Introduction
Spinal cord injury (SCI) is a serious neurological
injury caused by traffic accidents, falls, or violence-associated
injuries, resulting in heavy financial and psychosocial burdens on
patients and society (1–4). SCI can lead to impaired sensory and
motor functions, autonomic nervous dysfunction and altered mental
health (1). To treat SCI more
effectively, scientific research has focused on its underlying
pathological mechanisms, which involve primary mechanical injury
and secondary injury. Secondary injuries are thought to lead to
further tissue damage, followed by permanent dysfunction (5). Treatments that reduce secondary
injuries can improve the survival rate of spinal cord tissue and
reserve the necessary anatomical matrix for functional recovery
(6). Due to the complexity of SCI
pathogenesis, effective treatments that lead to full recovery have
not yet been identified (7). Thus,
investigating the mechanism of SCI is invaluable for SCI treatment
or recovery. Many pathophysiological events triggered by SCI are
tightly regulated by the expression levels of specific genes.
Previous studies suggested that changes in gene expression are
regulated by microRNAs (miRNAs), a family of short non-coding RNA
molecules that repress target mRNA translation (8–10).
miRNAs are non-coding endogenous RNA molecules of
about 18–24 nucleotides long that can play vital roles in
translational regulation and the suppression of specific mRNAs
(11,12). Thus, miRNAs effectively regulate
post-transcriptional gene expression in various tissues and the
development of several diseases (13–15).
In addition, changes in functional gene expression have been found
to play an important role in secondary damage progression in some
degenerative diseases (16–18).
As a result, abnormally expressed miRNAs may become new targets for
the treatment of various diseases, including traumatic injury,
cardiovascular disease or cancer, based on the specific
interactions between miRNAs and their target genes (19,20).
Moreover, many miRNAs have been demonstrated to play key roles in
regulating gene expression, proliferation, inflammation and
apoptosis (21). Various
neurological diseases have been observed to involve miRNAs
(21,22). Furthermore, an increasing number of
studies have reported that miRNAs are involved in SCI development
(8,9). For example, a previous study revealed
that a miR-219-5p inhibitor served a protective role in SCI by
regulating the LRH-1 (also known as NR5A2)/Wnt/β-catenin signaling
pathway (10). The knockdown of
miR-21 significantly reduced the inflammatory response at the
damaged spinal cord site and promoted motor function recovery
(23). A recent study demonstrated
that miR-21 was upregulated in neurons following SCI and could
reduce neuronal sensitivity to apoptosis by targeting programmed
cell death 4 (23). Moreover,
miR-137 inhibited inflammatory responses and apoptosis after SCI by
targeting MAPK activated protein kinase 2 (24). Another study indicated that
miR-199a-5p might protect the spinal cord against
ischemia-reperfusion induced injury by negatively regulating
endothelin converting enzyme 1 (25). Taken together, these previous
studies emphasized that both positive and negative regulators of
tissue regeneration were required for optimal control of overall
gene expression. Recent findings regarding miRNAs and their
specific role in SCI diseases have great significance for the
development of efficient treatments and novel specific drugs.
Several novel therapeutic approaches are currently
being used for the treatment of patients with SCI, including stem
cell therapy and electroacupuncture (EA) (26–28).
Both animal experiments and pain investigation have shown that EA
has an analgesic effect on chronic pain (29–32).
In addition, several previous studies suggested that EA at
different frequencies may exhibit varying degrees of analgesia
(33–35). To date, EA treatment at the Dazhui
and Mingmen acupuncture points have been evaluated for their impact
on SCI recovery (36). However,
the underlying mechanism of EA in SCI treatment remains unclear.
The expression profile of miRNAs in SCI rats and the possible
mechanism affected by EA remains to be elucidated.
To the best of the authors' knowledge, the present
study is the first to report a comprehensive analysis of the
differential expression profile of microRNAs in SCI rats treated
with EA. Specific miRNAs were selected and their possible functions
and enriched signaling pathways in SCI were systematically
analyzed. The present study provides a basis for better
understanding the effect of EA on SCI repair and examines how
changes in miRNA expression are involved in the molecular
mechanisms underlying EA-associated SCI recovery.
Materials and methods
Animals
All experiments were approved by The Institutional
Animal Care and Use Committee of The Second Affiliated Hospital of
Nanchang University (Nanchang, China) and were performed according
to the guidelines of The National Institutes of Health Guide for
the Care and Use of Laboratory Animals. All efforts were made to
minimize the number of animals used and their suffering. A total of
18 male Sprague-Dawley (SD) rats weighing 180–200 g (age, 8 weeks)
were obtained from Xipuer-Bikai Laboratory Animals Co., Ltd. All
rats were placed in plastic chambers at 22–24°C and were provided a
commercial diet and water under a 12-h reversed light-dark cycle
before the study. Health and behavior were monitored twice daily.
Following a 7-day adaptation, the rats were divided into three
groups (n=6 each), a sham group, an SCI model group, and an
electroacupuncture group (SCI+EA) to evaluate the effects of EA
treatment on SCI. Furthermore, to investigate the miRNA expression
profiles using next-generation sequencing (NGS), following
pathological assessment of half spinal cords of each group rats
[hematoxylin-eosin (HE) staining and ELISA], the other half spinal
cords from each rat of the model group (SCI; n=3) or
electroacupuncture group (SCI+EA; n=3) were used to perform
NGS.
Model establishment and EA
treatment
The rats were anesthetized by intraperitoneal
injection of 100 g/l chloral hydrate (350 mg/kg). Adequate
anesthesia was verified based on a lack of response to a
nociceptive stimulus. None of the rats exhibited signs of
peritonitis, pain or discomfort. Anesthetized rats were placed in a
prone position with a lumbar cushion with fixed limbs and head.
After shaving and sterilizing the back, cavitation was performed,
and the skin was cut along the back median line after routine
disinfection. On both sides of the spine, the muscles were
separated, and the spinous process was exposed. After determining
the location, the spinous process of L4 was removed with fine
bone-biting forceps, and the dura mater between the L4 and 5
segments was exposed. Under direct vision, 22G beveled needles were
used to pierce the L4-5 spinal cord segments toward the head along
the center of the spinal cord at a 60° angle to the end of the
spine. After 2 sec, the needles were removed, and the muscle,
subcutaneous fascia and skin were sutured layer-by-layer. An
effective puncture injury was determined by tail motion in rats.
Spinal cord puncture was not performed in the sham group.
Three days after the injury operation, the SCI+EA
group received EA therapy twice daily for three weeks. For EA
treatment, GV14 (Dazhui, the large vertebra), GV4 (Mingmen, the
‘vital door’) and Jiaji points were stimulated. EA stimulation
occurred for 20 min at 60 Hz and an alternating pulse width of 1.05
sec or 2.85 sec, which was sufficient to elicit slight twitching in
the hind limbs. For the sham group, acupuncture needles were
inserted bilaterally at a point lateral to the aforementioned
acupoints without any electrical stimulation. The total duration of
the SCI model experiment was 31 days. The animals were then
anesthetized by intraperitoneal injection of pentobarbital sodium
(200 mg/kg) after Basso, Beattie and Bresnahan (BBB) score
evaluation (37). The anesthetized
rats were then decapitated for assuring death. No animals died
during the modeling process, and all animals were successfully
modeled according to the BBB assessment and HE staining. Tissue
samples of the L4-5 spinal cord segments were removed from 6 rats
per group after EA stimulation for histological staining and stored
at −80°C.
BBB score evaluation and tissue
preparation
The BBB method was employed to examine the
functional deficits of rats following SCI. The sham group of normal
male rats without functional deficits (BBB score, 21) was selected
as a control. Hindlimb motor function of rats was assessed at 1, 3,
7 and 21 days after SCI. Following spinal cord surgery, trained
observers, who were blind to the experimental conditions, evaluated
the grade of each rat according to the BBB open field locomotion
test (38). Rats were placed in an
open basin, and hindlimb movement, ankle joint walking, trunk
movement and coordination were observed for 5 min to determine the
BBB score of each animal. A random selection of 6 rats per group
were sacrificed and quickly dissected at 3 weeks after SCI, and the
impaired spinal cords were harvested for real-time qPCR, HE
staining, and NGS. The tissues were frozen in liquid nitrogen and
stored at −80°C.
HE staining
After removal, using the aforementioned procedure,
tissue samples were fixed in 4% polyformaldehyde for 30 min at room
temperature and embedded in paraffin. The 5 µm paraffin-embedded
sections were placed in a 65°C-constant temperature oven for 30 min
and subsequently dewaxed with xylene I and xylene II for 15 min at
room temperature. The following steps were at room temperature: The
dewaxed slices were soaked in 100, 95, 85 and 75% ethanol for 5 min
each and washed with tap water for 10 min. The sections
subsequently underwent staining in hematoxylin for 5 min, followed
by color separation with ammonium hydroxide for several seconds.
Next, the sections were rinsed with water for 15 min and dehydrated
with 70 and 90% ethanol for 10 min each. Then sections were then
stained with eosin for 1–2 min, hydrated in 100% ethanol for 10
min, cleared with xylene, and finally mounted with neutral gum. The
spinal cord tissue structure was observed under a light microscope,
at ×200 magnification.
ELISA. The assessment of the IL-1β and TNF-α levels
in the spinal cord tissues was performed by ELISA, using a rat
IL-1β and TNF-α ELISA kit (cat. no. EK0393, BosterBiotech),
Briefly, 96-well plates were added the 100 µl sample and standard
for 3 duplicate samples and incubated for 90 min at room
temperature. Then 100 µl biotin-labeled antibodies were added and
incubated for 60 min at room temperature. The plates were washed
with PBS for three times, and avidin-biotin-complex added and
incubated for 30 min at room temperature. After further washing,
3,3′,5,5′-tetramethylbenzidine substrate solution was added and the
plates were incubated in the dark for 20 min. The reaction was
stopped by the addition of termination reagent, and the absorbance
at 450 and 570 nm was measured using a microplate reader.
Preparation of small RNA library and
NGS
Rats from the SCI and SCI+EA groups (n=3 in each
group) were randomly selected and sacrificed 21 days after SCI, and
the impaired spinal cords were harvested for NGS and
reverse-transcription-quantitative (RT-q)PCR. The tissues were
frozen in liquid nitrogen and stored at −80°C. Total RNA was
isolated from the spinal cords using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). RNA concentration and
quality were evaluated using a Nanodrop™ instrument (Thermo Fisher
Scientific, Inc.) and 1% gel electrophoresis, respectively.
Purified RNA samples were subjected to adapter ligation, cDNA
synthesis, PCR amplification, and construction of RNA libraries
using the miRNA Library Prep Kit (New England BioLabs, Inc.)
according to the manufacturer's protocol. RNA sequencing was then
performed on a Hiseq 2500 (Illumina, Inc.). Datasets were generated
with a sequencing depth of 50 million reads from the multiplexed
samples. The raw data were refined with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/;
version 0.11.2) (39) to filter
out short (<15 nucleotides) reads by evaluating the quality of
sequencing data (including base mass value distribution, mass value
location distribution and GC content) and low-quality reads
according to base quality value, including a phred quality mean
score >30 and reads between 18 and 40 nt in length. High-quality
reads were screened using miRBase (http://www.mirbase.org/) database to identify known
miRNAs.
Analysis of differentially expressed
(DE) miRNAs and their target gene prediction
Differential expression of miRNAs in the SCI+EA
group and the SCI group, was assessed using the Ebseq 2.0 package
(40) (P<0.05; |log2
FC|>1, where FC indicates fold change). MiRanda (http://miranda.org.uk/) (41) and RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid)
(42) software was used to predict
the target genes of the DE miRNAs. Overlapping genes from RNAhybrid
(energy <-25) and MiRanda (score ≥150; energy <-20) were
considered as potential targets of miRNAs. Finally, an integrated
mRNA-miRNA interaction network was constructed using Cytoscape
software (43) (version
3.5.1).
Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) analysis
GO (http://geneontology.org/) analysis, including
molecular function, biological process, and cellular component
assignment, was conducted to evaluate the functions of the screened
targets. Pathway enrichment analysis of the miRNA targets was
carried out using KEGG (http://www.genome.jp/kegg/) analysis. P<0.05 was
considered to indicate a statistically significant difference.
RT-qPCR
Total RNA was isolated from the spinal cords of rats
in the SCI and SCI+EA groups 21 days after SCI using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse transcribed into cDNA using the RevertAid™
First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.),
as per the manufacturer's instructions. Gene-specific RT primers
were used for each miRNA.
RT-qPCR was performed for 45 amplification cycles
with the following conditions: 95°C for 10 sec, 60°C for 60 sec,
and 95°C for 5 sec. FastStart Universal SYBR Green Master mix
(Thermo Fisher Scientific, Inc.) was used to amplify cDNA using the
ABI Q6 detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). All reactions were set up in triplicate.
Relative quantification and calculations were assessed using the
comparative threshold cycle method (2−ΔΔCq) (44). All primers were from Yingbiotech.
Primer sequences and are listed in Table I. GAPDH and U6 were used as the
internal control for mRNA and miRNA, respectively.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer name | Sequence
(5′-3′) |
|---|
| U6 F |
CGATACAGAGAAGATTAGCATGGC |
| U6 R |
AACGCTTCACGAATTTGCGT |
| rno-miR-219a-5p
RT |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAGAATTG |
| rno-miR-486 RT |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCTCGGGG |
| rno-miR-128-3p
RT |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAAAGAGA |
| rno-miR-136-5p
RT |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCCATCA |
| rno-miR-7b RT |
GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACACAACAA |
| rno-miR-219a-5p
F |
GCGGGTGATTGTCCAAACG |
| rno-miR-486 F |
GCGGGTCCTGTACTGAGCTG |
| rno-miR-136-5p
F |
CGCAGACTCCATTTGTTTTGA |
| rno-miR-128-3p
F |
GCGCAGTCACAGTGAACCG |
| rno-miR-7b F |
CGCAGTGGAAGACTTGTGATT |
| All miRNA-R |
AGTGCGTGTCGTGGAGTCG |
| GAPDH F |
TGGAGAAACCTGCCAAGTATGAT |
| GAPDH R |
TCAAAGGTGGAAGAATGGGAGT |
| Sema3a F |
ACATTTTTAAACTGCAGGACTCACA |
| Sema3a R |
GAGTCGTGCTGCTCGGTCC |
| Vangl2 F |
TTCTGCTGGAGCTGCGTCA |
| Vangl2 R |
AGGAGGGCGGGGTTGTAGA |
| Gng3 F |
GGATAAAGGTGTCCAAGGCAGC |
| Gng3 R |
GTGGGCACAGGAGTGATGAGG |
| TNF-α F |
GCCACCACGCTCTTCTGTCTA |
| TNF-α R |
GGGCTACGGGCTTGTCACT |
| IL-1β F |
ACAGACCCCAAAAGATTAAGGATT |
| IL-1β R |
CCACGGGCAAGACATAGGTAG |
Cell culture
The BV2 microglial cell line was obtained from
Procell Life Science & Technology Co., Ltd. Cells were cultured
in minimum essential medium (MEM) medium supplemented with 10%
fetal bovine serum and 1% penicillin-streptomycin (All from Gibco;
Thermo Fisher Scientific) at 37°C and 5% CO2, and
passaged twice a week.
Cell transfection
The miR-136-5p mimics (5′-ACUCCAUUUGUUUUGAUGAUGGA-3′
and 5′-CAUCAUCAAAACAAAUGGAGUUU-3′) and mimics negative control (NC;
5′-UUCUCCGAACGUGUCACGUTT-3′ and 5′-ACGUGACACGUUCGGAGAATT-3′) were
purchased from Shanghai GenePharma Co., Ltd.
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used to transfect BV2 microglial cells with
the 100 nM rno-miR-136-5p mimics or mimics NC. After a 48-h
incubation, cells at 80–90% confluence were harvested for RNA
extraction and reverse transcription. The levels of rno-miR-136-5p
expression were detected by RT-qPCR.
Cell Counting Kit-8 (CCK-8) assay
For cell proliferation assays, 2×104 BV2
cells per well were seeded in a 96-well plate. The cells were
incubated with 10 µl of CCK-8 reagent (Beyotime Institute of
Biotechnology) at 37°C for 2 h at the indicated time point for 0,
24, 48, 72 and 96 h. The absorbance at 450 nm was measured with
Infinite M1000 instrument (Tecan Group, Ltd.).
Apoptosis analysis
An Annexin V-FITC Early Apoptosis Detection kit
(cat. no. C1062; Beyotime Institute of Biotechnology) was used to
detect cell apoptosis. Briefly, 48 h after cell transfection, BV2
microglial cells were harvested with trypsin, re-suspended in
annexin V-FITC/propidium iodide, and then incubated for 10–15 min
at room temperature in the dark. A BD FACSVerse™ flow cytometer (BD
Biosciences) was used to assess cell apoptosis rates in different
groups according to the instrument's operating protocol. Data were
analyzed using WinMDI version 2.5 (Purdue University Cytometry
Laboratories; http://www.cyto.purdue.edu/flowcyt/software/Catalog.htm).
Statistical analysis
Data were analyzed using SPSS version 19.0 (SPSS,
Inc.) and are presented as the mean ± SD. Two-group comparisons
were performed using unpaired Student's t-test. Multi-group
comparisons were analyzed using one-way ANOVA, followed by Tukey's
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Effects of EA on behavior in SCI
rats
In the present study, the potential efficacy of EA
treatment in a rat model of SCI was investigated. Motor dysfunction
in the lower limbs was evaluated 1, 3, 7 and 21 days after SCI
injury, using the BBB scoring criteria. To determine the time
course of the changes in SCI injury, baseline measurements were
obtained prior to surgery. The baseline BBB scores on both hind
paws did not differ among the three groups (data not shown). After
surgery, rats exhibited various degrees of motor dysfunction,
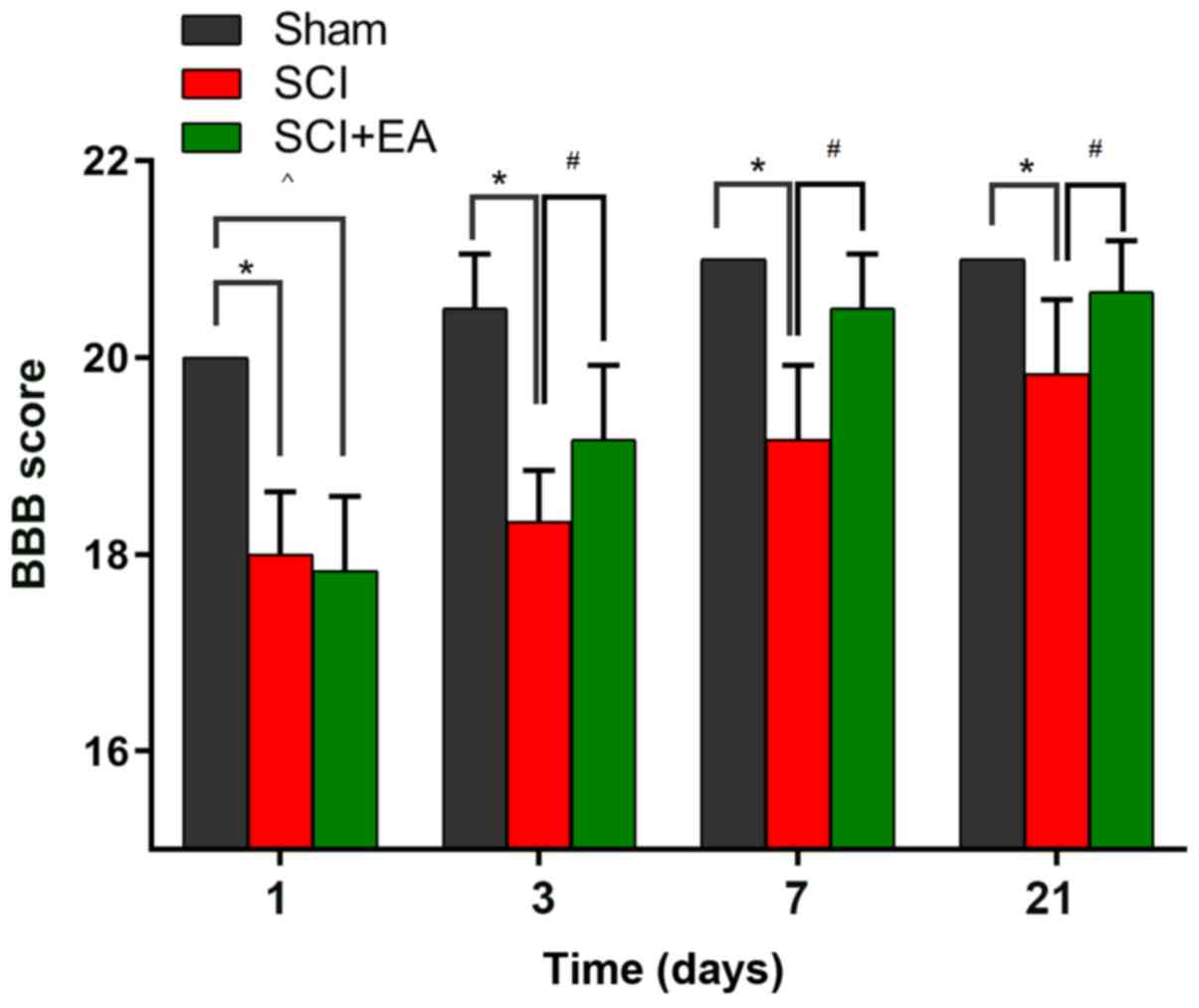
indicating successful establishment of the rat SCI model (Fig. 1). In the SCI group, BBB scoring was
markedly reduced from 20.00±0.00 to 18.00±0.63, compared with the
values in the sham group (P<0.05). On day 21 following SCI, the
BBB score was evaluated across all groups. Rats that received EA
treatment exhibited significantly higher BBB scores compared with
untreated rats following SCI. These results suggested that EA may
promote spinal recovery following SCI in rats.
Effects of EA on micromorphology of
spinal cords tissues
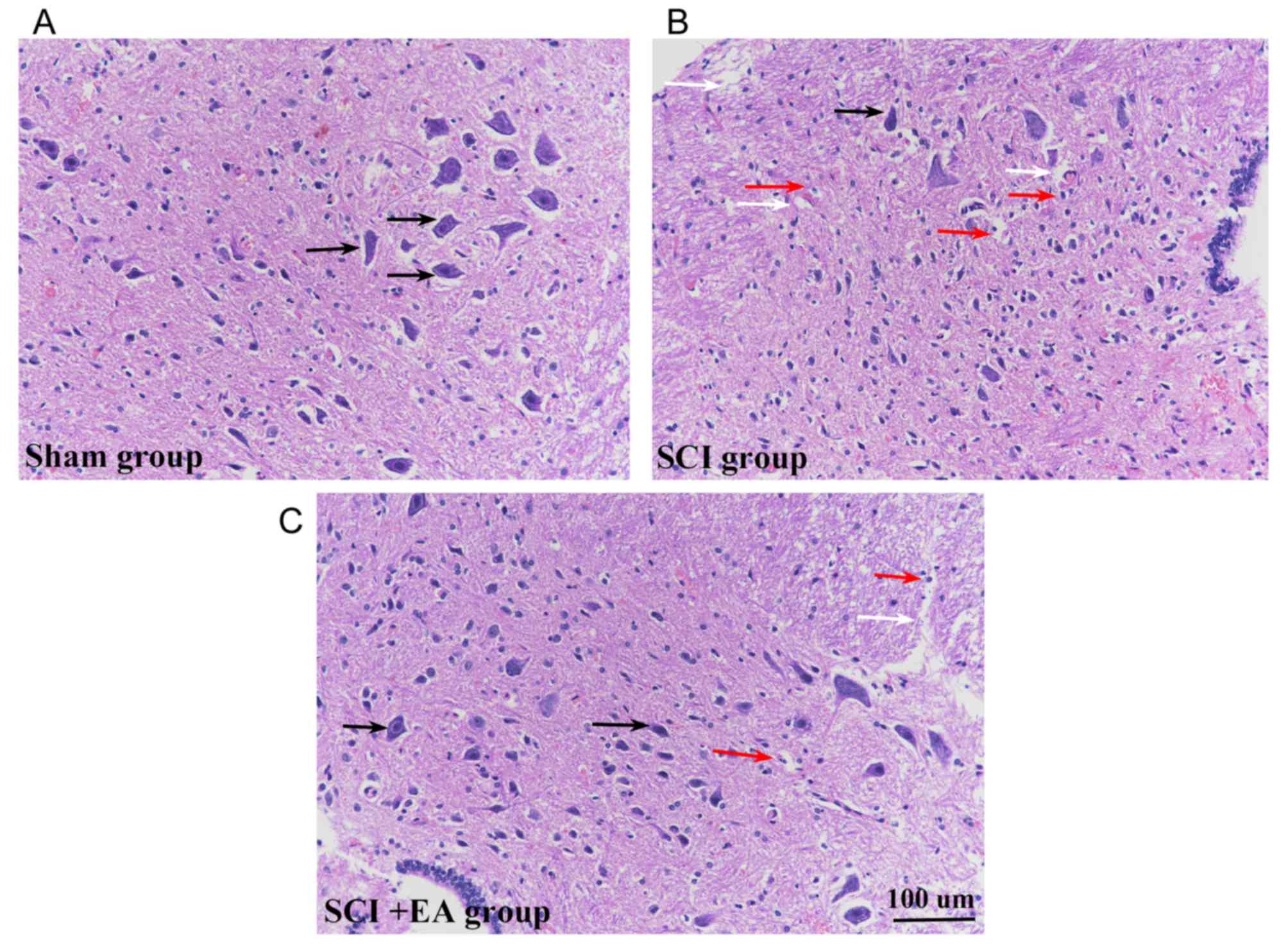
Morphological analysis of HE-stained sections
suggested that spinal cord tissue structures in the sham group were
clear and integrated, and that the cells were well-arranged, with
large round nuclei and abundant cytoplasm (Fig. 2A). Following the surgical SCI
procedure, the structural integrity of the spinal cord was severely
compromised, and the contusion injury site showed signs of
significant tissue compression. At 21 days post-surgery, HE-stained
sections revealed hemorrhage, edema, a progressive increase in
inflammatory cells, vacuolar degeneration at the injury site and
neuronal damage (Fig. 2B). Signs
of hemorrhage and inflammatory cell infiltration remained in the
spinal cord tissue, whereas SCI+EA group tissue sections exhibited
the most substantial restoration of neuronal morphology with axonal
regeneration (Fig. 2C). While both
normal and necrotic cells were present in the sham and SCI+EA
groups, the magnitude of necrosis appeared to be decreased in these
two groups, compared with the SCI group.
Effects of EA on tissue inflammatory
factors
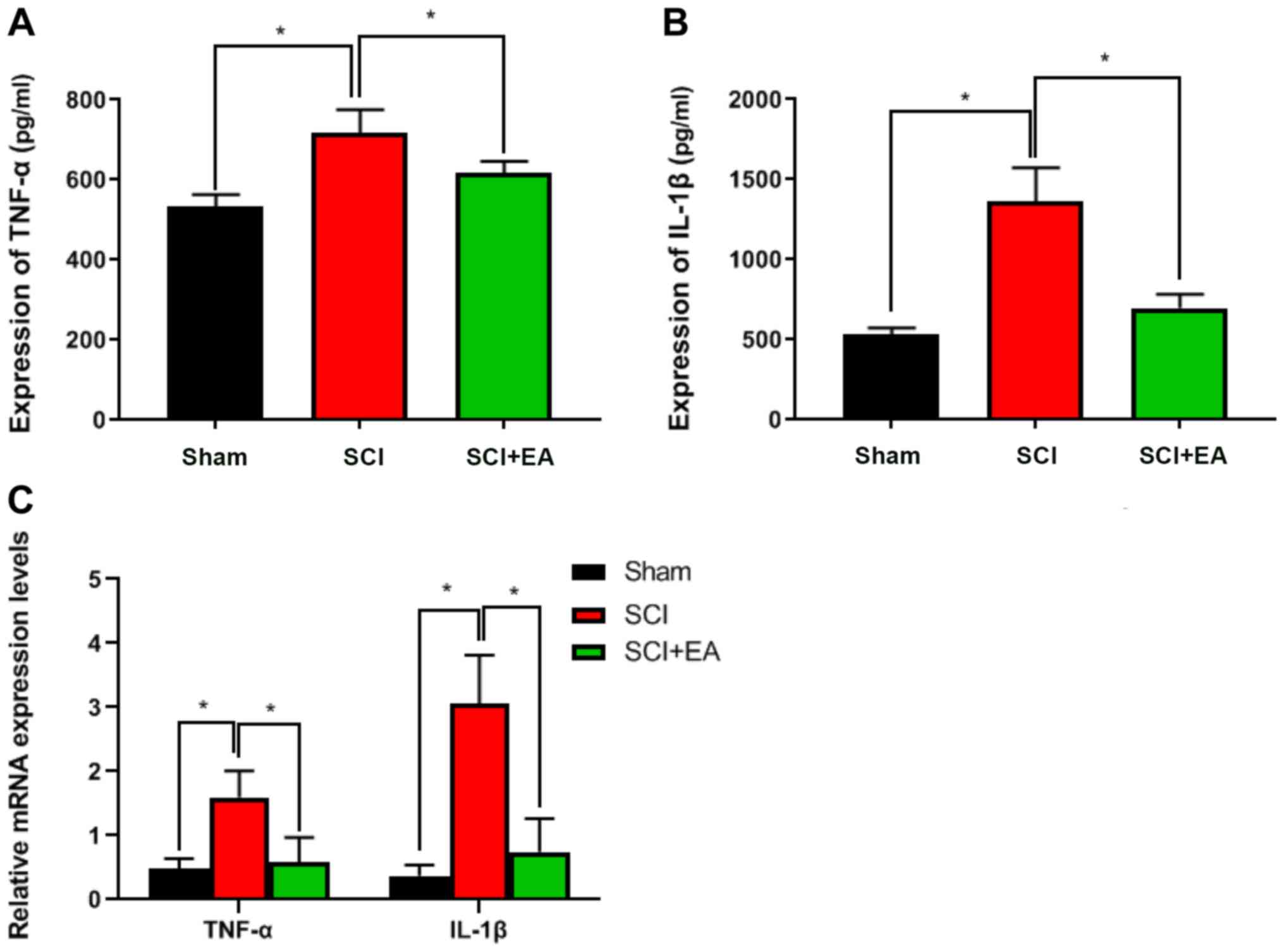
To observe the effect of EA treatment on tissue
inflammatory factor expression in SCI rats, TNF-α and IL-1β
expression levels were detected by ELISA. Both of these were
significantly lower in the SCI+EA treatment group than in the SCI
group (P<0.05; Fig. 3A and B).
RT-qPCR suggested similar results. Compared with the SCI group, the
relative expression levels of TNF-α and IL-1β in the sham and
SCI+EA treatment groups were significantly decreased (P<0.05;
Fig. 3C). These results
demonstrated that EA treatment could decrease inflammatory factors
and inflammation in the spinal cord tissue of rats.
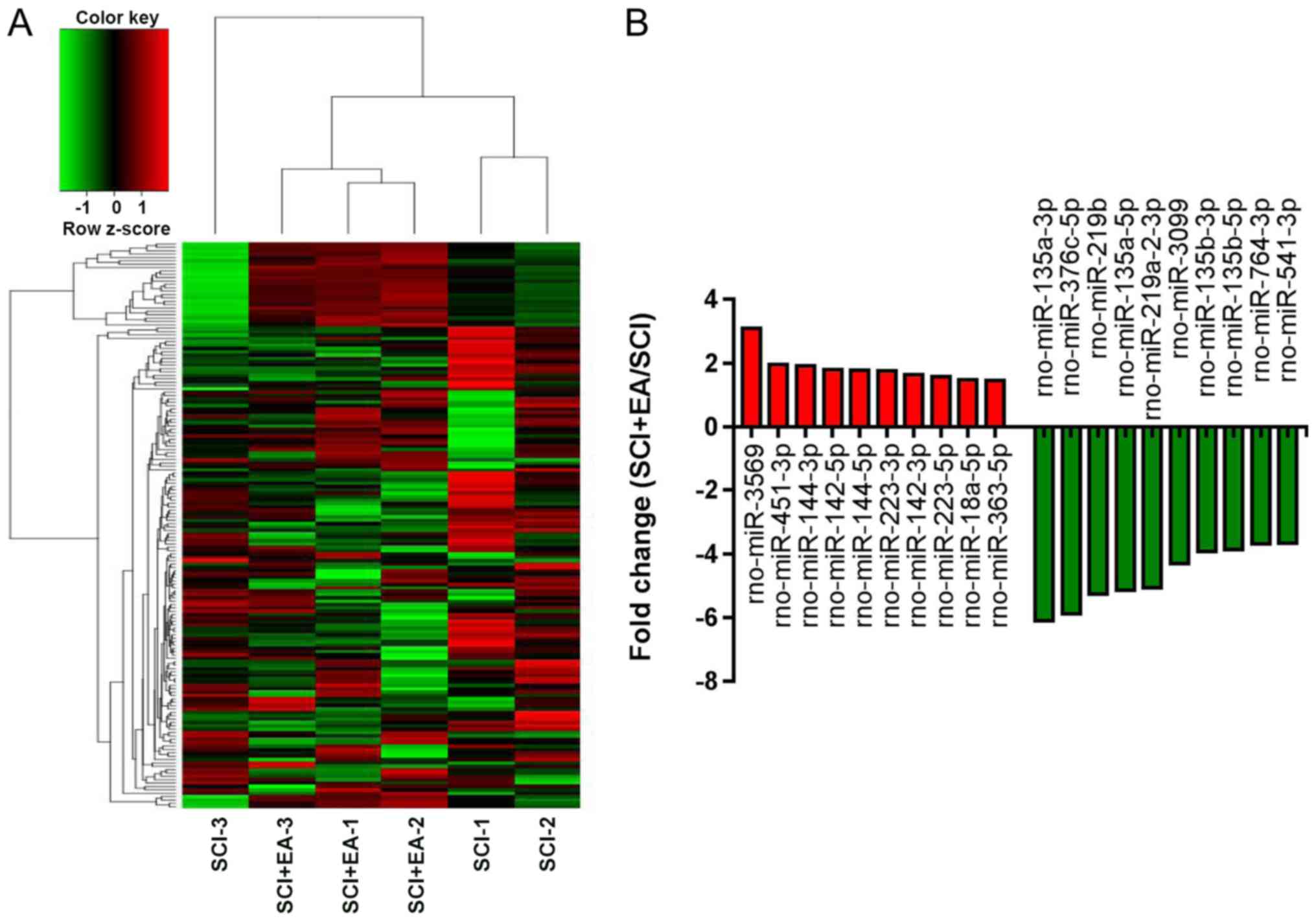
Identification of differentially
expressed (DE) miRNAs in SCI rats
To observe the effect of EA treatment on miRNA
expression profiles in SCI rats and determine the miRNAs involved
in EA treatment, NGS was used to obtain a profile of the miRNAs in
spinal cord tissue. In total, 764 miRNAs were identified from the
six samples. Among them, 168 miRNAs showed significant differential
expression between the SCI+EA and SCI groups. Of the DE miRNAs, 139
were downregulated and 29 were upregulated in the EA+SCI group,
compared with the SCI group. Statistical significance was defined
by an adjusted P<0.05 and |log2 FC|>1. A heatmap
of the 168 DE miRNAs with log2 FC values ranked by the
P-value indicated that the SCI+EA and SCI group rats could be
differentiated based on DE miRNAs (Fig. 4A). The top 20 up- and downregulated
miRNAs are shown in Fig. 4B.
Function and pathway analysis of
predicted target genes of DE miRNAs
Based on the target gene prediction, the potential
mRNAs of DE miRNAs were subjected to GO and KEGG enrichment
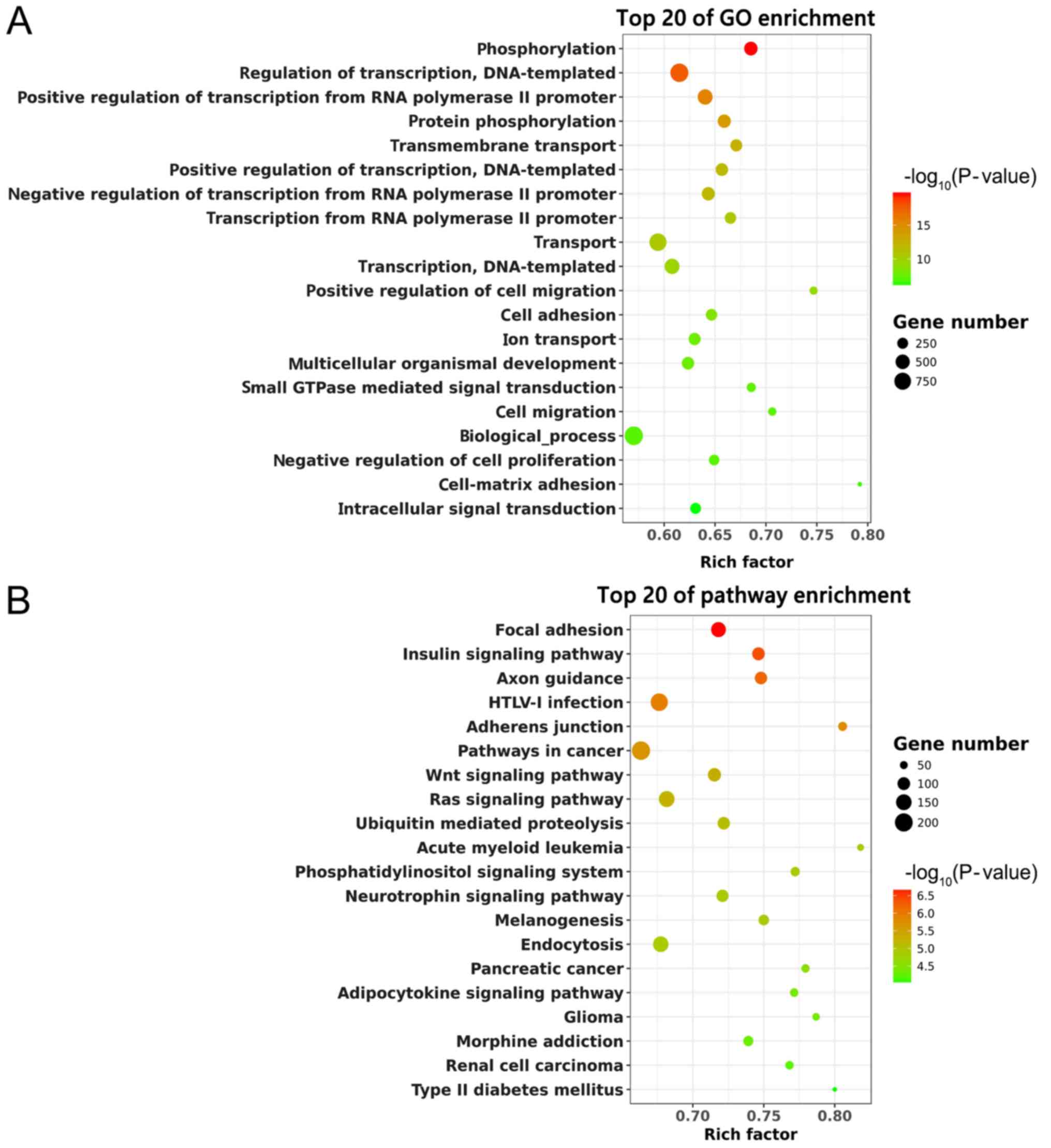
analysis. The top 20 GO terms are shown in Fig. 5A. ‘Phosphorylation’ and ‘Regulation
of transcription, DNA-templated’ were significantly enriched. KEGG
analysis suggested that these predicted target genes were enriched
in the ‘Focal adhesion’, ‘Wnt signaling pathway’ and ‘Ras signaling
pathway’ (Fig. 5B). Thus, these
pathways might have potential significance in SCI recovery with EA
treatment.
Validation of candidate miRNA
expression by RT-qPCR
To confirm the NGS results, five miRNAs,
rno-miR-219a-5p, rno-miR-486, rno-miR-136-5p, rno-miR-128-3p and
rno-miR-7b, with high fold change and high abundance were selected
as candidate miRNAs and were screened as they or their target mRNAs
were involved in SCI recovery (45–47).
Compared with the SCI group, rno-miR-219a-5p, rno-miR-128-3p, and
rno-miR-136-5p were significantly downregulated (P<0.05) in the
sham and SCI+EA groups. There was no significant difference in
rno-miR-486 and rno-miR-7b between the Sham, SCI and SCI+EA groups
(Fig. 6). The RT-qPCR validation
results were generally consistent with the sequencing results
(Table II), suggesting that the
latter were reliable.
 | Table II.Candidate miRNAs in the SCI and
SCI+EA group. |
Table II.
Candidate miRNAs in the SCI and
SCI+EA group.
| miRNA |
log2FC | P-value | FDR | Trend |
|---|
|
rno-miR-219a-5p | −2.194697258 | 0.000086252 | 0.000756832 | Down |
| rno-miR-486 | 1.123062605 | 0.010721736 | 0.038169381 | Up |
| rno-miR-136-5p | −1.824091331 | 0.000020324 | 0.000238909 | Down |
| rno-miR-128-3p | −1.776468827 | 0.000472567 | 0.003271216 | Down |
| rno-miR-7b | −3.300169904 | 0.000000009 | 0.000000364 | Down |
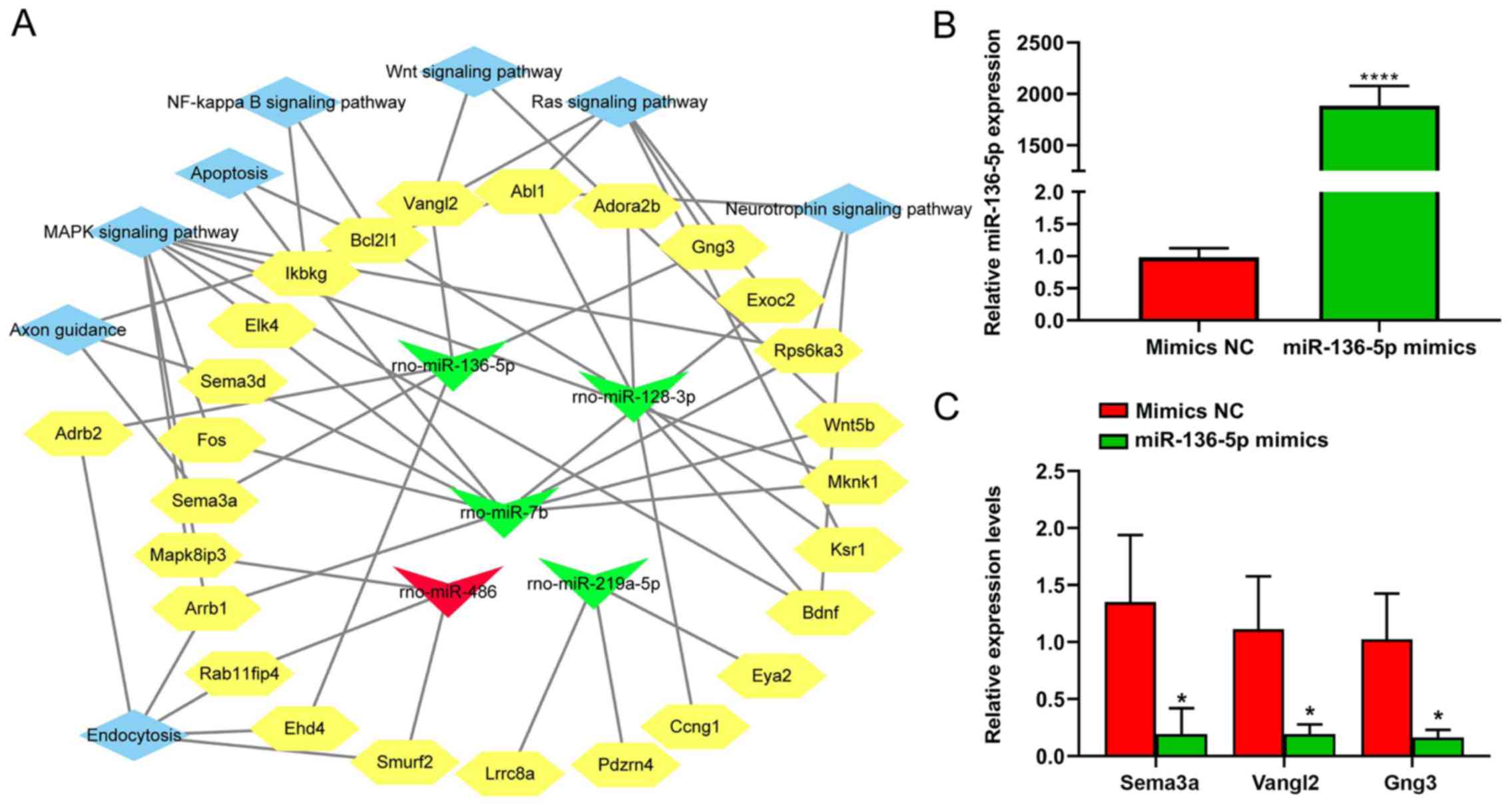
Analysis of the integrated
miRNA-mRNA-pathway network
Five candidate miRNAs, namely rno-miR-219a-5p,
rno-miR-486, rno-miR-136-5p, rno-miR-128-3p, and rno-miR-7b, and
their respective target genes associated with the top enriched
pathways were assembled in an integrated miRNA-mRNA pathway
network. The terms ‘Wnt signaling pathway’, ‘neurotrophin signaling
pathway’, ‘NF-κB signaling pathway’ and ‘MAPK signaling pathway’
were identified as enriched pathways using this analysis (Fig. 7A). From the network, miR-136-5p
targeted ‘Wnt signaling pathway’ through Van Gogh Drosophila-like
planar cell polarity protein 2 (Vangl2), the ‘axon guidance’
through semaphoring 3 A (Sema3a), and the ‘Ras signaling pathway’
through Vangl2 and G protein subunit γ 3 (Gng3). Moreover,
miR-136-5p was selected for further study as it showed a
significant difference in expression between the SCI and SCI+EA
groups. The expression of three candidate target genes of
miR-136-5p, including Sema3a, Vangl2 and Gng3, was assessed by
RT-qPCR in BV2 microglial cells transfected with miR-136-5p mimics.
Compared with the mimics-NC, the expression of Sema3a, Vangl2 and
Gng3 was significantly downregulated in microglial cells
transfected with the miR-136-5p mimics (Fig. 7B and C).
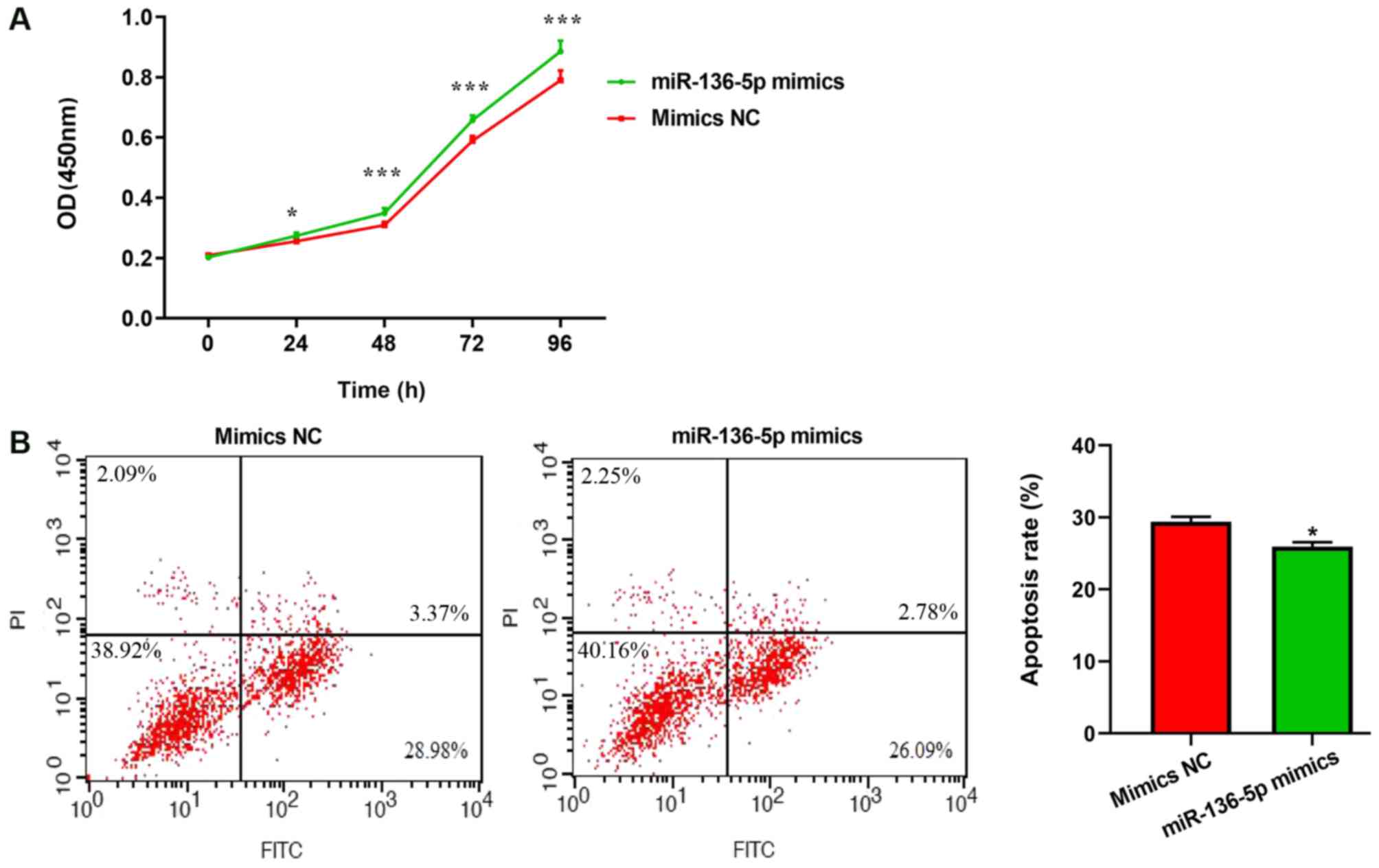
Effects of miR-136-5p on microglial
cell proliferation and apoptosis
To observe the function of miR-136-5p on
proliferative and apoptotic abilities, microglial cells were
transfected with miR-136-5p. Cell proliferation was quantified
using the CCK-8 method. Transfection with miR-136-5p mimics
significantly promoted microglial cell proliferation, compared with
mimics-NC (Fig. 8A). The effects
of miR-136-5p on cell apoptosis were then analyzed by flow
cytometry. Compared with the mimics-NC group, the number of
apoptotic cells was significantly decreased in BV2 cells
transfected with miR-136-5p mimics (P<0.01; Fig. 8B). These results indicated that
miR-136-5p enhanced proliferation and inhibited the apoptosis of
microglia cells.
Discussion
miRNAs are non-coding RNA molecules of 18–24
nucleotides that regulate gene expression by interacting with
specific sequences of target mRNAs or promoters (48,49).
To date, several studies have analyzed the effect of miRNAs in
injury and neuroprotection (50–52).
Previous studies also demonstrated that the circulating microRNA
profile may serve as a latent diagnosis biomarker and new target in
the molecular treatment of SCI (23,24).
Other previous studies suggested that EA treatment improved
hindlimb motor function of SCI rats (36,53).
However, analysis of the miRNA profile and functions affected by EA
treatment in SCI rats has not been investigated.
In the present study, the effects of EA on miRNA
expression following SCI were evaluated in rats. The present
results suggested that EA treatment on rats with SCI led to miRNA
dysregulation, thus affecting multiple processes that in many cases
are associated with secondary damage from SCI and recovery of rats.
The observed changes in expression mainly included an increased
number of downregulated miRNAs. However, few miRNAs were
upregulated. Progressive dysregulation of miRNAs has been reported
in rats with SCI (54). In
addition, Strickland et al (55) observed a similar increase in the
number of downregulated miRNAs in the 14 days after SCI. Previous
studies described the functional roles of the miRNAs that are
dysregulated in rats with SCI, which are potentially regulated by
co-expressed miRNAs (56,57). These analyses indicated that
changes in miRNA expression could affect numerous biological
functions known to be altered in SCI rats.
A previous study demonstrated that miR-223 promoted
neutrophil-mediated inflammation and aggravated SCI in the early
stage after SCI (58).
Furthermore, miRNA-136-5p upregulates p-nuclear factor κB (p-NF-κB)
expression by downregulating A20 expression, which causes
astrocytes to produce inflammatory factors and chemokine factors,
thus aggravating SCI (59).
Recently, Deng et al (45)
found that IL-1β, IL-6, TNF-α, interferon-α, inhibitor of nuclear
factor kappa B kinase subunit β, and NF-κB in SCI rats were
upregulated, while A20 was downregulated following miR-136-5p
overexpression. Under these conditions, inflammatory cell
infiltration into the rat spinal cord increases, significantly
aggravating injury. Silencing of miR-136-5p significantly reduces
these changes in protein expression and ameliorates the
inflammatory cell infiltration and spinal cord damage. Therefore,
miR-136-5p might be a new target for the treatment of SCI (45). In the present study, miR-136-5p
expression was decreased in EA-treated SCI rats, compared with the
SCI group. In addition, Zhang et al (60) observed that miRNA-127 can regulate
inflammation by activating the JNK and NF-κB pathways. NF-κB has
different states (phosphorylated and dephosphorylated) and activity
levels in different types of cells and tissues. NF-κB is a
multidirectional transcription factor as well as the converging
point of many signal transduction pathways. It plays an important
role in immunity, inflammation, cell cycle regulation, cell
proliferation and differentiation, and apoptosis (61).
Inflammatory reactions play an important role in SCI
progression. The present study demonstrated that EA significantly
improved inflammatory cell infiltration and inflammatory factor
expression. Moreover, EA-induced DE miRNAs were mainly enriched in
inflammation-related pathways, such as the aforementioned NF-κB
pathway. Additionally, microglial cells are the main inflammatory
cells in the brain and spinal cord. Previous studies used
microglial cells to study the functional recovery of SCI (62,63).
Therefore, the contribution of miR-136-5p to EA-induced alleviation
of inflammation was also evaluated in microglial cells in the
present study. miR-136-5p enhanced proliferation and inhibited
apoptosis of microglial cells, suggesting that miR-136-5p might be
involved in EA treatment by modulating inflammation in microglial
cells.
In conclusion, the present study provided an
analysis of DE miRNAs in SCI rats treated with EA using
high-throughput sequencing and described their functional
interaction network, therefore providing an understanding of the
mechanism and function of miRNAs in SCI rats. However, how miRNAs
target mRNA to participate in the regulation of EA treatment
through various signaling pathways remains to be elucidated.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FD and JL designed and funded the present study. ZZ,
HJL, HCL, JZ, KF, CC, FD and JL performed the experiments. ZZ, HJL,
KF and CC analyzed the data. ZZ, HCL, JZ and KF conducted
literature search. All authors prepared and revised the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All experiments were approved by The Institutional
Animal Care and Use Committee of the Second Affiliated Hospital of
Nanchang University and were performed according to the guidelines
of The National Institutes of Health Guide for The Care and Use of
Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McDonald JW and Sadowsky C: Spinal-cord
injury. Lancet. 359:417–425. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sharif-Alhoseini M and Rahimi-Movaghar V:
Animal models in traumatic spinal cord injury. Top Paraplegia.
2014.
|
|
3
|
Gao L, Sun Y, Li J, Bai F and Li P:
Effects of electroacupuncture in different time on variations of
fractional anisotropy mean value of diffusion tensor tractogra-phy
in spinal cord injured rats. Chin J Rehabil Theory Prac.
20:728–733. 2014.
|
|
4
|
Majdan M, Plancikova D, Nemcovska E,
Krajcovicova L, Brazinova A and Rusnak M: Mortality due to
traumatic spinal cord injuries in Europe: A cross-sectional and
pooled analysis of population-wide data from 22 countries. Scand J
Trauma Resusc Emerg Med. 25:642017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blight AR, Leroy EC Jr and Heyes MP:
Quinolinic acid accumulation in injured spinal cord: Time course,
distribution, and species differences between rat and guinea pig. J
Neurotrauma. 14:89–98. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hall ED and Springer JE: Neuroprotection
and acute spinal cord injury: A reappraisal. NeuroRx. 1:80–100.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tator CH: Update on the pathophysiology
and pathology of acute spinal cord injury. Brain Pathol. 5:407–413.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong J, Lu M, He X, Xu J, Qin J, Cheng Z,
Liang B, Wang D and Li H: Identifying the role of microRNAs in
spinal cord injury. Neurol Sci. 35:1663–1671. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ning B, Gao L, Liu RH, Liu Y, Zhang NS and
Chen ZY: MicroRNAs in spinal cord injury: Potential roles and
therapeutic implications. Int J Biol Sci. 10:997–1006. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li J, Li L and Shen Y: Protective role of
microRNA-219-5p inhibitor against spinal cord injury via liver
receptor homolog-1/Wnt/β-catenin signaling pathway regulation. Exp
Ther Med. 15:3563–3569. 2018.PubMed/NCBI
|
|
11
|
Bhalala OG, Srikanth M and Kessler JA: The
emerging roles of microRNAs in CNS injuries. Nat Rev Neurol.
9:328–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng Q, Zhang D, Yang YU, Cui X, Sun J,
Liang C, Qin H, Yang X, Liu S and Yan Q: MicroRNA-200c impairs
uterine receptivity formation by targeting FUT4 and
1,3-fucosylation. Cell Death Differ. 24:2161–2172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zeng Y, Liu JX, Yan ZP, Yao XH and Liu XH:
Potential microRNA biomarkers for acute ischemic stroke. Int J Mol
Med. 36:1639–1647. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hinkel R, Penzkofer D, Zühlke S, Fischer
A, Husada W, Xu QF, Baloch E, Van RE, Zeiher AM, Kupatt C and
Dimmeler S: Inhibition of microRNA-92a protects against
ischemia/reperfusion injury in a large-animal model. Circulation.
128:1066–1075. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
van Rooij E, Sutherland LB, Liu N,
Williams AH, McAnally J, Gerard RD, Richardson JA and Olson EN: A
signature pattern of stress-responsive microRNAs that can evoke
cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA.
103:18255–18260. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Y, Ransom JF, Li A, Vedantham V, von
Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ and
Srivastava D: Dysregulation of cardiogenesis, cardiac conduction,
and cell cycle in mice lacking miRNA-1-2. Cell. 129:303–317. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bala S, Marcos M and Szabo G: Emerging
role of microRNAsin liver diseases. World J Gastroenterol.
15:5633–5640. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krutzfeldt J, Rajewsky N, Braich R, Rajeev
KG, Tuschl T, Manoharan M and Stoffel M: Silencing of microRNAs in
vivo with ‘antagomirs’. Nature. 438:685–689. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jackson AL, Burchard J, Leake D, Reynolds
A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et
al: Position-specific chemical modification of siRNAs reduces
‘off-target’ transcript silencing. RNA. 12:1197–1205. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rao P, Benito E and Fischer A: MicroRNAs
as biomarkers for CNS disease. Front Mol Neurosci. 6:392013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang W, Kwon EJ and Tsai LH: MicroRNAs in
learning, memory, and neurological diseases. Learn Mem. 19:359–368.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang T, Ni SF, Luo Z, Lang Y, Hu J and Lu
H: The protective effect of microRNA-21 in neurons after spinal
cord injury. Spinal Cord. 57:141–149. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gao L, Dai C, Feng Z, Zhang L and Zhang Z:
MiR-137 inhibited inflammatory response and apoptosis after spinal
cord injury via targeting of MK2. J Cell Biochem. 119:3280–3292.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bao N, Fang B, Lv H, Jiang Y, Chen F, Wang
Z and Ma H: Upregulation of miR-199a-5p protects spinal cord
against ischemia/reperfusion-induced injury via downregulation of
ECE1 in rat. Cell Mol Neurobiol. 38:1293–1303. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Obermair FJ, Schröter A and Thallmair M:
Endogenous neural progenitor cells as therapeutic target after
spinal cord injury. Physiology (Bethesda). 23:296–304.
2008.PubMed/NCBI
|
|
27
|
Yan Q, Ruan JW, Ding Y, Li WJ, Li Y and
Zeng YS: Electro-acupuncture promotes differentiation of
mesenchymal stem cells, regeneration of nerve fibers and partial
functional recovery after spinal cord injury. Exp Toxicol Pathol.
63:151–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang SH, Tu WZ, Zou EM, Hu J, Wang S, Li
JR, Wang WS, He R, Cheng RD and Liao WJ: Neuroprotective effects of
different modalities of acupuncture on traumatic spinal cord injury
in rats. Evid Based Complement Alternat Med. 2014:4315802014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Park JH, Han JB, Kim SK, Park JH, Go DH,
Sun B and Min BI: Spinal GABA receptors mediate the suppressive
effect of electroacupuncture on cold allodynia in rats. Brain Res.
1322:24–29. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Aloe L and Manni L: Low-frequency
electro-acupuncture reduces the nociceptive response and the pain
mediator enhancement induced by nerve growth factor. Neurosci Lett.
449:173–177. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Park JH, Kim SK, Kim HN, Sun B, Koo S,
Choi SM, Bae H and Min BI: Spinal cholinergic mechanism of the
relieving effects of electroacupuncture on cold and warm allodynia
in a rat model of neuropathic pain. J Physiol Sci. 59:291–298.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Min YJ, Cheng LH and Gao J: Comparative
observations on three-unblocking acupuncture for the treatment of
spinal cord injury in convalescent patients with paraplegia.
Shanghai Zhenjiu Zazhi. 32:1010–1013. 2013.
|
|
33
|
Huang C, Wang Y, Han JS and Wan Y:
Characteristics of electroacupuncture-induced analgesia in mice:
Variation with strain, frequency, intensity and opioid involvement.
Brain Res. 945:20–25. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lao L, Zhang RX, Zhang G, Wang X, Berman
BM and Ren K: A para-metric study of electroacupuncture on
persistent hyperalgesia and Fos protein expression in rats. Brain
Res. 1020:18–29. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin JG, Lo MW, Wen YR, Hsieh CL, Tsai SK
and Sun WZ: The effect of high and low frequency electroacupuncture
in pain after lower abdominal surgery. Pain. 99:509–514. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang JF, Li SS and Wu YC: Recovery of
spinal cord injury following electroacupuncture in rats through
enhancement of Wnt/β-catenin signaling. Mol Med Rep. 16:2185–2190.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Filipp ME, Travis BJ, Henry SS, Idzikowski
EC, Magnuson SA, Loh MY, Hellenbrand DJ and Hanna AS: Differences
in neuroplasticity after spinal cord injury in varying animal
models and humans. Neural Regen Res. 14:7–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Basso DM, Beattie MS and Bresnahan JC: A
sensitive and reliable locomotor rating scale for open field
testing in rats. J Neurotrauma. 12:1–21. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ramayo-Caldas Y, Mach N, Esteve-Codina A,
Corominas J, Castelló A, Ballester M, Estellé J, Ibáñez-Escriche N,
Fernández AI, Pérez-Enciso M and Folch JM: Liver transcriptome
profile in pigs with extreme phenotypes of intramuscular fatty acid
composition. BMC Genomics. 13:5472012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wright GW and Simon RM: A random variance
model for detection of differential gene expression in small
microarray experiments. Bioinformatics. 19:2448–2455. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Turner DA: Miranda: A non-strict
functional language with polymorphic types. Proc of a conference on
functional programming languages and computer architecture. 1–16.
1985. View Article : Google Scholar
|
|
42
|
Krüger J and Rehmsmeier M: RNAhybrid:
MicroRNA target prediction easy, fast and flexible. Nucleic Acids
Res. 34((Web Server issue)): W451–W454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Deng G, Gao Y, Cen Z, He J, Cao B, Zeng G
and Zong S: miR-136-5p regulates the inflammatory response by
targeting the IKKβ/NF-κB/A20 pathway after spinal cord injury. Cell
Physiol Biochem. 50:512–524. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Knierim E, Hirata H, Wolf NI,
Morales-Gonzalez S, Schottmann G, Tanaka Y, Rudnik-Schöneborn S,
Orgeur M, Zerres K, Vogt S, et al: Mutations in subunits of the
activating signal cointegrator 1 complex are associated with
prenatal spinal muscular atrophy and congenital bone fractures. Am
J Human Genet. 98:473–489. 2016. View Article : Google Scholar
|
|
47
|
Matsuda M, Kanno H, Sugaya T, Yamaya S,
Yahata K, Handa K, Shindo T, Shimokawa H, Ozawa H and Itoi E:
Low-energy extracorporeal shock wave therapy promotes BDNF
expression and improves functional recovery after spinal cord
injury in rats. Exp Neurol. 328:1132512020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Saugstad JA: MicroRNAs as effectors of
brain function with roles in ischemia and injury, neuroprotection,
and neurodegeneration. J Cereb Blood Flow Metab. 30:1564–1576.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yip PK, Bowes AL, Hall JCE, Burguillos MA,
Ip THR, Baskerville T, Liu ZH, Mohamed MAEK, Getachew F, Lindsay
AD, et al: Docosahexaenoic acid reduces microglia phagocytic
activity via miR-124 and induces neuroprotection in rodent models
of spinal cord contusion injury. Human Mol Genet. 28:2427–2448.
2019. View Article : Google Scholar
|
|
52
|
Yan L, Shi E, Jiang X, Shi J, Gao S and
Liu H: Inhibition of microRNA-204 conducts neuroprotection against
spinal cord ischemia. Ann Thorac Surg. 107:76–83. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Min YJ, Ding LLQ, Cheng LH, Xiao WP, He
XW, Zhang H, Min ZY and Pei J: Effect of electroacupuncture on the
mRNA and protein expression of Rho-A and Rho-associated kinase II
in spinal cord injury rats. Neural Regen Res. 12:110–116. 2017.
|
|
54
|
Yunta M, Nieto-Díaz M, Esteban FJ,
Caballero-López M, Navarro-Ruíz R, Reigada D, Pita-Thomas DW, del
Águila A, Muñoz-Galdeano T and Maza RM: MicroRNA dysregulation in
the spinal cord following traumatic injury. PLoS One. 7:e345342012.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Strickland ER, Hook MA, Balaraman S, Huie
JR, Grau JW and Miranda RC: MicroRNA dysregulation following spinal
cord contusion: Implications for neural plasticity and repair.
Neuroscience. 186:146–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Xing SM, Wang J, He X, Lai J, Shen L, Chen
D, Fu K and Tan J: Identification of disease-related miRNAs based
on co-expression network in spinal cord injury. Int Neurosci.
125:270–276. 2015. View Article : Google Scholar
|
|
57
|
Wei H, Wang C, Zhang C, Li P, Wang F and
Zhang Z: Comparative profiling of microRNA expression between
neural stem cells and motor neurons in embryonic spinal cord in
rat. Int J Dev Neurosci. 28:545–551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Izumi B, Nakasa T, Tanaka N, Nakanishi K,
Kamei N, Yamamoto R, Nakamae T, Ohta R, Fujioka Y, Yamasaki K and
Ochi M: MicroRNA-223 expression in neutrophils in the early phase
of secondary damage after spinal cord injury. Neurosci Lett.
492:114–118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
He J, Zhao J, Peng X, Shi X, Zong S and
Zeng G: molecular mechanism of MiR-136-5p targeting NF-κB/A20 in
the IL-17-mediated inflammatory response after spinal cord injury.
Cell Physiol Biochem. 44:1224–1241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zhang Z, Wan F, Zhuang Q, Zhang Y and Xu
Z: Suppression of miR-127 protects PC-12 cells from LPS-induced
inflammatory injury by downregulation of PDCD4. Biomed
Pharmacother. 96:1154–1162. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ahn KS, Sethi G and Aggarwal BB: Nuclear
factor-kappa B: From clone to clinic. Curr Mol Med. 7:619–637.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wang C, Wang Q, Lou Y, Xu J, Feng Z, Chen
Y, Tang Q, Zheng G, Zhang Z, Wu Y, et al: Salidroside attenuates
neuroinflammation and improves functional recovery after spinal
cord injury through microglia polarization regulation. J Cell Mol
Med. 22:1148–1166. 2018.PubMed/NCBI
|
|
63
|
Zhang Y, Liu Z, Zhang W, Wu Q, Zhang Y,
Liu Y, Guan Y and Chen X: Melatonin improves functional recovery in
female rats after acute spinal cord injury by modulating
polarization of spinal microglial/macrophages. J Neurosci Res.
97:733–743. 2019. View Article : Google Scholar : PubMed/NCBI
|