Introduction
Spinal cord injury (SCI) covers various types of
damage to the spinal cord. According to the severity of injury, the
symptoms may vary, ranging from pain to complete loss of movement
and sensory function. SCI affects millions of people worldwide,
usually for life (1). To date, there
is no effective treatment. Therefore, finding new treatment methods
for patients with SCI is crucial. However, exploring the
pathogenesis of SCI and finding effective treatment strategies has
been a great challenge for researchers.
Previous studies have suggested that, in injured
spinal cords, the inflammasome can activate inflammatory caspases
and cytokines of the interleukin (IL)-1 family (IL-1β and IL-18) by
identifying host-derived damage-associated molecular patterns
(2–4). VX-765, also known as Belnacasan, is an
inhibitor of IL-1-converting enzyme (caspase-1), which controls the
generation of IL-1β and IL-18 (5–7). VX-765
has been shown to inhibit acute seizures and chronic epilepsy in
preclinical models (8). Therefore,
using VX-765 to inhibit caspase-1, the common converting enzyme of
these two inflammatory factors, in the acute stage of SCI might be
an effective anti-inflammatory intervention. However, the exact
mechanism is not entirely clear. The aim of the present study was
to use VX-765 8 h after SCI, in order to analyze the transcription
of the local genes, using RNA-sequencing (RNA-Seq). Next, through
bioinformatics analysis and reverse transcription-quantitative PCR
(RT-qPCR), key molecular and signaling pathways were screened and
identified, providing a new theoretical and experimental basis for
SCI clinical treatment.
Materials and methods
Animals
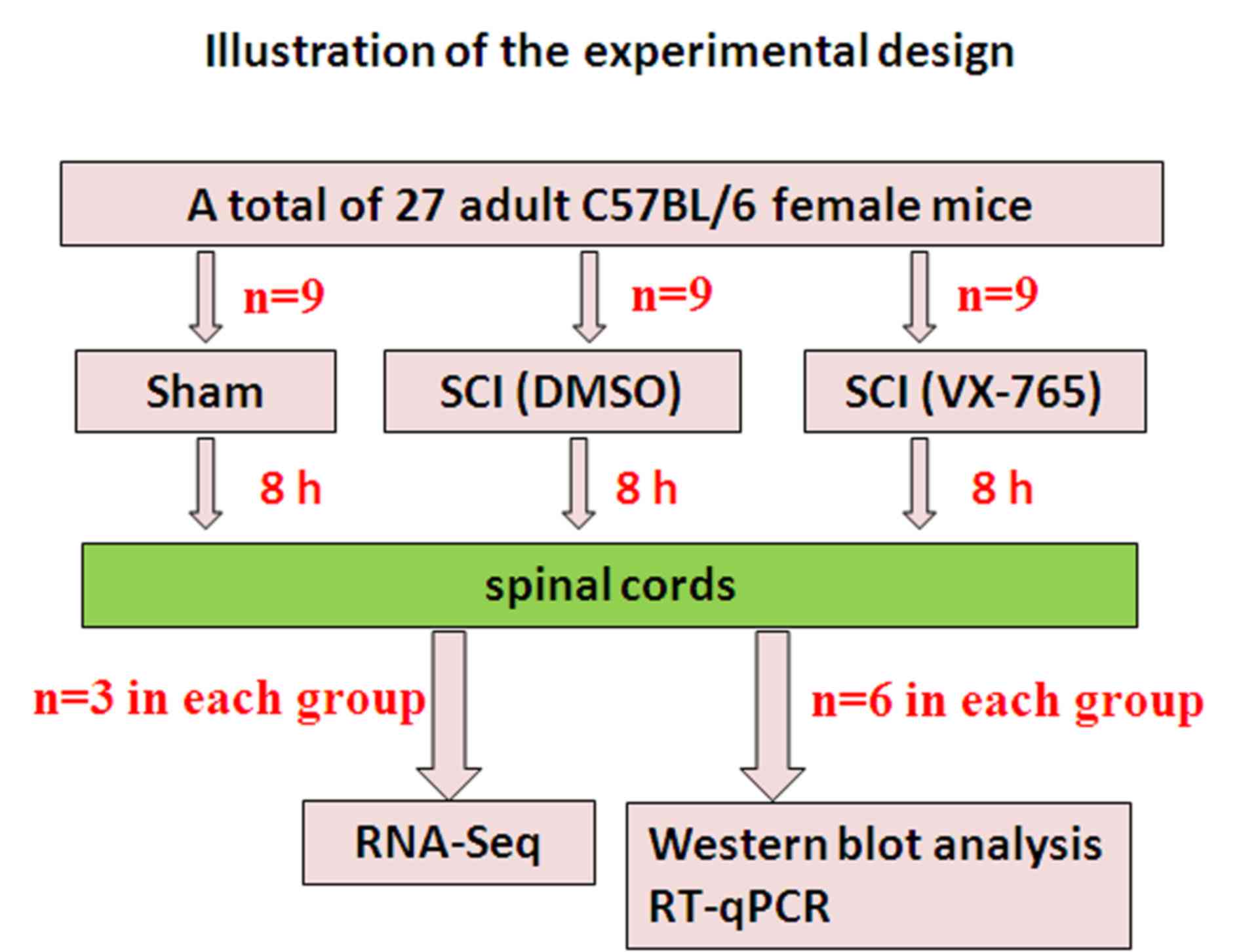
A total of 27 healthy and clean C57BL/6 female mice
(weight, 18–20 g; age, 8 weeks old; Chang Zhou Cavens Laboratory
Animal Ltd.) were used in this study (Fig. 1). Animal care following surgery
complied with the regulations for the management of experimental
animals (revised by the Ministry of Science and Technology of China
in June 2004). The study was approved by the Institutional
Committee on Animal Care, Use and Research of the Bengbu Medical
College (approval no. 2017037).
Contusive SCI and drug injection
An Infinite Horizon impactor (Precision Systems
& Instrumentation) was used to perform contusive SCI, as
previously described (9–11). The mice were first anesthetized with
pentobarbital sodium (50 mg/kg, intraperitoneally) and then the T9
lamina was excised. The spine was stabilized by clamping the T7 and
T11 spinous processes, and then a moderate SCI model was created
using a rod (1.3 mm in diameter) with a force of 50 Kdynes and a
dwell time of 0 sec. Sham-operated (sham) mice only received a
laminectomy without contusive injury.
The spinal cord-injured mice were randomly assigned
to the DMSO control or VX-765 injection groups (9 mice in every
group). Mice were intraperitoneally injected with DMSO or VX-765
(100 mg/kg prepared in DMSO) immediately following injury. Since
the aim was to investigate the effect of VX-765 on local gene
transcription in the acute stage of SCI, all specimens were
collected 8 h after SCI.
Identification of the effect of VX-765
on caspase-1 and 3 activities in injured spinal cords by western
blot analysis
A total of 8 h following surgery, the mice were
euthanized referring to previous references (12,13) with
an overdose of pentobarbital sodium (80 mg/kg, intraperitoneally)
and perfused with 10 ml PBS. An overdose of pentobarbital sodium
can make the mice lose consciousness within 2 min with minimal pain
and distress. Subsequent PBS perfusion can make animals die (no
spontaneous breathing and blink reflex) within 2–3 min with no pain
and distress, while spinal cord samples were taken (0.5 cm,
including the injury center, n=6 in every group). Total protein was
extracted from the spinal cords using a mammalian protein
extraction kit (cat. no. C600589; Sangon Biotech Co., Ltd.) and
western blot analysis was performed as previously described
(14). For the caspase-3 analysis,
the primary antibodies used were rabbit anti-β-actin (1:2,000; cat
no. BL005B; Biosharp) and rabbit anti-caspase-3 antibody (1:1,000;
cat no. ab13847; Abcam). The secondary antibody used was
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG
(1:10,000; cat. no. BL003A; Biosharp). As the molecular weights of
β-actin and pro-caspase-1 are similar (15), when western blot analysis was
performed, rabbit monoclonal anti-caspase-1+p10+p12 antibody
(1:1,000; cat. no. ab179515; Abcam) was first used to incubate for
12 h at 4°C and detect protein levels, then the restore western
blot stripping buffer (cat. no. 21059; Thermo Fisher Scientific,
Inc.) was used to wash off the antibody on the membrane. For the
stripping, the blot was placed in 20 ml restore western blot
stripping buffer and incubated for 15 min at 37°C. Then, the blot
was removed and washed 5 min with PBS three times. To test for
complete removal of the HRP labeled secondary antibody, the
membrane was incubated with a SuperSignal West Working solution
(cat. no. 34095; Thermo Fisher Scientific, Inc.) for 5 min at room
temperature and exposed to film. If no signal is detected using a
5-min exposure, the HRP-conjugated secondary antibody had been
successfully removed. To test for complete removal of the primary
antibody, the membrane was incubated with the HRP-labeled secondary
antibody, followed by washing 5 min in 1 X TBST buffer containing
0.01 M Tris, 0.15 M NaCl and 0.1% Tween-20 (cat. no. C520009;
Sangon Biotech Co., Ltd.) three times. Then, membrane was incubated
in SuperSignal West Working Solution for 5 min at room temperature
and exposed to film again. If no signal is detected with a 5-min
exposure, the primary antibody has been successfully removed from
the antigen. After determining that the membrane was properly
stripped, the second immunoprobing experiment of anti-β-actin
antibody was performed as previously described (14).
RNA isolation, quantification and
qualification
A total of 8 h following surgery, mice were
euthanized with an overdose of pentobarbital sodium (80 mg/kg,
intraperitoneally) (12,13) and perfused with 10 ml PBS, and the
spinal cords (0.5 cm, including the injury center) were removed.
Total RNA was extracted from spinal cords and purified, as
previously described (16).
Library preparation and transcriptome
sequencing
The sequencing libraries were produced using
NEBNext® Ultra™ RNA Library Prep kit for
Illumina® (New England Biolabs) as previously described
(13). Finally, the 125-bp/150-bp
paired-end reads were obtained and sequenced on an Illumina Hiseq
platform. The sequence data have been deposited into Sequence Read
Archive (https://www.ncbi.nlm.nih.gov/sra/PRJNA548970).
Differentially expressed gene (DEG)
analysis
Prior to DEG analysis, the gene expression
statistics were analyzed using RSEM software (v1.3.1; http://deweylab.biostat.wisc.edu/rsem/)
to convert the read count numbers to Fragments Per Kilobase of
transcript per Million fragments mapped (FPKM) and Principal
Component Analysis (PCA) was made to determine the similarities and
differences in the data. Differential gene expression in the three
groups was analyzed as previously described (16), using the DESeq software (http://www.bioconductor.org/). Benjamini and
Hochberg's approach was used to control the false discovery rate
and adjust the P-values. An adjusted P<0.05 was defined as a
standard for significant differences in gene expression.
Gene ontology (GO) and kyoto
encyclopedia of genes and genomes (KEGG) enrichment analysis of
DEGs
GO and KEGG analysis was performed using GOseq R
package and KOBAS software, as previously described (16).
RT-qPCR
To validate RNA-Seq results, nine DEGs were randomly
selected and verified by RT-qPCR, as previously described (16). PCR primer sequences are listed in
Table I. The relative quantitative
results of each group (n=6) of genes were calculated according to
the 2−ΔΔCq formula (17).
 | Table I.PCR primers used in the study. |
Table I.
PCR primers used in the study.
| Gene | GenBank Accession
no. | Forward primer
5′→3′ | Reverse primer
5′→3′ |
|---|
| Caspase-1 | NM_009807.2 |
CGTACACGTCTTGCCCTCAT |
GGGCAGGCAGCAAATTCTTT |
| IL-1 | NM_008361.4 |
ACAACTGCACTACAGGCTCC |
TGGGTGTGCCGTCTTTCATT |
| LCN2 | NM_008491.1 |
ACAACCAGTTCGCCATGGTAT |
AAGCGGGTGAAACGTTCCTT |
| Nlrp3 | NM_145827 |
GACCGTGAGGAAAGGACCAG |
GGCCAAAGAGGAATCGGACA |
| Pecam1 | XM_021175967.1 |
GTACCAATCCAGGTGTGCGA |
TTTTCGGACTGGCAGCTGAT |
| CD34 | NM_133654.3 |
ACCACAGACTTCCCCAACTG |
CATATGGCTCGGTGGGTGAT |
| Gbp7 | BK005760 |
GGACGTGTCATCACAGCAGA |
CCAACTGGTCCTCTGGCATT |
| Icosl | NM_015790.3 |
GAACCCACAGGAAACCCACA |
GTATAGCTTCGGTGGGGACG |
| Hpdg | XM_021170107.1 |
CTTCGAAGCACGGCATCATC |
TGGCAATGGTTGATGGGTGTA |
| Ccl9 | NM_011338.2 |
CAGGCCGGGCATCATCTTTA |
TGGCAGTTCACACCCTTCTC |
| Rel | NM_009044.2 |
TACTCGGCCTCTGAGTGTGA |
GGCCTAGCCTGGCATTACAT |
Statistical analysis
Statistical values of western blot analysis and
RT-qPCR (n=6/group) were presented as mean ± standard deviation.
The data were analyzed using one-way analysis of variance followed
by Student-Newman-Keuls tests. P<0.05 was considered to indicate
a statistically significant difference. All tests were performed
using SPSS 16.0 (SPSS, Inc.).
Results
Effect of VX-765 on caspase-1 activity
in injured spinal cords
To verify the effect of VX-765 on caspase-1 activity
in injured spinal cords, the homogenate extracts obtained from DMSO
and VX-765-treated spinal cords were detected by western blot
analysis. As the molecular weights of β-actin and pro-caspase-1 are
similar (14), when western blot
analysis was carried out, anti-caspase-1+p10+p12 antibody was first
used to incubate and detect protein levels, and then stripping
buffer was used to wash off the antibody on the membrane. Then,
anti-β-actin antibody incubation and detection was carried out. As
shown in Fig. 2A and B, no
significant differences were observed in the 43 and 45 kDa
pro-caspase-1 bands and 31 kDa pro-caspase-3 band. However, the 10
and 12 kDa caspase-1 bands (Fig. 2A)
and the 17 and 12 kDa caspase-3 bands (Fig. 2B) in the SCI (DMSO) group were
significantly increased compared with those in the sham group
(P<0.05). In VX-765 group, these bands (Fig. 2A and B) were significantly decreased
compared with those in the DMSO control group (P<0.05). Fig. 2C and D show a significant difference
among the three groups (n=6, P<0.01 or 0.05). These results
showed that SCI can induce the activities of caspase-1 and 3, and
VX-765 can inhibit their activities in injured spinal cords.
Identification of expressed
transcripts in the mouse spinal cords
For the quality assessment of sequencing data, nine
cDNA libraries were established, including Sham (Sham1, Sham2 and
Sham3), SCI (DMSO; SCI_C1, SCI_C2 and SCI_C3) and SCI (VX-765;
SCI_V1, SCI_V2 and SCI_V3). RNA-Seq produced 48,440,020-62,676,868
raw reads for each sample. After filtering out the low-quality
reads, the clean reads were from 47,736,336-61,993,808 (97.6–3.92%;
Table II).
 | Table II.Summary of sequence assembly after
Illumina sequencing. |
Table II.
Summary of sequence assembly after
Illumina sequencing.
| Sample name | Raw reads | Clean reads | Clean bases | Error rate (%) | Q20 (%) | Q30 (%) | GC content (%) |
|---|
| Sham1 | 56509230 | 55796658 | 8.37G | 0.03 | 97.73 | 93.95 | 51.23 |
| Sham2 | 48848744 | 48226002 | 7.23G | 0.03 | 97.60 | 93.67 | 51.71 |
| Sham3 | 58228350 | 57459748 | 8.62G | 0.03 | 97.67 | 93.78 | 51.42 |
| SCI_C1 | 56857568 | 56047834 | 8.41G | 0.03 | 97.80 | 94.08 | 50.96 |
| SCI_C2 | 52518750 | 51674904 | 7.75G | 0.03 | 97.80 | 94.11 | 50.99 |
| SCI_C3 | 62676868 | 61993808 | 9.30G | 0.03 | 97.80 | 94.09 | 51.09 |
| SCI_V1 | 48440020 | 47736336 | 7.16G | 0.02 | 97.92 | 94.39 | 51.23 |
| SCI_V2 | 55927984 | 54923592 | 8.24G | 0.03 | 97.86 | 94.31 | 51.33 |
| SCI_V3 | 59647724 | 59047622 | 8.86G | 0.03 | 97.90 | 94.30 | 51.09 |
In order to identify the source of variation in the
original data, PCA analysis was conducted. As shown in Fig. 3, PC1, PC2 and PC3 were 45.22, 24.13
and 6.20%, respectively, demonstrating that the data could be used
for the next analysis.
Effect of VX-765 treatment on gene
expression
FPKM and DESeq were used to analyze the gene
expression level and differential expression profiles,
respectively. The results showed that compared with the sham group
there were 4,476 DEGs in the SCI (DMSO) group, including 2,899 up-
and 1,577 downregulated genes (Fig.
4A and Table SI). As compared
with the SCI (DMSO) group, there were 2,899 DEGs in the SCI
(VX-765) group, 1,137 of which were up- and 1,762 were
downregulated (Fig. 4B and Table SI).
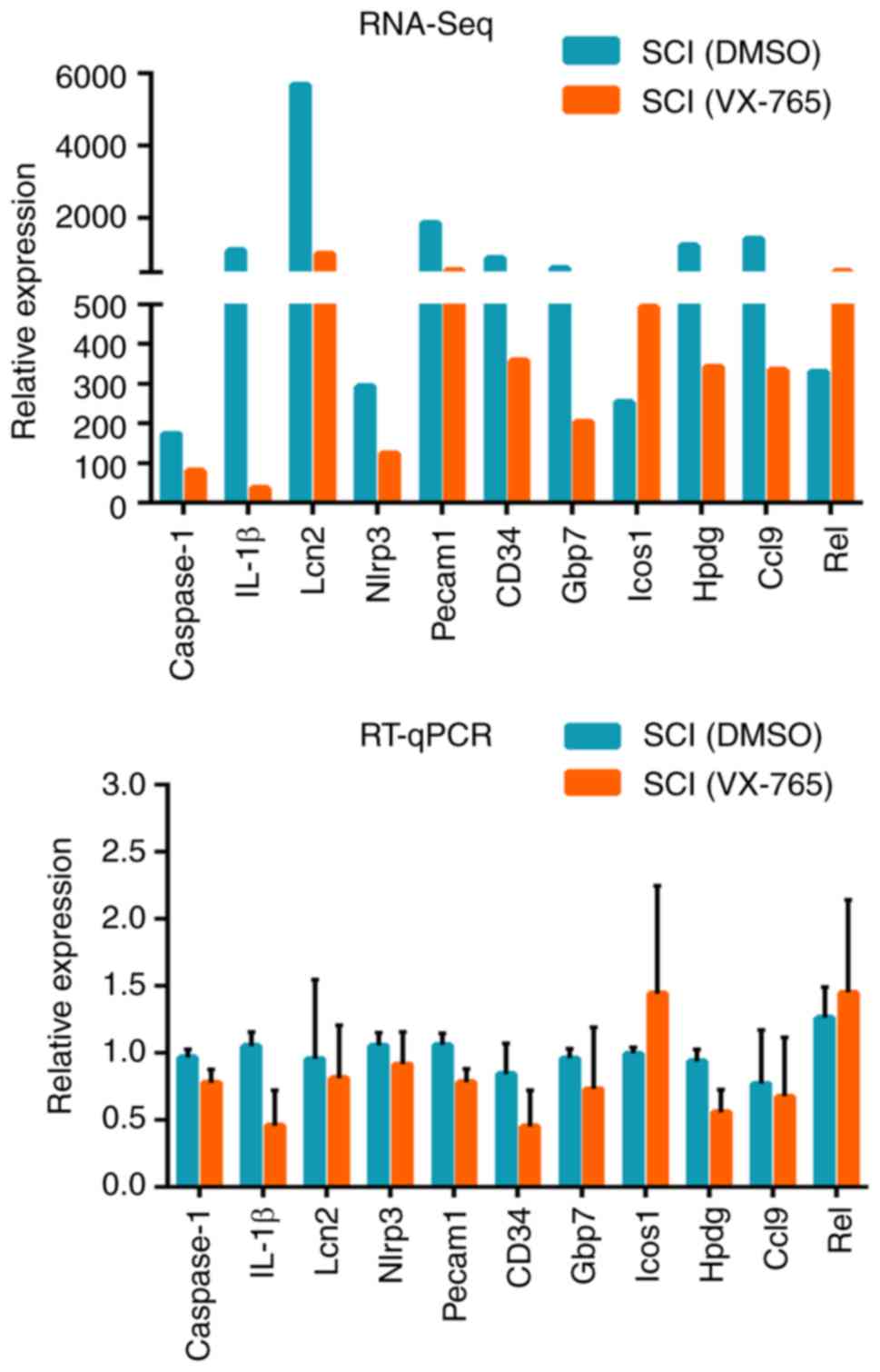
RT-qPCR identification of DEGs
In order to verify the RNA-Seq results, 11 DEGs were
randomly selected from the SCI (VX-765) group, as compared with the
SCI (DMSO) group, namely caspase-1, IL-1β, Lipocalin-2 (Lcn2),
nucleotide-binding oligomerization domain, leucine-rich repeat and
pyrin domain-containing 3 (Nlrp3), platelet endothelial cell
adhesion molecule (Pecam1), CD34, guanylate binding protein 7
(Gbp7), inducible T-cell COStimulator (Icos1),
15-hydroxyprostaglandin dehydrogenase (Hpdg), chemokine (C-C motif)
ligand 9 (Ccl9) and Rel. The RNA-Seq and RT-qPCR results indicated
that the expression patterns of these DEGs were similar (Fig. 5).
DEG cluster analysis
The DEGs from different groups were analyzed using
FPKM hierarchical cluster analysis. As shown in Fig. 6, DEGs were classified into different
expression clusters by hierarchical clustering. These clusters
contained up- or downregulated DEGs. Most upregulated DEGs in the
SCI (DMSO) group compared with the sham group were in the middle
cluster, while downregulated genes were observed in the upper and
lower clusters. Compared with the sham group, most upregulated DEGs
in the SCI (VX-765) group were in the middle and upper-lower
clusters, while downregulated genes were mainly observed in the
lower cluster. Compared with the SCI (DMSO) group, some upregulated
DEGs in the SCI (VX-765) group were observed in the upper cluster,
while downregulated DEGs were observed in the lower cluster; there
were also clusters with no significant differences at the
bottom.
GO enrichment analysis of DEGs
Compared with the sham group, 91 GO terms in
upregulated DEGs (Fig. 7A, Table SII) and 18 GO terms in downregulated
DEGs (Fig. 7B, Table SIII) were found in the SCI (DMSO)
group. In the SCI (VX-765) group, one GO term in upregulated DEGs
(Fig. 7C, Table SII) and ten GO terms in
downregulated DEGs (Fig. 7D,
Table SIII) were found, compared
with the SCI (DMSO) group. In the SCI (DMSO) group, the
downregulated DEGs were most enriched in protein binding,
extracellular-glutamate-gated ion channel activity, transmembrane
transporter activity, compared with the sham group. The upregulated
DEGs were most enriched in anion binding, ribonucleotide binding,
purine ribonucleoside triphosphate binding, intracellular signal
transduction and chemokine activity. In the SCI (VX-765) group, the
downregulated DEGs were most related to binding, signal
transduction, transferase, sialyltransferase and NAD+
ADP-ribosyltransferase activity, compared with the SCI (DMSO)
group. The upregulated DEGs were only enriched in one term, which
was binding.
KEGG enrichment analysis of DEGs
Scatter plots were used to express the KEGG
enrichment analysis results for the DEGs. As compared with the sham
group, the upregulated DEGs in the SCI (DMSO) group were most
enriched in focal adhesion, apoptosis, tumor necrosis factor,
nuclear factor (NF)-κB, Toll-like receptor, phosphatidylinositol
3-kinase (PI3K) protein kinase B (Akt), NOD-like receptor, mitogen
associated protein kinase and p53 signaling pathway (Fig. 8A, Table
SIV). The downregulated DEGs were most enriched in
glutamatergic synapse, endocytosis, Rap1, Hippo and the Ras
signaling pathway (Fig. 8B, Table SV). In the SCI (VX-765) group, no
enriched signaling pathways were found in the upregulated DEGs
(Fig. 8C, Table SIV), compared with the SCI (DMSO)
group. The downregulated DEGs were most enriched in focal adhesion,
cytokine-cytokine receptor interaction, leukocyte transendothelial
migration, extracellular matrix (ECM)-receptor interaction,
PI3K-Akt, Rap1 and hypoxia inducible factor (HIF)-1 signaling
pathway (Fig. 8D, Table SV).
Discussion
It has been proved that the activation of caspase-1
needs the recruitment of pro-caspase-1 into the inflammasome and
the initiation of its self-cleavage. Activated caspase-1 can
further cleave and activate IL-1 family cytokines (such as IL-1β
and IL-18), leading to downstream inflammatory cascades (18,19).
VX-765 is reported to be a potent and selective inhibitor of
caspase-1 (5–7). Although the mechanism is still unclear,
this process must be related to inhibiting pro-caspase-1
recruitment to the inflammasome and self-cleavage. Previous studies
have shown that the activation of caspase-1 and the resulting
expression and activation of IL-1β and IL-18 occur at the site of
SCI (2,20,21).
Since it is a potent and selective inhibitor, the present study
hypothesized that using VX-765 to inhibit caspase-1 in the acute
stage of SCI might be an effective anti-inflammatory and
anti-apoptotic intervention. To prove this hypothesis, RNA-Seq was
used in the present study as a detection method to explore the
effects of VX-765 on genome-wide transcription in SCI. To identify
the effectiveness of VX-765 on caspase-1, western blot analysis was
used to detect the pro-caspase-1 and activated caspase-1. The
results showed that VX-765 can inhibit caspase-1 activity in
injured spinal cords. At the same time, caspase-3, an essential
regulator of programmed cell death through apoptosis, was also
detected. The results showed that the inhibition of caspase-1
activity can indeed play an anti-apoptotic role. However, these
studies are limited and require a high-throughput experiment to
confirm these hypotheses. Next, the effects of VX-765 on the local
gene transcription of injured spinal cords were characterized by
RNA-Seq. Before analyzing the data, the cDNA library quality was
examined. >97% of the tags were clean and PCA analysis showed
that the variation was low. These showed that the cDNA library
could be used for the next functional analysis.
The results of RNA-Seq showed that, as compared with
the sham group, there were 4,476 DEGs in the SCI (DMSO) group,
including 2,899 upregulated and 1,577 downregulated. These were
consistent with the present study and other previous reports
(16,22), suggesting that the results of this
experiment are reliable. As compared with the SCI (DMSO) group,
there were 2,899 DEGs in the SCI (VX-765) group, 1,137 of which
were upregulated and 1,762 downregulated. To further verify the
RNA-seq results, 11 DEGs (caspase-1, IL-1β, Lcn2, Nlrp3, Pecam1,
CD34, Gbp7, Icos1, Hpdg, Ccl9 and Rel) were selected for RT-qPCR.
The results showed that the expression patterns of these genes
detected by these two methods were similar. These demonstrated that
the present RNA-seq results are reliable and can be used for
subsequent analysis. These also confirmed that VX-765 can inhibit
the expression and activation of caspase-1 and its downstream
inflammatory factors. In theory, to be a potent and selective
inhibitor of interleukin-converting enzyme/caspase-1, VX-765 can't
directly inhibit other types of proteins such as matrix
metalloproteinases (MMPs). However, in the present RNA-Seq results,
1,762 downregulated genes including MMPs (MMP3, MMP8, MMP11 and
MMP16) were found. Although, the possibility of VX-765 inhibiting
other types of proteins can't be excluded in this study, it
suggests that VX-765 may indirectly inhibit the expression and
activity of these proteins by inhibiting caspase-1. This is an
interesting topic, which deserves further discussion.
In order to further analyze the DEGs effected by
VX-765, GO enrichment analysis was used to determine the
distribution of DEGs and enriched cell component, molecular
function and biological process GO terms (23). In the SCI (VX-765) group, the
downregulated DEGs were most enriched in binding, signal
transduction, transferase, sialyltransferase, NAD+
ADP-ribosyltransferase activity, compared with the SCI (DMSO)
group. The upregulated DEGs were only enriched in one term, which
was binding. Next, KEGG was used to analyze signaling pathways
associated with treatment of SCI with VX-765. In THE SCI (VX-765)
group, no enriched signaling pathways were found in the upregulated
DEGs, as compared with the SCI (DMSO) group. The downregulated DEGs
were most enriched in focal adhesion, cytokine-cytokine receptor
interaction, leukocyte transendothelial migration, ECM-receptor
interaction, PI3K-Akt, Rap1 and HIF-1 signaling pathway.
Focal adhesions are specialized intracellular sites
in which aggregated integrin receptors interact with extracellular
matrices, while extracellular matrices interact with the
intracellular actin cytoskeleton (24,25).
Focal adhesions are the result of cell-ECM interactions (24,26). The
ECM plays an important role in tissue and organ morphogenesis
(27,28), as well as the control of cellular
activities such as adhesion, migration, differentiation,
proliferation and apoptosis (29).
Cytokines are important regulators and mobilizing factors between
cells, which are involved in inflammation, defense, cell growth,
differentiation, death and repair processes (30,31).
Following VX-765 treatment, the cytokine receptor interaction was
inhibited in the sequencing results, involving 56 related genes,
such as Csf3, Ifngr1, Ltbr, Tgfbr2 and Il6st. As expected,
leukocyte transendothelial migration was also inhibited. During
leukocyte division, leukocytes bind to endothelial cell adhesion
molecules and then migrate between vascular endothelial cells
(32). The migration or infiltration
of white blood cells from the blood into tissues is critical to
immune surveillance and inflammation (33–35). ECM
downregulated in SCI following the injection of VX-765, indicating
that VX-765 improves SCI by inhibiting adhesion, migration,
differentiation, proliferation and apoptosis through the
downregulation of caspase-1 activity.
It has been reported that Akt signaling mediates a
variety of extracellular and intracellular signal transduction
pathways that regulate macrophage biology, including the production
of pro-inflammatory cytokines, phagocytosis, autophagy and
homeostasis (36). The PI3K-Akt
signaling pathway was downregulated in SCI following the injection
of VX-765, indicating that VX-765 can improve SCI by regulating
macrophages and inhibiting inflammatory pathways.
The Rap1 signaling pathway plays an important role
in regulating cell-cell and cell-matrix interactions by regulating
the function of adhesion molecules (37,38). In
the present study, the Rap1 signaling pathway was enriched in
downregulated DEGs of SCI following the injection of VX-765,
suggesting that VX-765 may inhibit cell adhesion and polarization
by inhibiting the Rap1 signaling pathway, thereby inhibiting
inflammation.
HIF-1α is a specific transcription factor that is
activated under hypoxic conditions and its regulated signaling
pathway is the backbone of hypoxia signaling (39,40).
Immune cells accumulate at the site of inflammation, leaving a
rapidly hypoxic environment, which, in turn, causes immune cells to
transcribe HIF. In addition, NF-κB positively regulates HIF-1α
expression. The signaling pathway of HIF-1α and the upstream NF-κB
was inhibited following VX-765 treatment, demonstrating that VX-765
improves the local hypoxic environment of SCI.
In conclusion, the present results demonstrated that
VX-765 can lead to gene expression inhibition in acutely injured
spinal cords by inhibiting caspase-1. These downregulated DEGs and
their associated signaling pathways, including focal adhesion,
cytokine-cytokine receptor interaction, leukocyte transendothelial
migration, ECM-receptor interaction, PI3K-Akt, Rap1 and HIF-1
signaling pathway, are mainly associated with the inflammatory
response, local hypoxia, macrophage differentiation, adhesion
migration and apoptosis in local cells. This suggests that the
application of VX-765 in the acute phase of SCI can improve the
local microenvironment of SCI by inhibiting caspase-1. However,
whether VX-765 can be used as a therapeutic drug for SCI requires
further exploration. Next, detailed research on this subject will
be conducted by combining animal models and clinical practice.
Supplementary Material
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81772321
and 81571194).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
HZL and JGH participated in the design of the study.
JC, SNW and YQC performed experimental procedures. FXD, YJS and SQD
conducted data analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Committee on Animal Care, Use and Research of the Bengbu Medical
College (Approval no. 2017037).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Friedli L, Rosenzweig ES, Barraud Q,
Schubert M, Dominici N, Awai L, Nielson JL, Musienko P, Nout-Lomas
Y, Zhong H, et al: Pronounced species divergence in corticospinal
tract reorganization and functional recovery after lateralized
spinal cord injury favors primates. Sci Transl Med. 7:302ra1342015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
de Rivero Vaccari JP, Dietrich WD and
Keane RW: Activation and regulation of cellular inflammasomes: Gaps
in our knowledge for central nervous system injury. J Cereb Blood
Flow Metab. 34:369–375. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krakauer T: Inflammasomes, autophagy, and
cell death: The trinity of innate host defense against
intracellular bacteria. Mediators Inflamm. 2019:24712152019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li L, Tang W and Yi F: Role of
inflammasome in chronic kidney disease. Adv Exp Med Biol.
1165:407–421. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stack JH, Beaumont K, Larsen PD, Straley
KS, Henkel GW, Randle JC and Hoffman HM: IL-converting
enzyme/caspase-1 inhibitor VX-765 blocks the hypersensitive
response to an inflammatory stimulus in monocytes from familial
cold autoinflammatory syndrome patients. J Immunol. 175:2630–2634.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y and Zheng Y: Effects and
mechanisms of potent caspase-1 inhibitor VX765 treatment on
collagen-induced arthritis in mice. Clin Exp Rheumatol. 34:111–118.
2016.PubMed/NCBI
|
|
7
|
Flores J, Noel A, Foveau B, Lynham J,
Lecrux C and LeBlanc AC: Caspase-1 inhibition alleviates cognitive
impairment and neuropathology in an Alzheimer's disease mouse
model. Nat Commun. 9:39162018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maroso M, Balosso S, Ravizza T, Iori V,
Wright CI, French J and Vezzani A: Interleukin-1β biosynthesis
inhibition reduces acute seizures and drug resistant chronic
epileptic activity in mice. Neurotherapeutics. 8:304–315. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu J, Zhao Z, Kumar A, Lipinski MM, Loane
DJ, Stoica BA and Faden AI: Endoplasmic reticulum stress and
disrupted neurogenesis in the brain are associated with cognitive
impairment and depressive-like behavior after spinal cord injury. J
Neurotrauma. 33:1919–1935. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Horiuchi H, Oshima Y, Ogata T, Morino T,
Matsuda S, Miura H and Imamura T: Evaluation of injured axons using
two-photon excited fluorescence microscopy after spinal cord
contusion injury in YFP-H line mice. Int J Mol Sci. 16:15785–15799.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Galvan MD, Luchetti S, Burgos AM, Nguyen
HX, Hooshmand MJ, Hamers FP and Anderson AJ: Deficiency in
complement C1q improves histological and functional locomotor
outcome after spinal cord injury. J Neurosci. 28:13876–13888. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu JQ, Yang D and Folz RJ: A novel
bronchial ring bioassay for the evaluation of small airway smooth
muscle function in mice. Am J Physiol Lung Cell Mol Physiol.
291:L281–L288. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Song XY, Zhou FH, Zhong JH, Wu LL and Zhou
XF: Knockout of p75(NTR) impairs re-myelination of injured sciatic
nerve in mice. J Neurochem. 96:833–842. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mehto S, Jena KK, Nath P and Chauhan S,
Kolapalli SP, Das SK, Sahoo PK, Jain A, Taylor GA and Chauhan S:
The Crohn's disease risk factor IRGM limits NLRP3 inflammasome
activation by impeding its assembly and by mediating its selective
autophagy. Mol Cell. 73:429–445.e27. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin YH, Wu Y, Wang Y, Yao ZF, Tang J, Wang
R, Shen L, Ding SQ, Hu JG and Lü HZ: Spatio-temporal expression of
Hexokinase-3 in the injured female rat spinal cords. Neurochem Int.
113:23–33. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shi LL, Zhang N, Xie XM, Chen YJ, Wang R,
Shen L, Zhou JS, Hu JG and Lü HZ: Transcriptome profile of rat
genes in injured spinal cord at different stages by RNA-sequencing.
BMC Genomics. 18:1732017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Martinon F, Burns K and Tschopp J: The
inflammasome: A molecular platform triggering activation of
inflammatory caspases and processing of proIL-beta. Mol Cell.
10:417–426. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Latz E, Xiao TS and Stutz A: Activation
and regulation of the inflammasomes. Nat Rev Immunol. 13:397–411.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mortezaee K, Khanlarkhani N, Beyer C and
Zendedel A: Inflammasome: Its role in traumatic brain and spinal
cord injury. J Cell Physiol. 233:5160–5169. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zendedel A, Monnink F, Hassanzadeh G,
Zaminy A, Ansar MM, Habib P, Slowik A, Kipp M and Beyer C: Estrogen
attenuates local inflammasome expression and activation after
spinal cord injury. Mol Neurobiol. 55:1364–1375. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen K, Deng S, Lu H, Zheng Y, Yang G, Kim
D, Cao Q and Wu JQ: RNA-seq characterization of spinal cord injury
transcriptome in acute/subacute phases: A resource for
understanding the pathology at the systems level. PLoS One.
8:e725672013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang Q, Wu LY, Wang Y and Zhang XS: GOMA:
Functional enrichment analysis tool based on GO modules. Chin J
Cancer. 32:195–204. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Burridge K: Focal adhesions: A personal
perspective on a half century of progress. FEBS J. 284:3355–3361.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
LaFlamme SE, Mathew-Steiner S, Singh N,
Colello-Borges D and Nieves B: Integrin and microtubule crosstalk
in the regulation of cellular processes. Cell Mol Life Sci.
75:4177–4185. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
De Pascalis C and Etienne-Manneville S:
Single and collective cell migration: The mechanics of adhesions.
Mol Biol Cell. 28:1833–1846. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Rabelink TJ, van den Berg BM, Garsen M,
Wang G, Elkin M and van der Vlag J: Heparanase: Roles in cell
survival, extracellular matrix remodelling and the development of
kidney disease. Nat Rev Nephrol. 13:201–212. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bonnans C, Chou J and Werb Z: Remodelling
the extracellular matrix in development and disease. Nat Rev Mol
Cell Biol. 15:786–801. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yue B: Biology of the extracellular
matrix: An overview. J Glaucoma. 23 (8 Suppl 1):S20–S23. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roy S, Rizvi ZA and Awasthi A: Metabolic
checkpoints in differentiation of helper T cells in tissue
inflammation. Front Immunol. 9:30362019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ridiandries A, Tan JTM and Bursill CA: The
role of chemokines in wound healing. Int J Mol Sci. 19(pii):
E32172018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang JG, Williams JC, Davis BK, Jacobson
K, Doerschuk CM, Ting JP and Mackman N: Monocytic microparticles
activate endothelial cells in an IL-1β-dependent manner. Blood.
118:2366–2374. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Prendergast CT and Anderton SM: Immune
cell entry to central nervous system-current understanding and
prospective therapeutic targets. Endocr Metab Immune Disord Drug
Targets. 9:315–327. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shechter R, London A and Schwartz M:
Orchestrated leukocyte recruitment to immune-privileged sites:
Absolute barriers versus educational gates. Nat Rev Immunol.
13:206–218. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Demeestere D, Libert C and Vandenbroucke
RE: Clinical implications of leukocyte infiltration at the choroid
plexus in (neuro)inflammatory disorders. Drug Discov Today.
20:928–941. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vergadi E, Ieronymaki E, Lyroni K,
Vaporidi K and Tsatsanis C: Akt signaling pathway in macrophage
activation and M1/M2 polarization. J Immunol. 198:1006–1014. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim C, Ye F and Ginsberg MH: Regulation of
integrin activation. Annu Rev Cell Dev Biol. 27:321–345. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pollan SG, Huang F, Sperger JM, Lang JM,
Morrissey C, Cress AE, Chu CY, Bhowmick NA, You S, Freeman MR, et
al: Regulation of inside-out β1-integrin activation by CDCP1.
Oncogene. 37:2817–2836. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dehne N, Fuhrmann D and Brune B:
Hypoxia-inducible factor (HIF) in hormone signaling during health
and disease. Cardiovasc Hematol Agents Med Chem. 11:125–135. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Risbud MV and Shapiro IM: Notochordal
cells in the adult intervertebral disc: New perspective on an old
question. Crit Rev Eukaryot Gene Expr. 21:29–41. 2011. View Article : Google Scholar : PubMed/NCBI
|