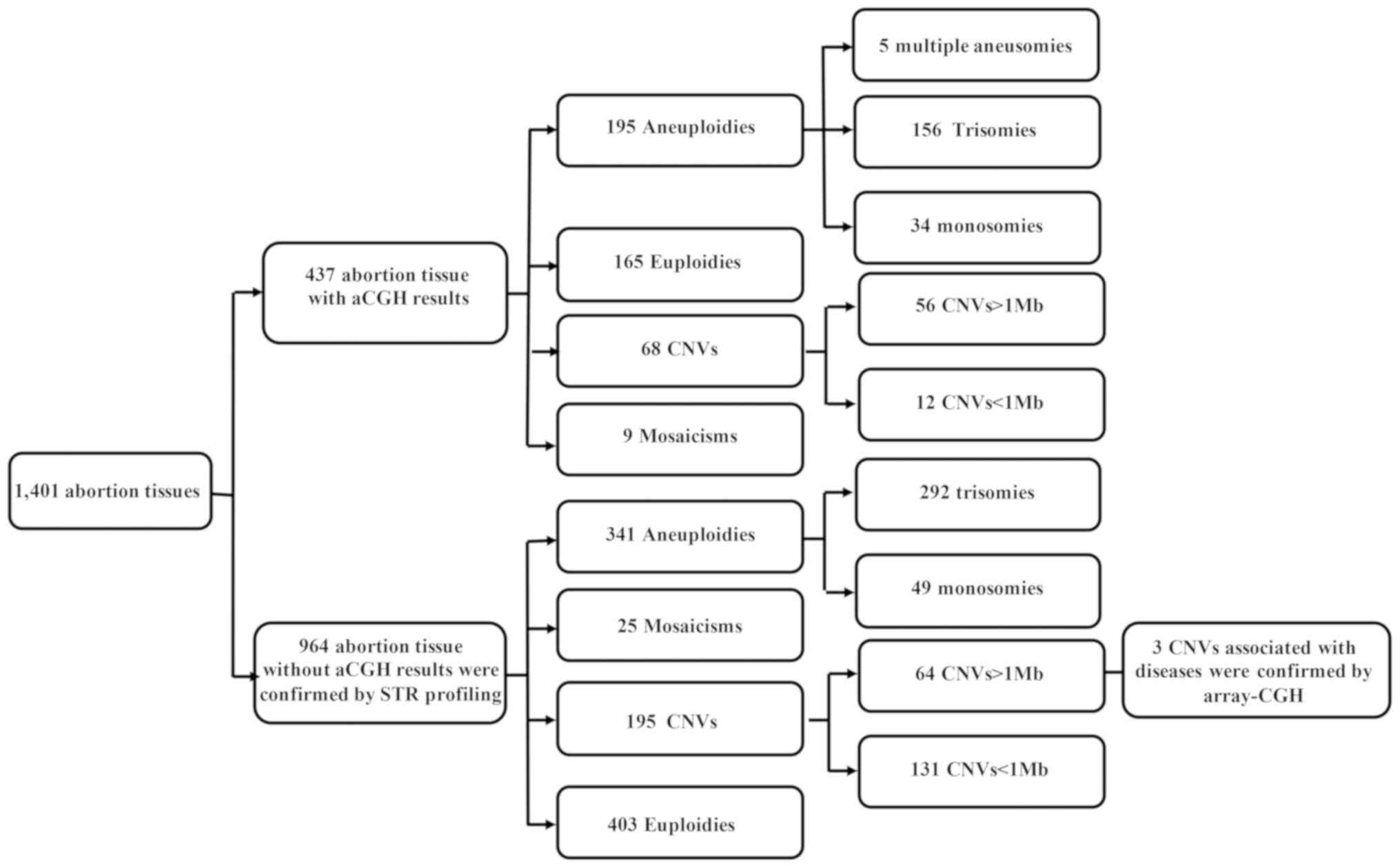
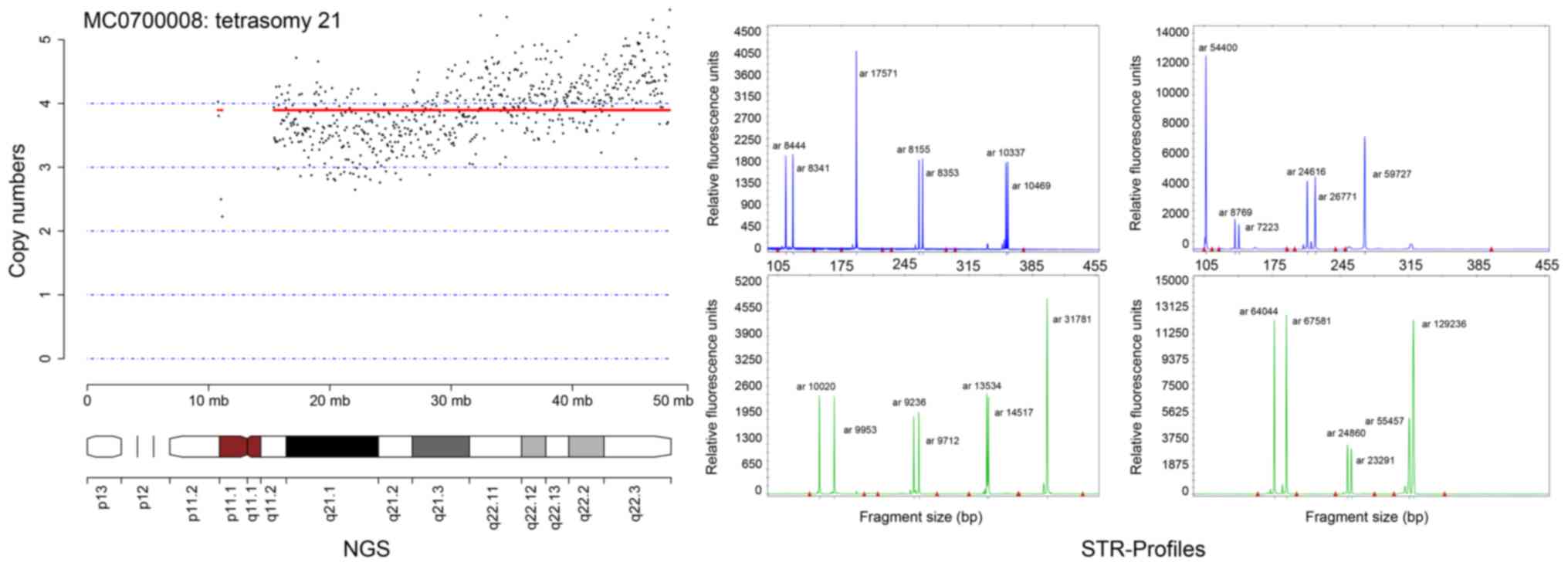
|
1
|
Liu S, Song L, Cram DS, Xiong L, Wang K,
Wu R, Liu J, Deng K, Jia B, Zhong M, et al: Traditional karyotyping
vs copy number variation sequencing for detection of chromosomal
abnormalities associated with spontaneous miscarriage. Ultrasound
Obstet Gynecol. 46:472–477. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
van den Berg MM, van Maarle MC, van Wely M
and Goddijn M: Genetics of early miscarriage. Biochim Biophys Acta.
1822:1951–1959. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Daniely M, Aviram-Goldring A, Barkai G and
Goldman B: Detection of chromosomal aberration in fetuses arising
from recurrent spontaneous abortion by comparative genomic
hybridization. Hum Reprod. 13:805–809. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim JW, Lee WS, Yoon TK, Seok HH, Cho JH,
Kim YS, Lyu SW and Shim SH: Chromosomal abnormalities in
spontaneous abortion after assisted reproductive treatment. BMC Med
Genet. 11:1532010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lomax B, Tang S, Separovic E, Phillips D,
Hillard E, Thomson T and Kalousek DK: Comparative genomic
hybridization in combination with flow cytometry improves results
of cytogenetic analysis of spontaneous abortions. Am J Hum Genet.
66:1516–1521. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lathi RB, Gustin SL, Keller J,
Maisenbacher MK, Sigurjonsson S, Tao R and Demko Z: Reliability of
46,XX results on miscarriage specimens: A review of 1,222
first-trimester miscarriage specimens. Fertil Steril. 101:178–182.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schaeffer AJ, Chung J, Heretis K, Wong A,
Ledbetter DH and Lese Martin C: Comparative genomic
hybridization-array analysis enhances the detection of aneuploidies
and submicroscopic imbalances in spontaneous miscarriages. Am J Hum
Genet. 74:1168–1174. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kimberly L, Case A, Cheung AP, Sierra S,
AlAsiri S, Carranza-Mamane B, Case A, Dwyer C, Graham J, Havelock
J, et al: Advanced reproductive age and fertility: No. 269,
November 2011. Int J Gynaecol Obstet. 117:95–102. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shen J, Wu W, Gao C, Ochin H, Qu D, Xie J,
Gao L, Zhou Y, Cui Y and Liu J: Chromosomal copy number analysis on
chorionic villus samples from early spontaneous miscarriages by
high throughput genetic technology. Mol Cytogenet. 9:72016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bagheri H, Mercier E, Qiao Y, Stephenson
MD and Rajcan-Separovic E: Genomic characteristics of miscarriage
copy number variants. Mol Hum Reprod. 21:655–661. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Foyouzi N, Cedars MI and Huddleston HG:
Cost-effectiveness of cytogenetic evaluation of products of
conception in the patient with a second pregnancy loss. Fertil
Steril. 98:151–155. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liang D, Peng Y, Lv W, Deng L, Zhang Y, Li
H, Yang P, Zhang J, Song Z, Xu G, et al: Copy number variation
sequencing for comprehensive diagnosis of chromosome disease
syndromes. J Mol Diagn. 16:519–526. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kearney HM, South ST, Wolff DJ, Lamb A,
Hamosh A and Rao KW; Working Group of the American College of
Medical Genetics, : American College of Medical Genetics
recommendations for the design and performance expectations for
clinical genomic copy number microarrays intended for use in the
postnatal setting for detection of constitutional abnormalities.
Genet Med. 13:676–679. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shaffer LG and Rosenfeld JA:
Microarray-based prenatal diagnosis for the identification of fetal
chromosome abnormalities. Expert Rev Mol Diagn. 13:601–611. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang MZ, Lin FQ, Li M, He D, Yu QH, Yang
XX and Wu YS: Semiconductor Sequencing Analysis of Chromosomal Copy
Number Variations in Spontaneous Miscarriage. Med Sci Monit.
23:5550–5557. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen J, Hanna CW, Martell S, Leung PC,
Lewis SM, Robinson WP, Stephenson MD and Rajcan-Separovic E:
Functional consequences of copy number variants in miscarriage. Mol
Cytogenet. 8:62015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang H, Dong Z, Zhang R, Chau MHK, Yang Z,
Tsang KYC, Wong HK, Gui B, Meng Z, Xiao K, et al: Low-pass genome
sequencing versus chromosomal microarray analysis: Implementation
in prenatal diagnosis. Genet Med. 22:500–510. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chiu RW, Chan KC, Gao Y, Lau VY, Zheng W,
Leung TY, Foo CH, Xie B, Tsui NB, Lun FM, et al: Noninvasive
prenatal diagnosis of fetal chromosomal aneuploidy by massively
parallel genomic sequencing of DNA in maternal plasma. Proc Natl
Acad Sci USA. 105:20458–20463. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Chen S, Xie W, Vogel I, Choy KW,
Chen F, Christensen R, Zhang C, Ge H, Jiang H, et al: PSCC:
Sensitive and reliable population-scale copy number variation
detection method based on low coverage sequencing. PLoS One.
9:e850962014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szatkiewicz JP, Wang W, Sullivan PF, Wang
W and Sun W: Improving detection of copy-number variation by
simultaneous bias correction and read-depth segmentation. Nucleic
Acids Res. 41:1519–1532. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Menasha J, Levy B, Hirschhorn K and Kardon
NB: Incidence and spectrum of chromosome abnormalities in
spontaneous abortions: New insights from a 12-year study. Genet
Med. 7:251–263. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Levy SE and Myers RM: Advancements in
Next-Generation Sequencing. Annu Rev Genomics Hum Genet. 17:95–115.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gu W, Miller S and Chiu CY: Clinical
Metagenomic Next-Generation Sequencing for Pathogen Detection. Annu
Rev Pathol. 14:319–338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bianchi DW, Parker RL, Wentworth J,
Madankumar R, Saffer C, Das AF, Craig JA, Chudova DI, Devers PL,
Jones KW, et al CARE Study Group, : DNA sequencing versus standard
prenatal aneuploidy screening. N Engl J Med. 370:799–808. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang J, Li J, Saucier JB, Feng Y, Jiang
Y, Sinson J, McCombs AK, Schmitt ES, Peacock S, Chen S, et al:
Non-invasive prenatal sequencing for multiple Mendelian monogenic
disorders using circulating cell-free fetal DNA. Nat Med.
25:439–447. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen M and Zhao H: Next-generation
sequencing in liquid biopsy: Cancer screening and early detection.
Hum Genomics. 13:342019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dong Z, Zhang J, Hu P, Chen H, Xu J, Tian
Q, Meng L, Ye Y, Wang J, Zhang M, et al: Low-pass whole-genome
sequencing in clinical cytogenetics: A validated approach. Genet
Med. 18:940–948. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duan J, Zhang JG, Deng HW and Wang YP:
Comparative studies of copy number variation detection methods for
next-generation sequencing technologies. PLoS One. 8:e591282013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luthra R, Chen H, Roy-Chowdhuri S and
Singh RR: Next-Generation Sequencing in Clinical Molecular
Diagnostics of Cancer: Advantages and Challenges. Cancers (Basel).
7:2023–2036. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Chen L, Zhou C, Wang L, Xie H,
Xiao Y, Zhu H, Hu T, Zhang Z, Zhu Q, et al: Prospective chromosome
analysis of 3429 amniocentesis samples in China using copy number
variation sequencing. Am J Obstet Gynecol. 219:287.e1–287.e18.
2018. View Article : Google Scholar
|
|
31
|
South ST, Lee C, Lamb AN, Higgins AW and
Kearney HM; Working Group for the American College of Medical
Genetics and Genomics Laboratory Quality Assurance Committee, :
ACMG Standards and Guidelines for constitutional cytogenomic
microarray analysis, including postnatal and prenatal applications:
Revision 2013. Genet Med. 15:901–909. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kearney HM, Thorland EC, Brown KK,
Quintero-Rivera F and South ST; Working Group of the American
College of Medical Genetics Laboratory Quality Assurance Committee,
: American College of Medical Genetics standards and guidelines for
interpretation and reporting of postnatal constitutional copy
number variants. Genet Med. 13:680–685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
33. Brothman AR, Dolan MM, Goodman BK,
Park JP, Persons DL, Saxe DF, Tepperberg JH, Tsuchiya KD, Van Dyke
DL, Wilson KS, et al: College of American Pathologists/American
College of Medical Genetics proficiency testing for constitutional
cytogenomic microarray analysis. Genet Med. 13:765–769. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Du Y, Chen L, Lin J, Zhu J, Zhang N, Qiu
X, Li D and Wang L: Chromosomal karyotype in chorionic villi of
recurrent spontaneous abortion patients. Biosci Trends. 12:32–39.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yuan SM, Liao C, Li DZ, Huang JZ, Hu SY,
Ke M, Zhong HZ and Yi CX: Chorionic villus cell culture and
karyotype analysis in 1,983 cases of spontaneous miscarriage.
Zhonghua Fu Chan Ke Za Zhi. 52:461–466. 2017.(In Chinese).
PubMed/NCBI
|
|
36
|
Benn P: Trisomy 16 and trisomy 16
Mosaicism: A review. Am J Med Genet. 79:121–133. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wapner RJ, Martin CL, Levy B, Ballif BC,
Eng CM, Zachary JM, Savage M, Platt LD, Saltzman D, Grobman WA, et
al: Chromosomal microarray versus karyotyping for prenatal
diagnosis. N Engl J Med. 367:2175–2184. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hanna JS, Shires P and Matile G: Trisomy 1
in a clinically recognized pregnancy. Am J Med Genet. 68:981997.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Dunn TM, Grunfeld L and Kardon NB: Trisomy
1 in a clinically recognized IVF pregnancy. Am J Med Genet.
99:152–153. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ohsaka A, Hisa T, Watanabe N, Kojima H and
Nagasawa T: Tetrasomy 21 as a sole chromosome abnormality in acute
myeloid leukemia. fluorescence in situ hybridization and spectral
karyotyping analyses. Cancer Genet Cytogenet. 134:60–64. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Slavotinek AM, Chen XN, Jackson A, Gaunt
L, Campbell A, Clayton-Smith J and Korenberg JR: Partial tetrasomy
21 in a male infant. J Med Genet. 37:E302000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Alkan G, Emiroglu MK and Kartal A:
DiGeorge Syndrome with Sacral Myelomeningocele and Epilepsy. J
Pediatr Neurosci. 12:344–345. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Siew JX and Yap F: Growth trajectory and
pubertal tempo from birth till final height in a girl with
Wolf-Hirschhorn syndrome. Endocrinol Diabetes Metab Case Rep.
2018:18–0001. 2018.PubMed/NCBI
|
|
44
|
Warner ME, Martin DP, Warner MA, Gavrilova
RH, Sprung J and Weingarten TN: Anesthetic Considerations for
Angelman Syndrome: Case Series and Review of the Literature. Anesth
Pain Med. 7:e578262017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hyde KJ and Schust DJ: Genetic
considerations in recurrent pregnancy loss. Cold Spring Harb
Perspect Med. 5:a0231192015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rull K, Nagirnaja L and Laan M: Genetics
of recurrent miscarriage: Challenges, current knowledge, future
directions. Front Genet. 3:342012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ogasawara M, Aoki K, Okada S and Suzumori
K: Embryonic karyotype of abortuses in relation to the number of
previous miscarriages. Fertil Steril. 73:300–304. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sullivan AE, Silver RM, LaCoursiere DY,
Porter TF and Branch DW: Recurrent fetal aneuploidy and recurrent
miscarriage. Obstet Gynecol. 104:784–788. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Larsen EC, Christiansen OB, Kolte AM and
Macklon N: New insights into mechanisms behind miscarriage. BMC
Med. 11:1542013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sugiura-Ogasawara M, Ozaki Y, Katano K,
Suzumori N, Kitaori T and Mizutani E: Abnormal embryonic karyotype
is the most frequent cause of recurrent miscarriage. Hum Reprod.
27:2297–2303. 2012. View Article : Google Scholar : PubMed/NCBI
|