identified
that the expression of miR-590-3p was significantly downregulated
(Fig. 6A). Subsequently, the
relative expression levels of lncRNA NORAD and miR-590-3p were
assessed using RT-qPCR following co-transfection. Cells
co-transfected with shNORAD and miR-590-3p inhibitor exhibited
decreased miR-590-3p expression levels, but increased NORAD
expression, compared with cells transfected with shNORAD and
inhibitor-NC (Fig. 6B). Therefore,
it was speculated that miR-590-3p negatively regulated NORAD, and
similar results were observed under hypoxia (Fig. 6C). Cell viability in the
transfected hypoxia-exposed HUVECs, in which NORAD and miR-590-3p
expression levels were downregulated, was significantly increased
compared with cells transfected with shNORAD and inhibitor-NC
(Fig. 6D). Furthermore,
downregulation of NORAD and miR-590-3p in the HUVECs also increased
migration and tube formation under hypoxia compared with the
control cells (Fig. 6E and F).
Thus, downregulation of both NORAD and miR-590-3p in HUVECs
reversed the decrease in angiogenic activity caused by knockdown of
NORAD alone, which resulted in enhanced angiogenic properties under
hypoxic conditions. Collectively, the present results suggested
that miR-590-3p lay downstream of the regulatory effects of lncRNA
NORAD on angiogenic activity in hypoxia-exposed HUVECs.
 | Figure 6.lncRNA NORAD regulates angiogenesis
of HUVECs under hypoxia via miR-590-3p. (A) Relative expression of
miR-590-3p in HUVECs transfected with miR-590-3p inhibitor and
inhibitor-NC. ***P<0.001 vs. inhibitor-NC. (B) HUVECs were
co-transfected with shNORAD and miR-590-3p inhibitor or shNORAD and
inhibitor-NC, and expression levels of lncRNA NORAD and miR-590-3p
were determined under atmospheric conditions. (C) Expression levels
of lncRNA NORAD and miR-590-3p were again assessed under hypoxic
condition after the same HUVEC co-transfection. (D) Cell viability,
(E) cell migration (magnification, ×200) and (F) tube formation
(magnification, ×100) were measured in HUVECs transfected with both
shNORAD and miR-590-3p inhibitor under hypoxia. *P<0.05,
**P<0.01 and ***P<0.001 vs. respective inhibitor-NC group.
lncRNA, long non-coding RNA; lncRNA NORAD, lncRNA non-coding RNA
activated by DNA damage; miR, microRNA; NC, negative control; sh,
short hairpin RNA; OD, optical density. |
Downstream targets of the lncRNA
NORAD/miR-590-3p axis in HUVECs under hypoxia
In total, 3 possible regulated genes were proposed,
VEGFA, FGF1 and FGF2, all of which are closely associated with
angiogenesis and cell proliferation (5,8).
After 24 h of hypoxia treatment, it was demonstrated that the mRNA
expression levels of all 3 genes were significantly decreased in
NORAD knockdown HUVECs, but significantly increased in cells where
NORAD and miR-590-3p were both downregulated (Fig. 7A). Moreover, ELISAs were performed
to assess the changes in the levels of all 3 proteins, and it was
identified that when NORAD was downregulated, the concentrations of
VEGFA, FGF1 and FGF2 were decreased under hypoxia. However, when
miR-590-3p was knocked down, the expression levels of these genes
were increased significantly under the same condition (Fig. 7B). Binding of miR-590-3p to these
genes was determined using a dual luciferase assay, which
identified that cells co-transfected with the 3′UTR of VEGFA, FGF1
or FGF2, and miR-590-3p mimics, exhibited lower luciferase activity
compared with cells transfected with their mutated 3′UTR and
miR-590-3p mimics (Fig. 7C).
Therefore, the present results suggested that a lncRNA
NORAD/miR-590-3p axis regulated the activity of the angiogenic
indicators VEGFA, FGF1 and FGF2, and that these genes were direct
downstream targets of miR-590-3p.
 | Figure 7.Downstream target genes of the lncRNA
NORAD/miR-590-3p axis in HUVECs under hypoxia. (A) Relative mRNA
expression levels of VEGFA, FGF1 and FGF2 were identified in HUVECs
transfected with shNORAD or shNC1, and co-transfected with shNORAD
and miR-590-3p inhibitor, or shNORAD and inhibitor-NC under hypoxia
for 24 h. (B) ELISA was performed to measure the concentration of
VEGFA, FGF1 and FGF2 in transfected HUVECs under hypoxic condition.
***P<0.001 in hypoxia + shNORAD vs. hypoxia + shNC1.
##P<0.01, ###P<0.001 in hypoxia +
shNORAD + miR-590-3p inhibitor vs. hypoxia + shNORAD +
inhibitor-NC. (C) Dual luciferase reporter assay was used to assess
binding of miR-590-3p with the proangiogenic factors VEGFA, FGF1
and FGF2. Luciferase activity was assessed in 293T cells following
co-transfection of miR-590-3p mimics with the 3′UTR of VEGFA, FGF1
and FGF2 or mutant 3′UTRs. **P<0.01 vs. targeted mutants +
miR-590-3p mimics. lncRNA, long non-coding RNA; lncRNA NORAD,
lncRNA non-coding RNA activated by DNA damage; miR, microRNA;
VEGFA, vascular endothelial growth factor A; FGF, fibroblast growth
factor; sh, short hairpin RNA; NC, negative control; 3′UTR, 3′
untranslated region; wt, wild-type; mut, mutant. |
Discussion
The aim of the present study was to determine
whether the lncRNA NORAD served a role in angiogenesis induced by
hypoxia, and the involvement of miR-590-3p in this process. lncRNA
NORAD is activated by DNA damage and maintains genomic stability
via isolating Pumilio proteins, which bind with mRNAs to suppress
their stability and translation activity (17). Thus, NORAD is crucial for cell
division and growth during cell development (29). In the present study, in short term
hypoxia (24 h), downregulation of NORAD in the HUVECs resulted in
decreased angiogenic properties, including cell viability, cell
migration and tube formation. Moreover, NORAD overexpression
resulted in opposite effects, suggesting that NORAD regulates the
angiogenic properties of HUVECs under hypoxic conditions. It was
identified that the overexpression of miR-590-3p caused a similar
decrease in tube formation in hypoxia-exposed HUVECs. The present
results observed in cells overexpressing miR-590-3p were consistent
with the effects of NORAD downregulation, and when NORAD was
downregulated, miR-590-3p expression was increased both in
vitro and in vivo. Several miRNAs, including miR-20,
miR-130 family and miR-199, are involved in angiogenesis under
hypoxic conditions by modulating the expression or activity of
proteins, including HIF-1 and VEGF (30). Based on the present results, it was
speculated that miR-590-3p may also be considered an additional
miRNA involved in angiogenesis under hypoxic conditions.
The present results suggested that NORAD directly
binds to miR-590-3p, and miR-590-3p directly targeted the 3′UTR of
VEGFA, FGF1 and FGF2. In addition, knockdown of both NORAD and
miR-590-3p could alleviate and reverse the decreased angiogenic
activities and expression levels of proangiogenic indicators caused
by knockdown of NORAD alone, leading to increased cell viability,
migration, tube formation and expression of associated
proangiogenic factors. Collectively, the present results indicated
the presence of a regulatory pathway in which lncRNA NORAD binds to
miR-590-3p to regulate angiogenesis in HUVECs, by affecting the
expression levels of downstream proangiogenic proteins, such as
VEGFA, FGF1 and FGF2, under hypoxic conditions. It is possible that
other positive factors involved in the regulation of angiogenesis
also serve a role in the NORAD/miR-590-3p axis under hypoxia, but
this was not investigated in the present study. TGF-β has been
shown to indirectly regulate angiogenesis and also be involved in a
miR-590 regulated pathway in cardiac diseases and NORAD-associated
functions in lung cancer (21–23,31,32).
Thus, TGF-β may also affect this NORAD/miR-590-3p regulated
hypoxia-induced angiogenesis, but further studies are required to
assess this.
The mechanisms regulating angiogenesis are
complicated and involve not only the regulation of angiogenic
indicators and chemokines, but also the participation of
endothelial cells, macrophages or pericytes during various
functions of organs under different conditions (5,12,33).
Hypoxia-induced diseases are primarily divided into two types:
Hypoxia-related ischemic diseases; and angiogenesis-caused cancer
types (5). Hypoxia/HIF-1-dependent
injuries or angiogenesis are highly associated with ischemic
diseases (5). A previous study
showed that ischemia/hypoxia-induced angiogenesis briefly improves
the blood supply to an ischemic heart, and that this phenomenon is
associated with innate immune system activation and macrophage
participation in the growing vessels, rather than upregulation of
traditional proangiogenic factors (12). However, the detailed mechanisms of
this effect have not been determined. Furthermore, a study
investigating the angiogenic response under hypoxia in vivo
showed that hypoxia-induced angiogenesis of endothelial cells was
modulated by a complex network of interactors, and included both
the regulation of angiogenic factors such as VEGF, and the
regulation of the expression of VEGFRs and VEGF inhibitors
(33). In the present study, the
regulatory mechanisms of hypoxia-induced angiogenesis in
endothelial cells via the NORAD/miR-590-3p axis in vitro
were assessed, thus identifying a novel pathway of regulation of
angiogenesis. However, in vivo studies are required to
confirm the relevance of this axis.
The present study identified the need to use
different strategies for treating hypoxia-induced diseases based on
the specifics of each case. Therapies targeting a single angiogenic
factor, such as VEGF, have been shown to be insufficient, which
highlights the requirement for the use of a combination of other
drugs to maintain or regulate angiogenesis (1,5,33).
Therefore, the hubs of hypoxia-induced angiogenesis should be
investigated further. The expression levels of several miRNAs have
been shown to be age-associated and cell type-specific (30,34),
which needs to be consideration clinically during angiogenic
therapy. While there are few previous studies on lncRNAs in
hypoxia-induced angiogenic regulation, the present results
suggested that lncRNA NORAD could be involved in angiogenic
activity of endothelial cells under hypoxic conditions, which may
be a potential target for the treatment of associated diseases,
such as ischemic heart disease.
In conclusion, the present results indicated that
lncRNA NORAD was upregulated, and miR-590-3p was downregulated, in
hypoxic HUVECs in vitro and MI left ventricular tissues
in vivo. Furthermore, downregulation of lncRNA NORAD in
HUVECs decreased cell viability and angiogenic activity, and
overexpression of miR-590-3p resulted in similar effects. It was
identified that knockdown of both NORAD and miR-590-3p reversed
this decrease, and resulted in enhanced angiogenic activity. NORAD
was also identified to target miR-590-3p and negatively regulate
its expression. It was demonstrated that miR-590-3p further
targeted the angiogenic factors VEGFA, FGF1 and FGF2. Therefore,
the present results suggested that a lncRNA NORAD/miR-590-3p axis
may be involved in the regulation of angiogenesis in HUVECs under
hypoxia, which highlights potential targets for treating
hypoxia-induced angiogenic diseases.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ designed the study, performed the experiments,
analysed and interpreted the data, and drafted the manuscript. XWe
and XWa performed the experiments and collected the data. GQ
designed and supervised the study, interpreted the data and
critically revised the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Animal experiments were performed in accordance with
the principles described in the Guide for The Care and Use of
Laboratory Animals, and were approved by The Ethics Committee of
The First Hospital of China Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Carmeliet P: Mechanisms of angiogenesis
and arteriogenesis. Nat Med. 6:389–395. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Risau W: Mechanisms of angiogenesis.
Nature. 386:671–674. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yancopoulos GD, Davis S, Gale NW, Rudge
JS, Wiegand SJ and Holash J: Vascular-specific growth factors and
blood vessel formation. Nature. 407:242–248. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferrara N: Vascular endothelial growth
factor: Molecular and biological aspects. Curr Top Microbiol
Immunol. 237:1–30. 1999.PubMed/NCBI
|
|
5
|
Zimna A and Kurpisz M: Hypoxia-inducible
factor-1 in physiological and pathophysiological angiogenesis:
Applications and therapies. Biomed Res Int. 2015:5494122015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greijer AE, van der Groep P, Kemming D,
Shvarts A, Semenza GL, Meijer GA, van de Wiel MA, Belien JA, van
Diest PJ and van der Wall E: Up-regulation of gene expression by
hypoxia is mediated predominantly by hypoxia-inducible factor 1
(HIF-1). J Pathol. 206:291–304. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Semenza GL: Angiogenesis in ischemic and
neoplastic disorders. Annu Rev Med. 54:17–28. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mori S, Tran V, Nishikawa K, Kaneda T,
Hamada Y, Kawaguchi N, Fujita M, Saegusa J, Takada YK, Matsuura N,
et al: A dominant-negative FGF1 mutant (the R50E mutant) suppresses
tumorigenesis and angiogenesis. PLoS One. 8:e579272013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Semenza GL: Oxygen sensing,
hypoxia-inducible factors, and disease pathophysiology. Annu Rev
Pathol. 9:47–71. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Go AS, Mozaffarian D, Roger VL, Benjamin
EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, et al:
Heart disease and stroke statistics-2013 update: A report from the
American heart association. Circulation. 127:e6–e245. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Murray CJ and Lopez AD: Mortality by cause
for eight regions of the world: Global burden of disease study.
Lancet. 349:1269–1276. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lavine KJ, Kovacs A, Weinheimer C and Mann
DL: Repetitive myocardial ischemia promotes coronary growth in the
adult mammalian heart. J Am Heart Assoc. 2:e0003432013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han P, Li W, Lin CH, Yang J, Shang C,
Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al: A long noncoding
RNA protects the heart from pathological hypertrophy. Nature.
514:102–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang K, Long B, Zhou LY, Liu F, Zhou QY,
Liu CY, Fan YY and Li PF: CARL lncRNA inhibits anoxia-induced
mitochondrial fission and apoptosis in cardiomyocytes by impairing
miR-539-dependent PHB2 downregulation. Nat Commun. 5:35962014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee S, Kopp F, Chang TC, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering PUMILIO proteins. Cell.
164:69–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu H, Li J, Koirala P, Ding X, Chen B,
Wang Y, Wang Z, Wang C, Zhang X and Mo YY: Long non-coding RNAs as
prognostic markers in human breast cancer. Oncotarget.
7:20584–20596. 2016.PubMed/NCBI
|
|
19
|
Li H, Wang X, Wen C, Huo Z, Wang W, Zhan
Q, Cheng D, Chen H, Deng X, Peng C and Shen B: Long noncoding RNA
NORAD, a novel competing endogenous RNA, enhances the
hypoxia-induced epithelial-mesenchymal transition to promote
metastasis in pancreatic cancer. Mol Cancer. 16:1692017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Miao Z, Guo X and Tian L: The long
noncoding RNA NORAD promotes the growth of gastric cancer cells by
sponging miR-608. Gene. 687:116–124. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kawasaki N, Miwa T, Hokari S, Sakurai T,
Ohmori K, Miyauchi K, Miyazono K and Koinuma D: Long noncoding RNA
NORAD regulates transforming growth factor-β signaling and
epithelial-to-mesenchymal transition-like phenotype. Cancer Sci.
109:2211–2220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ekhteraei-Tousi S, Mohammad-Soltani B,
Sadeghizadeh M, Mowla SJ, Parsi S and Soleimani M: Inhibitory
effect of hsa-miR-590-5p on cardiosphere-derived stem cells
differentiation through downregulation of TGFβ signaling. J Cell
Biochem. 116:179–191. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jafarzadeh M and Soltani BM:
Hsa-miR-590-5p interaction with SMAD3 transcript supports its
regulatory effect on The TGFβ signaling pathway. Cell J. 18:7–12.
2016.PubMed/NCBI
|
|
24
|
Nallamshetty S, Chan SY and Loscalzo J:
Hypoxia: A master regulator of microRNA biogenesis and activity.
Free Radic Biol Med. 64:20–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kumar MM and Goyal R: LncRNA as a
therapeutic target for angiogenesis. Curr Top Med Chem.
17:1750–1757. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rehmsmeier M, Steffen P, Hochsmann M and
Giegerich R: Fast and effective prediction of microRNA/target
duplexes. RNA. 10:1507–1517. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Guide for the Care and Use of Laboratory
Animals. Institute for Laboratory Animal Research. National
Academies Press. (Washington, DC). 2011.
|
|
29
|
Yang Z, Zhao Y, Lin G, Zhou X, Jiang X and
Zhao H: Noncoding RNA activated by DNA damage (NORAD): Biologic
function and mechanisms in human cancers. Clin Chim Acta. 489:5–9.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Madanecki P, Kapoor N, Bebok Z, Ochocka R,
Collawn JF and Bartoszewski R: Regulation of angiogenesis by
hypoxia: The role of microRNA. Cell Mol Biol Lett. 18:47–57. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Folkman J and Shing Y: Angiogenesis. J
Biol Chem. 267:10931–10934. 1992.PubMed/NCBI
|
|
32
|
Geng L, Chaudhuri A, Talmon G, Wisecarver
JL and Wang J: TGF-Beta suppresses VEGFA-mediated angiogenesis in
colon cancer metastasis. PLoS One. 8:e599182013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aplin AC and Nicosia RF: Hypoxia
paradoxically inhibits the angiogenic response of isolated vessel
explants while inducing overexpression of vascular endothelial
growth factor. Angiogenesis. 19:133–146. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Noren Hooten N, Abdelmohsen K, Gorospe M,
Ejiogu N, Zonderman AB and Evans MK: microRNA expression patterns
reveal differential expression of target genes with age. PLoS One.
5:e107242010. View Article : Google Scholar : PubMed/NCBI
|