Introduction
Osteosarcoma (OS) is a type of primary malignant
tumor of the bone that is characterized by the production of
osteoid or immature bone by malignant cells (1,2). OS
is an uncommon tumor type. In total, ~750-900 new cases are
diagnosed each year in the USA, of which 400 arise in children and
adolescents <20 years of age (3). Despite its rarity, OS is the most
common primary malignancy of bone in children and adolescents, and
the fifth most common malignancy among adolescents and young adults
aged 15–19 years (3,4).
It has been shown that effective chemotherapy may
improve the survival of patients with OS in numerous reports
(5). The combination of surgical
resection and multiple chemotherapy, including neoadjuvant therapy,
has been standardized for the clinical treatment of patients with
OS since 1970 (6). This regimen
markedly ameliorates symptoms and extends the overall survival time
of patients with OS (6). It has
been reported that the combination of three types of chemotherapy,
including methotrexate, doxorubicin and cisplatin (CDDP), has
achieved a good outcome, particularly for children and young adults
(5). As such, reducing drug
resistance and improving drug sensitivity are significant issues
for chemotherapy in OS.
Lee et al (7) first discovered microRNAs (miRNAs),
endogenous small RNA molecules that can regulate >30% of all
human genes, and which have been shown to exhibit regulatory
functions (8,9). miRNAs bind to the 3′-untranslated
region perfectly or imperfectly and lead to translational
suppression or the degradation of various target mRNAs (10). Furthermore, it has been reported
that miRNAs have regulatory roles in gene expression at the
post-transcriptional level (11).
miRNAs have been found to serve regulatory roles in tumorigenesis
and chemosensitivity in various types of cancer (12). An increasing number of studies have
concentrated on the role of miRNAs in OS drug resistance, with the
aim of providing possibilities for improving the quality of life of
patients with OS. Numerous reports have argued that miRNAs serve a
significant role in OS drug resistance (13). miRNAs are associated with OS drug
resistance through the DNA damage response, apoptosis avoidance,
autophagy induction, activation of cancer stem cells and alteration
of signaling pathways (14–17).
Notably, it has been found that apoptosis is associated with death,
while autophagy acts as a double-edged sword in malignant tumor
cells (18). The role of autophagy
in OS drug-resistance requires further investigation.
miRNA-22 (miR-22) is located on the 17p13 chromosome
and serves crucial roles in tumor suppression in numerous
malignancies, including breast and prostate cancers (19–21).
A previous study found that miR-22 suppresses the proliferation and
promotes the sensitivity of OS cells by inhibiting metadherin
(MTDH)-mediated autophagy (22).
MTDH upregulates multidrug resistance gene 1 expression levels and
is associated with autophagy and chemoresistance (23). A recent report demonstrated that
miR-22 inhibits the proliferation and migration of OS cells, as
well as increasing their sensitivity to CDDP (24). However, the mechanisms of action of
miR-22 in OS drug resistance, particularly with regards to
drug-resistance, require further study. Furthermore, the
association between apoptosis and autophagy, which are associated
with miR-22 in OS drug resistance, requires defining, and more
target genes associated with miR-22 need to be investigated. This
present study aimed to investigate the roles of miR-22-regulated
apoptosis and autophagy in OS CDDP resistance.
Materials and methods
Cell lines
Human OS cell lines, including MG-63, U2OS, Saos2
were obtained from The Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences, Shanghai, China. OS9901 cells
(25–28) were donated by the Department of
Orthopedics, Tangdu Hospital, the Fourth Military Medical
University, Xi'an, China. The cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; Hyclone; GE Healthcare) containing
10% FBS (HyClone; GE Healthcare Life Sciences), 100 U/ml penicillin
(HyClone; GE Healthcare Life Sciences) and 100 µg/ml streptomycin
(HyClone; GE Healthcare Life Sciences), and incubated at 37°C with
5% CO2. CDDP (2 µM) was added to the cell lines for 24
h. Next, reverse transcription-quantitative polymerase chain
reaction (RT-qPCR) assays and western blotting were performed to
investigate the expression levels of cleaved caspase-3 and
microtubule-associated protein 1A/1B-light chain 3 (LC3).
Cell culture and transfection
The drug resistant cells (MG-63/CDDP) were obtained
by growing MG-63 cells in a 6-well plate and adding 2 µM CDDP for
24 h. Dead cells were removed by phosphate-buffered saline (PBS).
When the cells had reached 80% confluence, 2 µM CDDP was added for
24 h. This was repeated until CDDP no longer led to any further
cell death. The final cells were deemed drug-resistant cells
(MG-63/CDDP).
Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.) was used for all transfection assays,
according to the manufacturer's protocol. MG-63 cells and
MG-63/CDDP cells were transiently transfected using negative
control (NC) or miR-22 mimic (Shanghai GenePharma Co., Ltd.). The
concentration of each miRNA transfected was 50 nM per well. A total
of 50 µl Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) was used
to dilute 50 nM NC or mimic. The diluted NC and mimic solutions
were mixed with 3 µl Lipofectamine® 3000 and incubated
for 20 min at room temperature. This solution was incubated with
the cells in a 6-well plate (100 µl liposome transfection mixture
per well). Following incubation for 6 h, the medium was replaced
with DMEM with 10% fetal bovine serum (HyClone; Cytiva). After 48 h
of incubation, the cells were harvested and centrifuged at 500 g
for 5 min at room temperature.
The 5′-3′ sequences of the NC and miR-22 mimic
constructs were as follows: NC sense, UUCUCCGAACGUGUCACGUTT and
antisense, ACGUGACACGUUCGGAGAATT; and miR-22 sense,
AAGCUGCCAGUUGAAGAACUGU and antisense, AGUUCUUCAACUGGCAGCUUUU.
Cell proliferation assays
MG-63 and MG-63/CDDP cells (5,000 cells/well) were
plated into 96-well plates, which were divided into MG-63 and
MG-63/CDDP groups. The MG-63 groups consisted of a control group, a
miR-22 group, a CDDP group and a CDDP + miR-22 group. The
MG-63/CDDP groups also consisted of a control group, a miR-22
group, a CDDP group and a CDDP + miR-22 group. Each group was
analyzed in triplicate. After the cells were adhered, the 2 µM CDDP
treatment was applied and the cells were incubated for 6, 12 or 24
h. A total of 10 µl MTT (5 mg/ml) was added to each well and the
plates were incubated for a further 3 h. Finally, ~150 µl dimethyl
sulfoxide (Sigma-Aldrich; Merck KGaA) was added to each well
following removal of the medium. An absorbance microplate reader
(Thermo Fisher Scientific, Inc.; Multiskan 51119000) was used to
measure the absorbance at 570 nm.
Apoptosis assays
The experimental cells were washed three times with
PBS at. Centrifugation was performed at 500 × g for 5 min at 4°C.
The cells were then bathed with 1 ml binding buffer and centrifuged
(500 × g, 5 min, 4°C). Binding buffer-suspended cells (100 µl), and
5 µm propidium iodide (eBioscience; Thermo Fisher Scientific, Inc.)
and 5 µl FITC-Annexin V (eBioscience; Thermo Fisher Scientific,
Inc.) were added. The cells were uniformly mixed and then incubated
at room temperature for 15 min in the dark. Binding buffer (400 µl)
was added to the mixture. The cells were immediately analyzed by
flow cytometry. Finally, the data and output reports were collected
and analyzed. Following incubation for 24 h, apoptosis assays was
performed.
Flow cytometry assays
Following incubation for 24 h, a flow cytometry
assay was performed. Changes in the expression levels of the
autophagy-related protein, LC3, were quantitatively evaluated using
flow cytometry (BD Calibur flow cytometer; BD Biosciences). Cells
were fixed with 4% paraformaldehyde for 10 min at 4°C, followed by
three washes with PBS (5 min each wash). PBS/1% BSA solution was
used for preparing a 1:500 dilution of the LC3 antibody.
Subsequently, the cells were incubated for 2 h with the primary
antibody (LC3, cat. no. ab229327; Abcam) at room temperature. The
Alexa Fluor-488-conjugated secondary antibody (cat. no. A11008;
Invitrogen; Thermo Fisher Scientific, Inc.) was dissolved in PBS/1%
BSA solution to 1:400 and then added to the fixed cells in the dark
at room temperature for 1 h. The treated cells were analyzed using
flow cytometry on a BD FACSCalibur platform (BD Biosciences).
Changes in cellular fluorescence were detected in the FL1 channel
(BD Caliber, FACSCalibur; BD Biosciences).
The isotype control was stained with rabbit IgG
(cat. no. ab172730; Abcam) under the aforementioned conditions. BSA
was used for blocking unspecific protein.
RT-qPCR
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to isolate total RNA from cells,
according to the manufacturer's protocol. The PrimeScript RT
Reagent kit (Takara Bio, Inc.) was used to convert RNA into cDNA.
The condition of RT was 15 min at 37°C and 5 sec at 85°C. The miRNA
Extraction kit (Guangzhou RiboBio Co., Ltd.) was used to convert
miRNA into cDNA. Quantification of the transcript expression levels
were obtained by performing qPCR using SYBR Premix Ex Taq (Takara
Bio, Inc.). The ABI StepOne Plus Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used for qPCR using
the SYBR-Green PCR kit (Takara Bio, Inc.). The qPCR protocol
consists of the following steps: i) Initial denaturation for 5 min
at 95°C; and ii) 40 cycles involving denaturation at 95°C for 10
sec, followed by annealing and extension at 60°C for 34 sec. Each
experiment was repeated three times. The miR-22 primers used were
from the Bulgeloop miRNA qRT-PCR kit (Guangzhou RiboBio Co., Ltd.).
Detailed information of the primers employed for the qPCR
experiments is presented in Table
I. The 2−ΔΔCq analysis method was used as
quantification method (29).
 | Table I.Detailed information of primers for
the quantitative polymerase chain reaction experiments. |
Table I.
Detailed information of primers for
the quantitative polymerase chain reaction experiments.
| Gene | Sense | Anti-sense | bp |
|---|
| GAPDH |
CTTTGGTATCGTGGAAGGACTC |
AGTAGAGGCAGGGATGATGT | 133 |
|
Caspase-3 |
CTCCACAGCACCTGGTTATT |
AAATTCAAGCTTGTCGGCATAC | 106 |
| Bcl-2 |
GGAGGATTGTGGCCTTCTTT |
GTTCAGGTACTCAGTCATCCAC | 113 |
| Bax |
GGAGCTGCAGAGGATGATTG |
AGTTGAAGTTGCCGTCAGAA | 98 |
| Beclin1 |
GCAGCTGGATAAGCTGAAGA |
CGACCCAGCCTGAAGTTATT | 99 |
| Atg5 |
TGACAAAGATGTGCTTCGAGAT |
AGTATGGTTCTGCTTCCCTTTC | 98 |
| LC3 |
GACATCTACGAGCAGGAGAAAG |
TCAGAAGCCGAAGGTTTCC | 78 |
Western blot analysis
The cells were washed with pre-chilled PBS and lysed
in 200 µl RIPA buffer (Beyotime Institute of Biotechnology) for 30
min on ice, followed by centrifugation at 13,500 × g at 4°C for 5
min. Protein quantification was performed using a bicinchoninic
acid protein quantification kit (Beyotime Institute of
Biotechnology). Next, 10% SDS-PAGE (Bio-Rad Laboratories, Inc.) was
used to separate the cellular proteins (30 µg/sample), followed by
blot transfer to a polyvinylidene difluoride membrane (EMD
Millipore). Post-blotting, the membrane was incubated with 5%
skimmed milk at 4°C for 1 h. To prepare 1:1,000 dilutions of the
primary antibodies, 5% BSA-containing TBS/Tween-20 (TBST) solution
was used, and the primary antibodies were incubated with the
membrane overnight at 4°C. The membranes were washed three times
with TBST at room temperature (10 min each wash). The secondary
antibody was diluted with TBST (1:5,000 dilution) and the membranes
were further incubated with the secondary antibody at room
temperature for 1 h, followed by three washes with TBST at room
temperature (10 min each wash). The chemiluminescence reaction was
performed using the Tanon chemiluminescence sensor system (Tanon
Science and Technology Co., Ltd.). GAPDH (cat. no. 200306-7E4;
Zen-BioScience) was used as the loading control. The antibodies
used in the western blot assays were as follows: Cleaved caspase-3
(cat. no. ab49822; Abcam), B-cell lymphoma-2 (Bcl-2; cat. no.
50599-2-Ig; ProteinTech Group, Inc.), Bcl-2-associated X protein
(Bax; cat. no. 12789-1-AP; ProteinTech), autophagy protein 5 (ATG5;
cat. no. ab108327; Abcam), beclin1 (cat. no. ab207612; Abcam), LC3
(cat. no. ab229327; Abcam) and MTDH (cat. no. ab227981; Abcam). The
secondary antibodies used were as follows: Horseradish peroxidase
(HRP)-conjugated goat anti-mouse IgG (H+L; cat. no. A0216, Beyotime
Institute of Biotechnology) and HRP goat anti-rabbit IgG (H+L; cat.
no. A0208; Beyotime Institute of Biotechnology). The LC3 protein
quantification was defined as the ratio of LC3 II to LC3 I.
Inoculation of tumor cells
Tumors used in the present study were the ones
obtained from our previous study (30). The detailed information is as
follows: To obtain the miR-22 overexpression vector, the pre-miR-22
hairpin was constructed in the pLV6-EF-1Af/puro lentivirus plasmid
(Shanghai GenePharma Co., Ltd.). The plasmid was co-transfected
into 293T cells using Lipofectamine® 2000 with the
pG-P1-VSVG, pG-P2-REV or pG-P3-RRE plasmids. Following culture for
48 h, the lentivirus particles were harvested and concentrated
using the lenti-X concentrator (Clontech Laboratories, Inc.),
according to the manufacturer's protocol. MG-63 and MG-63/CDDP
cells were plated into 6-well plates. Following culture for 8 h,
the lentivirus particles were added to the plate to treat cells
with a multiplicity of infection of 1:50. The stable clones were
selected using 1 µg/ml puromycin for ~1 week (fluid was changed
every three days).
All animal experiments were performed following the
approval of the Inner Mongolia Medical University Animal Ethics
Committee and according to the Guidelines for the Care and Use of
Laboratory Animals. After 7 days of adaptive feeding, 48 male nude
mice (19.51±2.71 g, 25°C, 12 h of light and 12 h of darkness
every day, 200 ml water per day, SPF-level animal house) aged
8-weeks (SCXK2015-0001) were randomly divided into the following 8
groups, with 6 mice in each group: i) MG63; ii) MG63 + miR-22; iii)
MG63 + CDDP; iv) MG63 + CDDP + miR-22; v) MG63/CDDP; vi) MG63/CDDP
+ miR-22; vii) MG63/CDDP + CDDP; and viii) MG63/CDDP + CDDP +
miR-22. Subcutaneous injections of 5×106 (100 µl)
cells in the right axilla were performed. The nude mice were fed
normally with access to food and water ad libitum and after
the appearance of small nodules (after 7 days of inoculation), the
MG63, MG63 + miR-22, MG63/CDDP and MG63/CDDP + miR-22 groups were
administered with a normal saline injection of 100 µl, twice
per week for 5 weeks. CDDP injections were administered (2 mg/kg)
twice per week for 5 weeks to the MG63 + CDDP, MG63 + CDDP +
miR-22, MG63/CDDP + CDDP and the MG63/CDDP + CDDP + miR-22 groups.
The Animal health and behavior were monitored every day. Time was
started when the forelimb underarm was injected with tumor cells
using a syringe. Subsequently, the diameter of the tumor was
measured twice a week, to ensure that the tumor did not exceed 1.5
cm length, and the nude mice were sacrificed after 6 weeks. No mice
exhibited more than one tumor and the maximum tumor volume were as
follows: 897.793 mm3 in the MG63 group, 490.469
mm3 in the MG63 + miR-22 group, 418.669 mm3
in the MG63 + CDDP group, 274.963 mm3 in the MG63 +
miR-22 + CDDP group, 964.291 mm3 in the MG63/CDDP group,
749.117 mm3 in the MG63/CDDP + miR-22 group, 456.618
mm3 in the MG63/CDDP + CDDP group and 303.634
mm3 in MG63/CDDP + miR-22 + CDDP group.
When nude mice were loaded with a tumor or injected
with cisplatin, the nude mice were anesthetized using isoflurane
gas. Following the operation, the nude mice were placed in the cage
for normal feeding. Samples were collected after 6 weeks of normal
feeding. A dose of 1.5% isoflurane was used for anesthesia. The
dose of pentobarbital administered was 100 mg/kg and the method of
euthanasia was intraperitoneal injection. After 10 min of
intraperitoneal administration of pentobarbital, the tumor tissue
was removed following the absence of respiration, heartbeat,
corneal reflex, muscle tone and mucosal color. Forty nude mice were
sacrificed 6 weeks after tumor cell tumor bearing, none of nude
mice died during the 6 weeks. The tumor growth was shown in our
previous study (30).
Immunohistochemistry
Samples were fixed with 4% paraformaldehyde at room
temperature for 48 h. Paraffin-embedded sections (4 µm) were placed
in an oven at 65°C for 2 h, dewaxed with water and washed with PBS
three times, for 5 min each wash. The dehydration box was put into
the lifting basket and dehydrated with 75% ethanol for 4 h, 85%
ethanol for 2 h, 90% ethanol for 2 h, 95% ethanol for 1 h,
anhydrous ethanol I for 30 min, anhydrous ethanol II for 30 min,
alcohol benzene for 5–10 min, -xylene I for 5–10 min, xylene II for
5–10 min, Wax I for 5–10 min, Wax II for 1 h and wax III for 1 h.
The slices were placed in EDTA buffer and microwaved for antigen
retrieval. The EDTA buffer was boiled over medium heat for, allowed
to cool for 10 min and then boiled again over low heat. The slices
were placed in 3% hydrogen peroxide solution (Sinopharm Chemical
Reagent Co., Ltd.) and incubated at room temperature for 10 min to
block endogenous peroxidase activity. PBS washes were performed
three times, each time for 5 min. The samples were blocked using 5%
BSA for 20 min at room temperature. BSA was then removed and 50
µl diluted primary antibodies (1:100) were added, ensuring
to cover the tissue on each slice, and samples were incubated
overnight at 4°C. Subsequently, the samples were washed with PBS
solution, followed by the addition of 50–100 µl secondary
antibody to each section and incubation at 4°C for 50 min. A total
of 50–100 µl fresh DAB (Sinopharm Chemical Reagent Co.,
Ltd.) was then added to each slice and the color development was
visualized under an Upright Metallurgical light microscope (Nikon
Corporation) at ×200 magnification. After the color development was
complete, the samples were rinsed with distilled water,
counterstained with hematoxylin (5 min, room temperature),
dehydrated with 1% hydrochloric acid alcohol (1 sec), rinsed with
tap water, turned back/blue with ammonia and then rinsed with
running water. The sections were subjected to a gradient of ethanol
(70–100%) for 10 min to dehydrate and dry the samples, followed by
the addition of xylene (Sinopharm Chemical Reagent Co., Ltd.). The
samples were then sealed with neutral gum (Sinopharm Chemical
Reagent Co., Ltd.).
The antibodies used in the immunohistochemistry
analyses were as follows: ATG5 (cat. no. ab108327; Abcam), beclin1
(cat. no. ab207612; Abcam), LC3 (cat. no. ab229327; Abcam) and MTDH
(cat. no. ab227981; Abcam).
Statistical analysis
Each experiment was performed three times and the
results are shown as the mean ± standard deviation. SPSS 17.0
software (SPSS, Inc.) was used for statistical analysis.
Differences among three or more groups were compared using one-way
analysis of variance followed by Tukey's post hoc test for
comparisons of the data between the groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of apoptosis and
autophagy-related genes is highest in the MG-63 cells, compared
with the other cell lines
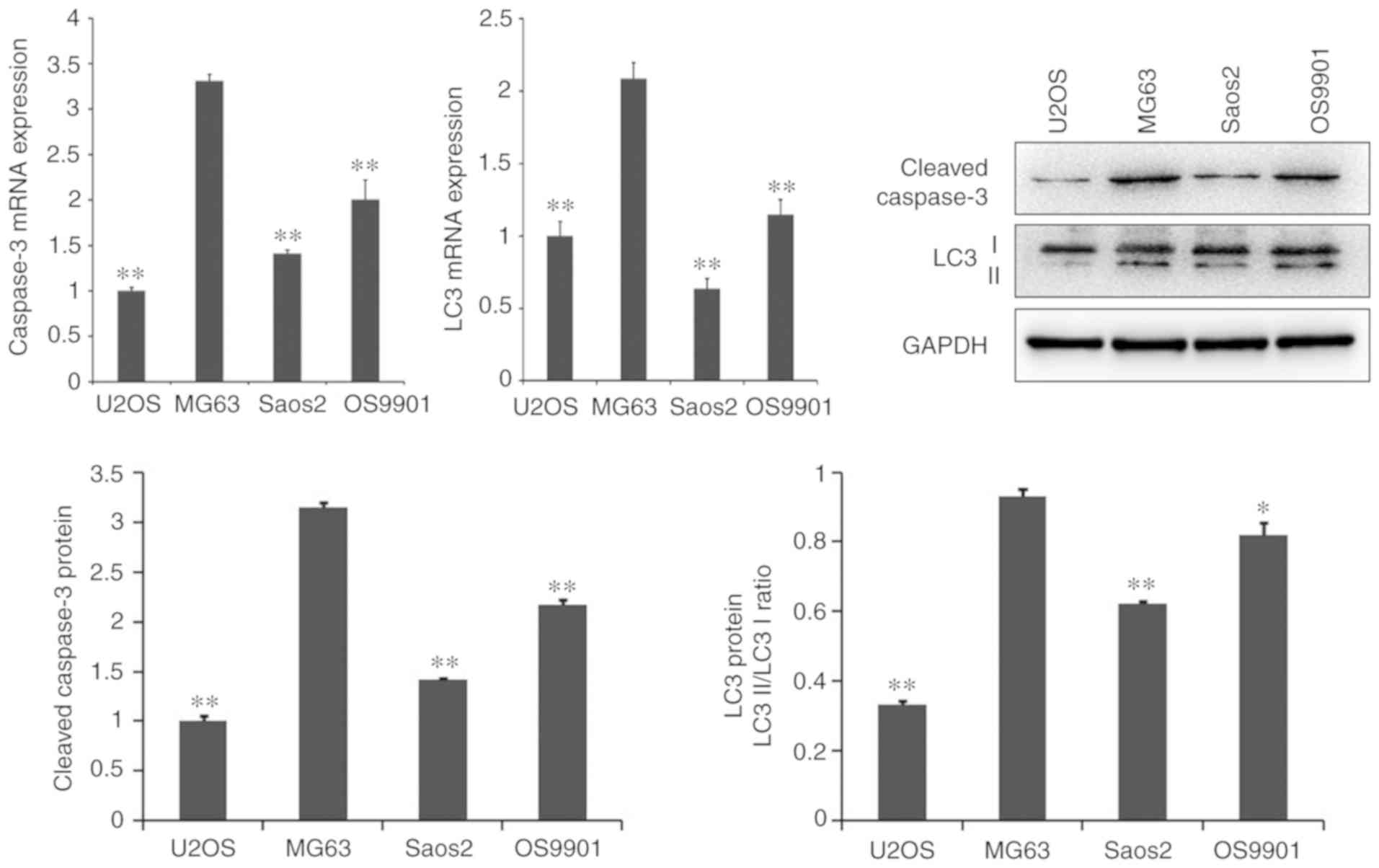
As presented in Fig.
1, the mRNA and protein expression levels of the
apoptosis-related gene, caspase-3, and the autophagy-related gene,
LC3, were significantly higher in MG-63 cells than in other cell
lines, including U2OS, Saos2 and OS9901 (P<0.05).
miR-22 inhibits the proliferation of
MG-63 cells and MG-63/CDDP cells and enhances the
anti-proliferative ability of CDDP
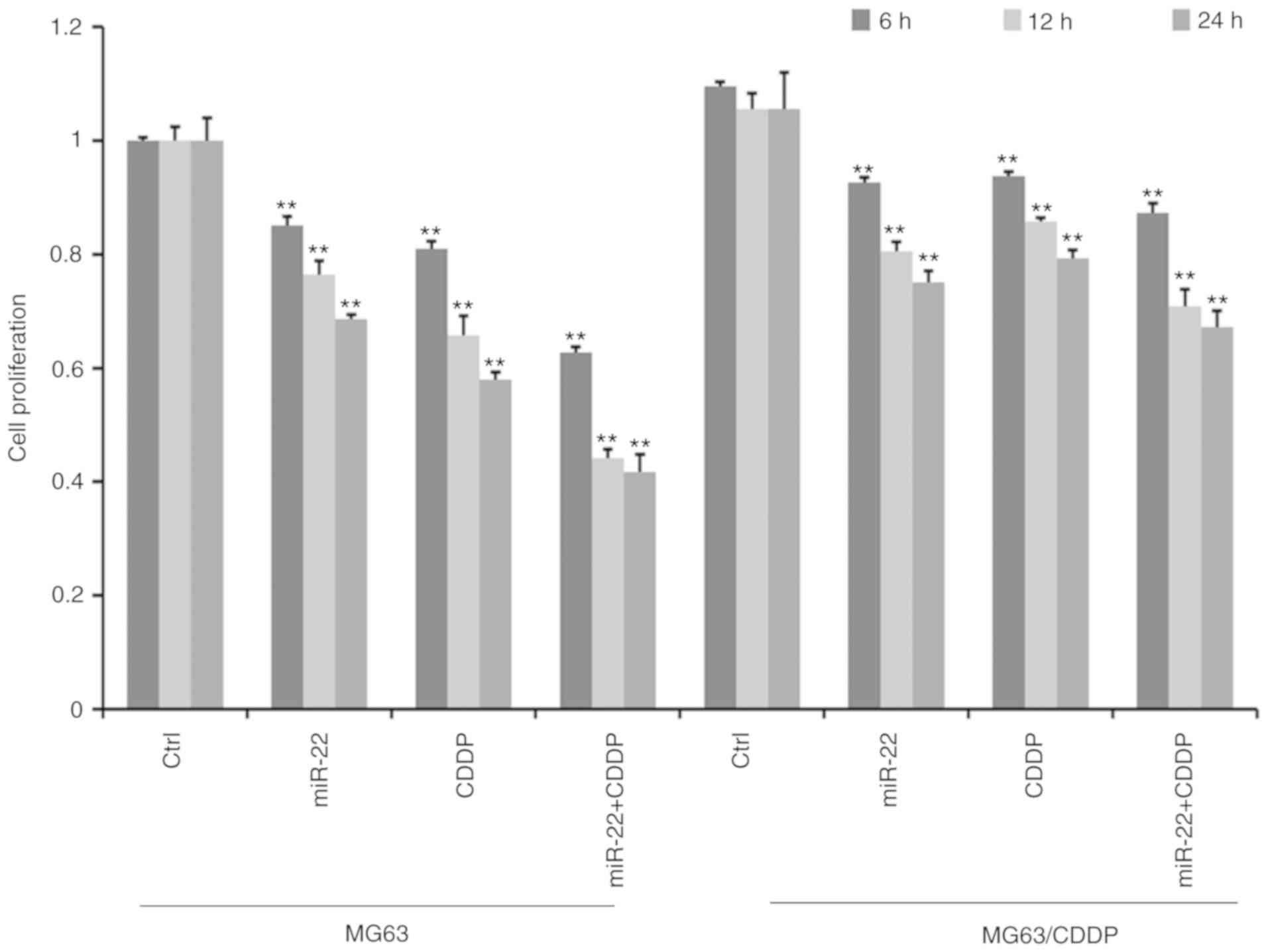
MTT assays revealed that the cell proliferation
ability decreased gradually with increasing treatment time in MG-63
cells and MG-63/CDDP cells (Fig.
2). Furthermore, cell proliferation was significantly
suppressed in the miR-22 transfected group compared with the
control group in both MG63 and MG63/CDDP cells (P<0.01). As
presented in Fig. 2, the miR-22 +
CDDP subgroup exhibited a significant decrease in proliferative
capacity compared with the other subgroups, both in MG-63 cells and
MG-63/CDDP cells at each time point of 6, 12 and 24 h (P<0.01),
which suggested that miR-22 enhanced CDDP sensitivity in MG-63
cells and that miR-22 decreased the drug resistance in MG-63/CDDP
cells.
miR-22 induces apoptosis of MG-63
cells and MG-63/CDDP cells
The results of the apoptosis assay are presented in
Fig. 3A and B. The apoptosis assay
was performed using an Annexin V-FITC kit. It was demonstrated that
miR-22-overexpression and CDDP treatment significantly induced cell
apoptosis, compared with the untreated cells in MG-63 cells and
MG-63/CDDP cells (P<0.01). In line with the results of the MTT
assays, the miR-22 + CDDP subgroup in the MG-63 cells and
MG-63/CDDP cells had the highest apoptosis rate at 24 h
(P<0.01). It was revealed that miR-22 enhanced CDDP sensitivity
and decreased drug resistance by inducing cell apoptosis.
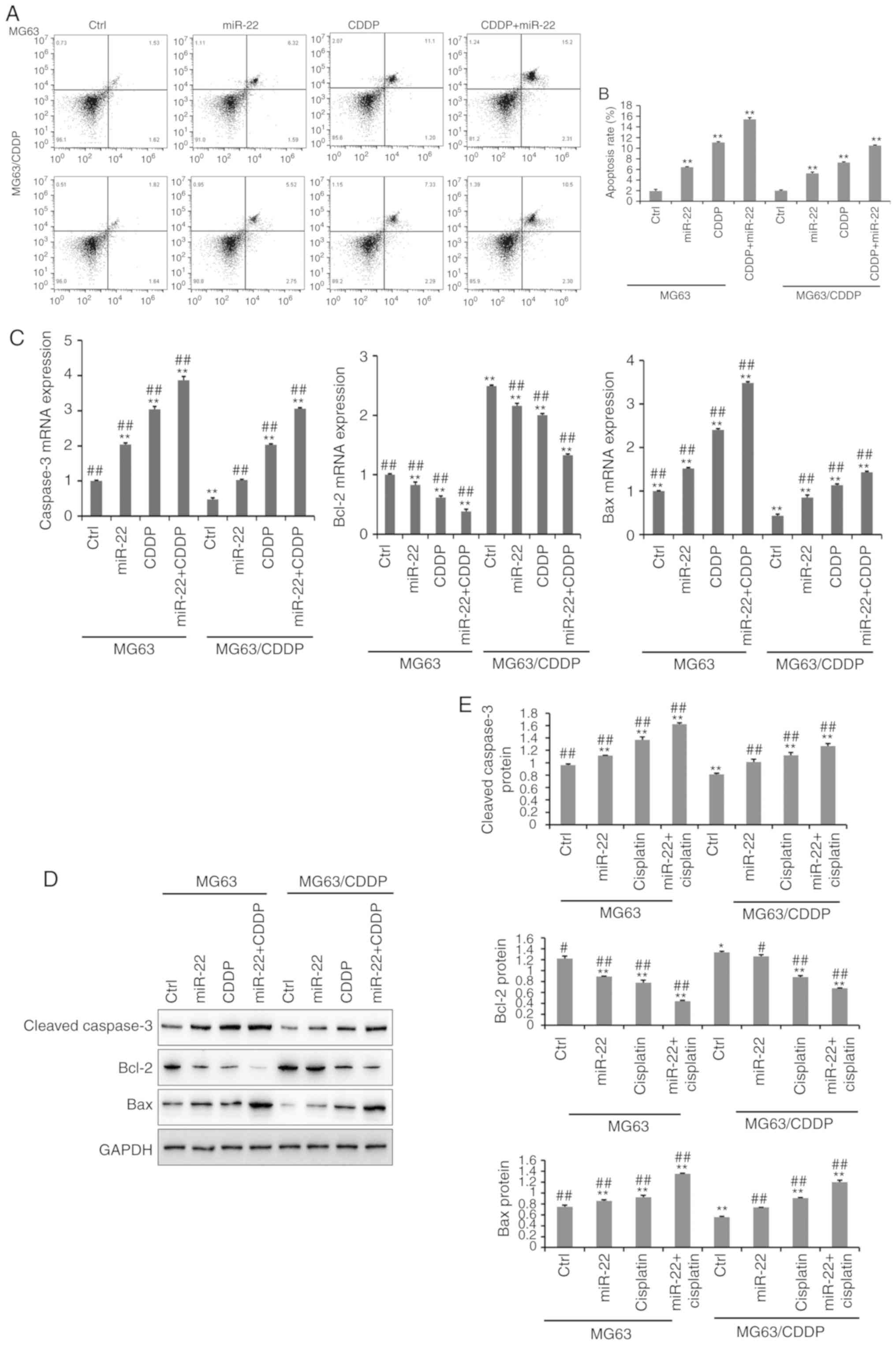 | Figure 3.The results of cell apoptosis (A-E).
(A) Cell apoptosis in MG-63 and MG-63/CDDP cells at 24 h. The upper
left quadrant represents dead cells, the lower left quadrant
represents normal living cells, the lower right quadrant represents
early apoptotic cells and the upper right quadrant represents late
apoptotic cells. The percentage of cells in the upper right
quadrant was calculated. (B) The rate of apoptosis is expressed as
the mean ± standard deviation. (C) The mRNA expression levels of
the apoptosis-related genes, caspase-3, Bcl-2 and Bax. (D and E)
The protein expression levels of the apoptosis-related genes,
cleaved caspase-3, Bcl-2 and Bax, as determined by western bolt
analyses. All protein bands were produced from the same membrane
and as a result they have the same loading control. The data are
presented as the mean ± standard deviation. *P<0.05, **P<0.01
vs. the MG-63 control group; #P<0.05,
##P<0.01 vs. the MG-63/CDDP control group. CDDP,
cisplatin; Bcl-2, B-cell lymphoma-2; Bax, Bcl-2-associated X
protein. |
Further experiments demonstrated that the expression
levels of apoptosis-related genes, including cleaved caspase-3,
Bcl-2 and Bax, at the mRNA and protein level (Fig. 3C-E); MG-63/CDDP cells exhibited an
upregulation of cleaved caspase-3 and Bax, and downregulation of
Bcl-2 (P<0.01). The expression levels of cleaved caspase-3 and
Bax increased gradually in the control, miR-22, CDDP and miR-22 +
CDDP subgroups (all P<0.01), while Bcl-2 was downregulated. The
results were the same in the MG-63 cells and MG-63/CDDP cells
(P<0.01). The combination of miR-22 and CDDP demonstrated the
most efficient ability to induce apoptosis by regulating the
aforementioned genes (P<0.01).
miR-22 inhibits autophagy of MG-63
cells and MG-63/CDDP cells
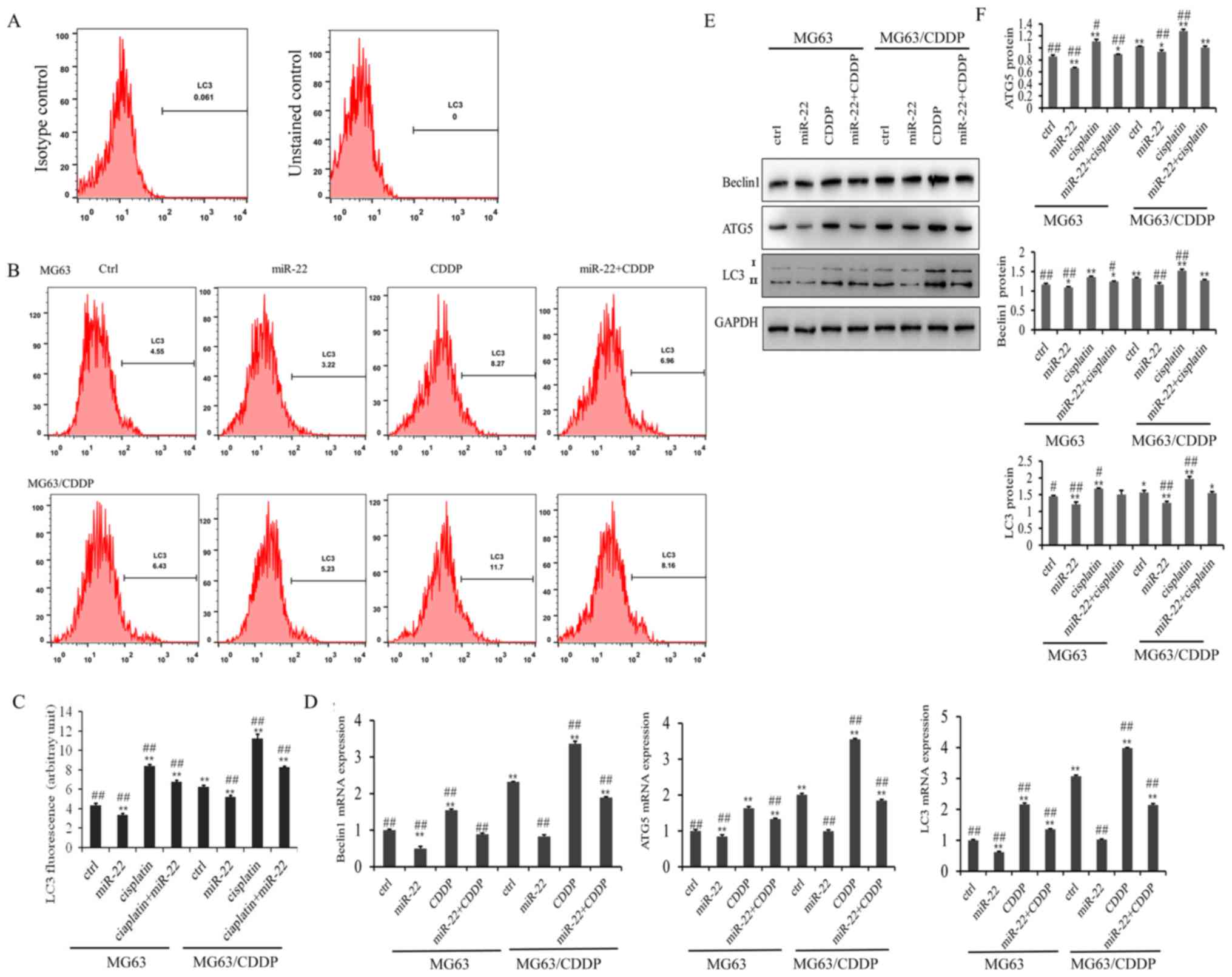
The results of the LC3 flow cytometry assay are
presented in Fig. 4A. As shown in
Fig. 4B, the LC3 fluorescence
value in the miR-22 group of the MG-63 and MG-63/CDDP cells was
smaller than in each control group, while the value was higher in
the CDDP group of the two cell lines, which revealed that miR-22
inhibited autophagy and that CDDP induced autophagy (P<0.01).
miR-22 decreased the autophagy induced by CDDP in MG-63 and
MG-63/CDDP cells (P<0.01).
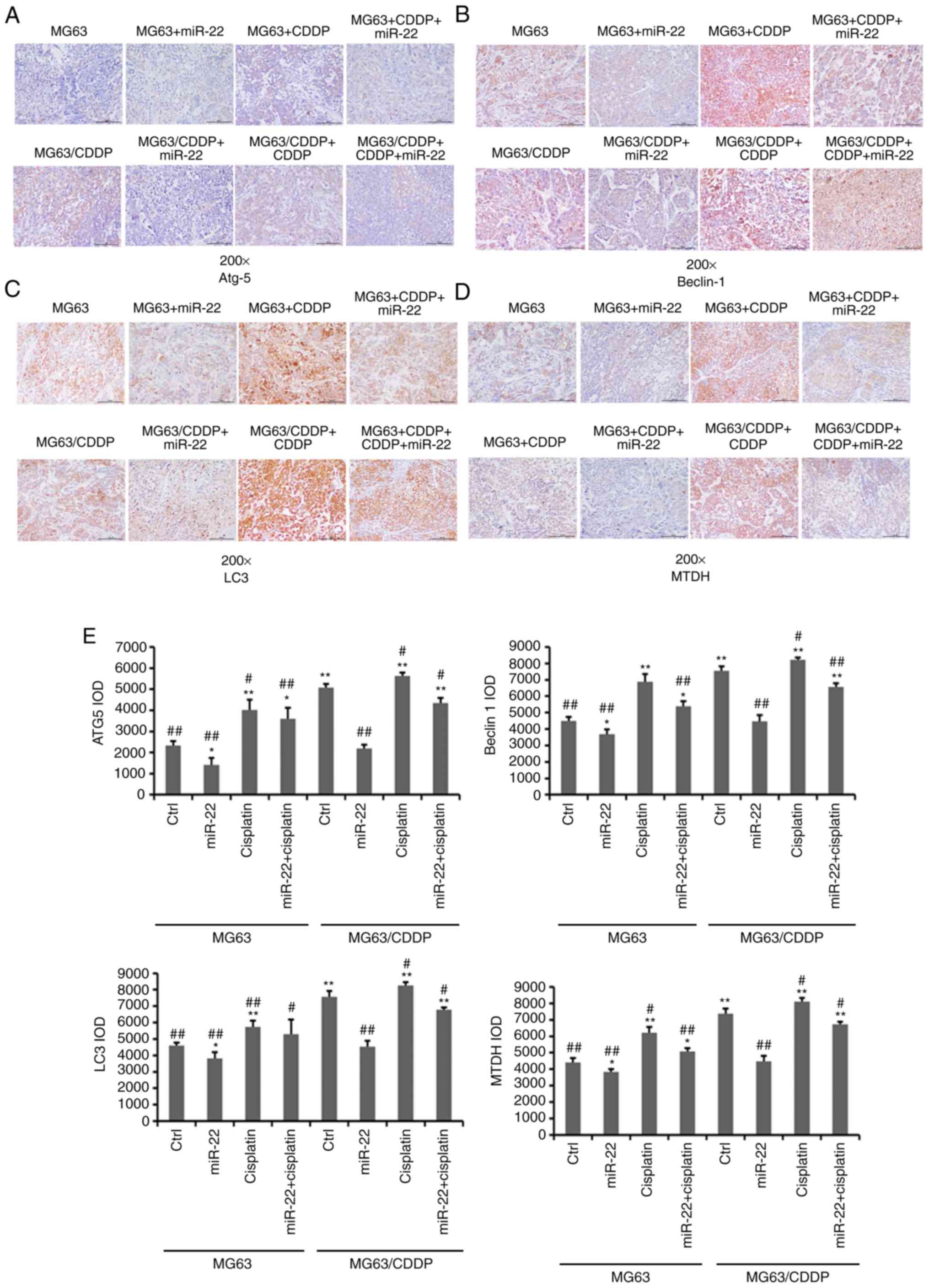
The mRNA and protein expression levels of the
autophagy-related genes, ATG5, beclin1 and LC3, are shown in
Fig. 4C-E. It was demonstrated
that miR-22-overexpression downregulated the mRNA and protein
expression levels of ATG5, beclin1 and LC3, while CDDP induced the
expression of ATG5, beclin1 and LC3 in the MG-63 cells and
MG-63/CDDP cells (P<0.01). Furthermore, miR-22 inhibited
CDDP-induced autophagy in the MG-63 cells and MG-63/CDDP cells.
These results suggested that miR-22 could enhance CDDP sensitivity
and decrease drug resistance through inhibiting cell autophagy.
miR-22 downregulates MTDH to improve
the sensitivity of CDDP and to decrease drug resistance
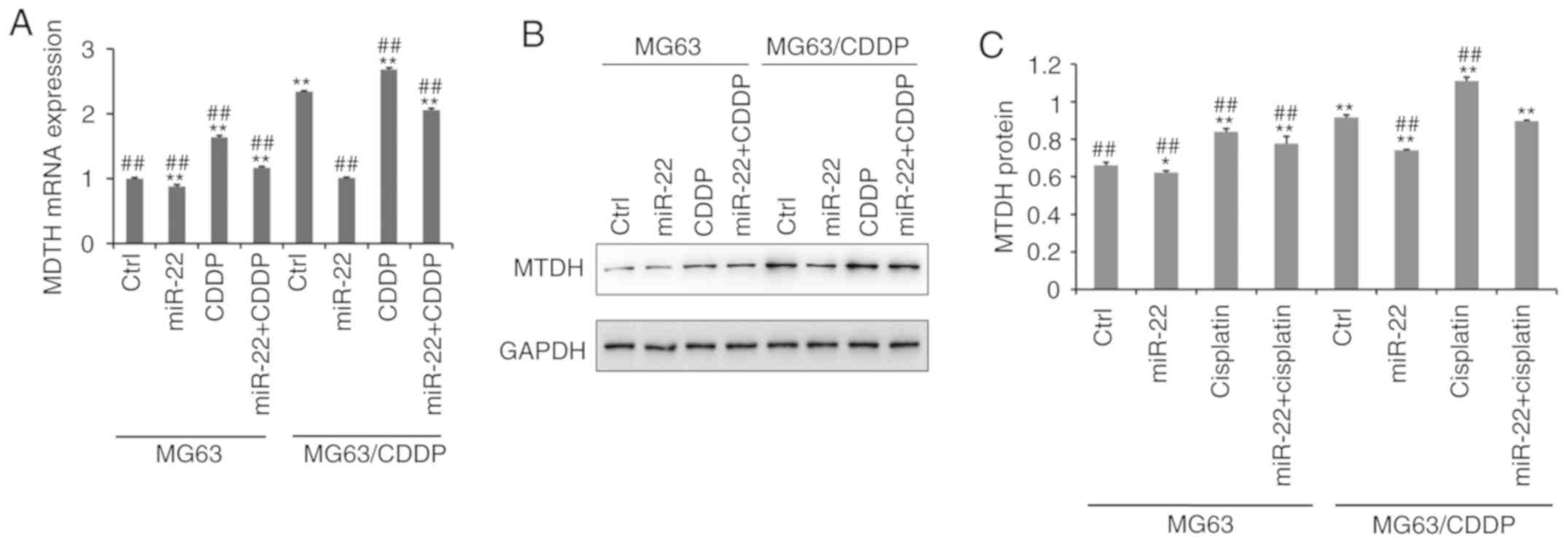
Fig. 5A
demonstrates that the mRNA expression level of MTDH was
downregulated following transfection with miR-22 mimic in MG-63
cells and MG-63/CDDP cells, whereas CDDP upregulated the expression
of MTDH (P<0.01), particularly in the drug-resistant cell line.
At the protein level, as shown in Fig.
5B and C, the expression level of MTDH was downregulated by
miR-22 mimic in MG-63 cells and MG-63/CDDP cells (P<0.05).
Additionally, at the mRNA and protein level, miR-22 may serve a
role in inhibiting CDDP-induced MTDH upregulation to increase the
sensitivity to CDDP and to decrease the drug resistance
(P<0.05).
miR-22 regulates the expression of
autophagy-related genes and MTDH in in vivo OS and OS drug
resistance models
As shown in Fig. 6,
the expression levels of Atg5, beclin1, LC3 and MTDH were
downregulated by miR-22 mimic in MG-63 and MG-63/CDDP cells
(P<0.05). The expression levels of these genes were further
downregulated in the MG-63 cells, compared with the MG-63/CDDP
cells (P<0.01). As in the in vitro model, in the in
vivo model, CDDP upregulated the expression levels of Atg5,
beclin1, LC3 and MTDH in the MG-63 and MG-63/CDDP groups
(P<0.05).
Discussion
Effective chemotherapy may improve the survival of
patients with OS. However, drug resistance is always a challenge
for treatment. In the present study, the role of miR-22 in CDDP
resistance was investigated. In the present study, it was
demonstrated that miR-22 could inhibit the proliferation of MG-63
cells and MG-63/CDDP cells and enhance the anti-proliferative
ability of CDDP treatment. miRNA decreased the drug-resistance
ability in OS through affecting cell proliferation. Zou et
al (31) reported that
miR-133b induces chemoresistance of OS cells to CDDP treatment by
promoting cell death, migration and invasion. Vanas et al
(32) revealed that miR-21 could
inhibit cell proliferation to increase chemosensitivity to
antitumor drugs by targeting sprouty2. The present results were
similar to a recent report by Zhou et al, which argued that
miR-22 inhibits the proliferation and migration, as well as
increasing the cisplatin-sensitivity of OS cells (24). In the present study, the mechanisms
of action of miR-22 in drug-resistance cells were investigated, and
it was demonstrated that miR-22 inhibited the proliferation of
MG-63/CDDP cells. According to the present results, it can be
verified that miR-22 may not only increase the CDDP sensitivity but
also reduce the resistance of OS cells.
Apoptosis is considered to predict the treatment
effect of anticancer drugs. Dysfunction of the normal apoptosis
pathways may lead to uncontrolled cell proliferation, which can be
seen in resistant cancer cells. Furthermore, the apoptotic process
has been deemed an effective target to reverse cancer drug
resistance (33). It has been
reported that numerous miRNAs are associated with OS through cell
apoptosis. Downregulation of miR-138 has been revealed in OS
tissues and cell lines, with further research demonstrating that
miR-138 transfection suppresses cell proliferation and induces cell
apoptosis, as well as increasing drug responsiveness by binding to
Enhancer Of Zeste Homolog 2 (34).
It has been demonstrated that Bcl-2 expression has a positive
correlation with miR-21, which inhibits apoptosis and induces
resistance to CDDP, while Bcl-2 siRNA may decrease the
Bcl-2-induced resistance to CDDP (35). Additionally, miR-22 has been
demonstrated to promote the apoptosis of OS cells through inducing
cell cycle arrest (36). In the
present study, it was revealed that miR-22 induced the apoptosis of
MG-63 cells and MG-63/CDDP cells. Further experiments suggested
that the expression levels of cleaved caspase-3 and Bax increased
gradually in the control, miR-22, CDDP and miR-22 + CDDP subgroups,
while Bcl-2 was shown to be downregulated. The present study
confirmed that miR-22 may enhance the CDDP sensitivity and decrease
the drug resistance through inducing cell apoptosis.
In the present study, the autophagy-related genes,
ATG5, beclin1 and LC3, were downregulated by miR-22 overexpression
in MG-63/CDDP cells, which was consistent with the conclusions
drawn from a previous study, which reported that miR-22 decreased
the expression of ATG5, beclin1 and LC3 in MG-63 cells (22). The present study further verified
that miR-22 could reduce CDDP resistance by inhibiting autophagy.
It has been reported that miR-22 may target HMGB1-suppressed
autophagy to reduce adriamycin and CDDP resistance (15,37).
MTDH has been reported to be associated with autophagy. The
overexpression of MTDH induces autophagy, while downregulation of
MTDH suppresses autophagy (38,39).
However, it has also been found that the simultaneous inhibition of
autophagy and overexpression of MTDH decreases the levels of P-gp
and inhibits 5-FU resistance (40). A previous study has reported that
miR-22-overexpressing OS cells treated with CDDP demonstrated a
significant decrease in the extent of autophagy from MDC staining.
In this present study, it was further verified that miR-22 directly
targeted the expression of MTDH in MG-63 cells. Furthermore, it was
revealed that miR-22 may serve a role in inhibiting CDDP-induced
MTDH upregulation, to increase the sensitivity to CDDP and to
decrease drug resistance, which may aid in providing a new target
gene to reduce CDDP resistance.
In conclusion, it was demonstrated in the present
study that miR-22 could regulate CDDP sensitivity and decrease CDDP
resistance by inhibiting autophagy and inducing apoptosis of OS
cells and drug-resistant cells. The mechanism of action behind
these effects may involve the inhibition of MG-63 and MG-63/CDDP
cell proliferation, as well as through enhancing the
anti-proliferative ability of CDDP treatment by apoptosis.
Furthermore, miR-22 inhibited CDDP-induced MTDH upregulation and
downregulated the expression levels of MTDH. The tumor formation
experiments in vivo, and the detailed autophagy pathways
associated with miR-22 in OS CDDP resistance require further study.
The present study provided new insights for anti-chemoresistance in
OS, contributing to the development of potential new therapeutic
drugs for treating OS.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81660440) and the
Natural Science Foundation of Inner Mongolia (grant no.
2018MS08031).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CYM, ZQZ, SBG, WF, CS and YXW participated in the
design of the study and drafted the manuscript. RB, LS and WZ
collected the data and performed the statistical analyses. CYM,
ZQZ, SBG and WF were major contributors to the design of this study
and revised the manuscript. All authors read and approved the
manuscript and agree to be accountable for all aspects of the
research in ensuring that the accuracy or integrity of any part of
the work are appropriately investigated and resolved.
Ethics approval and consent to
participate
All animal experiments were performed following the
approval of the Inner Mongolia Medical University Animal Ethics
Committee and according to the Guidelines for the Care and Use of
Laboratory Animals.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Atg5
|
autophagy protein 5
|
|
CDDP
|
cisplatin
|
|
LC3
|
microtubule-associated protein
1A/1B-light chain 3
|
|
MDC
|
monodansylcadaverine staining
|
|
miRNA/miR
|
microRNA
|
|
MTDH
|
metadherin
|
|
OS
|
osteosarcomas
|
References
|
1
|
Sissons HA: The WHO classification of bone
tumors. Recent Results Cancer Res. 54:104–108. 1976.
|
|
2
|
Mckenna R: Sarcomata of the osteogenic
series (osteosarcoma, fibrosarcoma, chondrosarcoma, parosteal
osteogenic sarcoma, and sarcomata arising in abnormal bone): An
analysis of 552 cases. J Bone Joint Surg Am. 48:1–26. 1966.
View Article : Google Scholar
|
|
3
|
Ries LAG, Smith MA, Gurney JG, Linet M,
Tamra T, Young JL and Bunin GR: Cancer incidence and survival among
children and adolescents: United States SEER Program, 1975–1995.
Bethesda, MD: 1999
|
|
4
|
Stiller CA, Bielack SS, Jundt G and
Steliarova-Foucher E: Bone tumours in European children and
adolescents, 1978–1997. Report from the automated childhood cancer
information system project. Eur J Cancer. 42:2124–2135. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Anninga JK, Gelderblom H, Fiocco M, Kroep
JR, Taminiau AH, Hogendoorn PC and Egeler RM: Chemotherapeutic
adjuvant treatment for osteosarcoma: Where do we stand? Eur J
Cancer. 47:2431–2445. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bentwich I, Avniel A, Karov Y, Aharonov R,
Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, et al:
Identification of hundreds of conserved and nonconserved human
microRNAs. Nat Genet. 37:766–770. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhong X, Coukos G and Zhang L: miRNAs in
human cancer. J Pathol. 223:102–115. 2015.
|
|
11
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xie L, Jing R, Qi J, Lin Z and Ju S: Drug
resistance-related microRNAs in hematological malignancies:
Translating basic evidence into therapeutic strategies. Blood Rev.
29:33–44. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Torreggiani E, Roncuzzi L, Perut F, Zini N
and Baldini N: Multimodal transfer of MDR by exosomes in human
osteosarcoma. Int J Oncol. 49:1892016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Duan Z, Gao Y, Shen J, Choy E, Cote G,
Harmon D, Bernstein K, Lozano-Calderon S, Mankin H and Hornicek FJ:
miR-15b modulates multidrug resistance in human osteosarcoma in
vitro and in vivo. Mol Oncol. 11:151–166. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Guo S, Bai R, Liu W, Zhao A, Zhao Z, Wang
Y, Wang Y, Zhao W and Wang W: miR-22 inhibits osteosarcoma cell
proliferation and migration by targeting HMGB1 and inhibiting
HMGB1-mediated autophagy. Tumor Biol. 35:7025–7034. 2014.
View Article : Google Scholar
|
|
16
|
Zhou J, Wu S, Chen Y, Zhao J, Zhang K,
Wang J and Chen S: microRNA-143 is associated with the survival of
ALDH1+CD133+ osteosarcoma cells and the chemoresistance of
osteosarcoma. Exp Biol Med. 240:867–875. 2015. View Article : Google Scholar
|
|
17
|
Zhao G, Cai C, Yang T, Qiu X, Liao B, Li
W, Ji Z, Zhao J, Zhao H, Guo M, et al: MicroRNA-221 induces cell
survival and cisplatin resistance through PI3K/Akt pathway in human
osteosarcoma. PLoS One. 8:e539062013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT,
Liu B and Bao JK: Programmed cell death pathways in cancer: A
review of apoptosis, autophagy and programmed necrosis. Cell
Prolif. 90:487–498. 2012. View Article : Google Scholar
|
|
19
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Knyazev EN, Samatov TR, Fomicheva KA,
Nyushko KM, Alekseev BY and Shkurnikov MY: MicroRNA hsa-miR-4674 in
hemolysis-free blood plasma is associated with distant metastases
of prostatic cancer. Bull Exp Biol Med. 161:112–115. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Damavandi Z, Torkashvand S, Vasei M,
Soltani BM, Tavallaei M and Mowla SJ: Aberrant expression of breast
development-related MicroRNAs, miR-22, miR-132, and miR-212, in
breast tumor tissues. J Breast Cancer. 19:148–155. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang P, Zhao ZQ, Guo SB, Yang TY, Chang
ZQ, Li DH, Zhao W, Wang YX, Sun C, Wang Y and Feng W: Roles of
microRNA-22 in suppressing proliferation and promoting sensitivity
of osteosarcoma cells via metadherin-mediated autophagy. Orthop
Surg. 11:285–293. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wan L, Hu G, Wei Y, Yuan M, Bronson RT,
Yang Q, Siddiqui J, Pienta KJ and Kang Y: Genetic ablation of
metadherin inhibits autochthonous prostate cancer progression and
metastasis. Cancer Res. 74:5336–5347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou X, Natino D, Zhai X, Gao Z and He X:
MicroRNA-22 inhibits the proliferation and migration, and increases
the cisplatin sensitivity, of osteosarcoma cells. Mol Med Rep.
17:7209–7217. 2018.PubMed/NCBI
|
|
25
|
Li Y, Geng P, Jiang W, Wang Y, Yao J, Lin
X, Liu J, Huang L, Su B and Chen H: Enhancement of radiosensitivity
by 5-Aza-CdR through activation of G2/M checkpoint response and
apoptosis in osteosarcoma cells. Tumour Biol. 35:4831–4839. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang TM, Guo SF, Chen CR, Zhang XY and Li
WG: Anti-Osteosarcom aeffects and mechanisms of 4-O-amino-
phenol-4′-demethylepipodophyllotoxin ether. J Pharm Pharmacol.
60:179–188. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tang J, Shen L, Yang Q and Zhang C:
Overexpression of metadherin mediates metastasis of osteosarcoma by
regulating epithelial-mesenchymal transition. Cell Prolif.
47:427–434. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ye H, Lin J, Yao X, Li Y, Lin X and Lu H:
Overexpression of long non-coding RNA NNT-AS1 correlates with tumor
progression and poor prognosis in osteosarcoma. Cell Physiol
Biochem. 45:1904–1914. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao S, Chen C, Chang K, Karnad A,
Jagirdar J, Kumar AP and Freeman JW: CD44 expression level and
isoform contributes to pancreatic cancer cell plasticity,
invasiveness, and response to therapy. Clin Cancer Res.
22:5592–5604. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meng CY, Zhao ZQ, Bai R, Zhao W, Wang YX,
Xue HQ, Sun L, Sun C, Feng W and Guo SB: MicroRNA-22 mediates the
cisplatin resistance of osteosarcoma cells by inhibiting autophagy
via the PI3K/Akt/mTOR pathway. Oncol Rep. 43:1169–1186.
2020.PubMed/NCBI
|
|
31
|
Zou Y, Yang J, Wu J, Luo C and Huang Y:
miR?133b induces chemoresistance of osteosarcoma cells to cisplatin
treatment by promoting cell death, migration and invasion. Oncol
Lett. 15:1097–1102. 2018.PubMed/NCBI
|
|
32
|
Vanas V, Haigl B, Stockhammer V and
Sutterlüty-Fall H: MicroRNA-21 increases proliferation and
cisplatin sensitivity of osteosarcoma-derived cells. PLoS One.
11:e01610232016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Karpel-Massler G, Shu C, Chau L, Banu M,
Halatsch ME, Westhoff MA, Ramirez Y, Ross AH, Bruce JN, Canoll P
and Siegelin MD: Combined inhibition of Bcl-2/Bcl-xL and Usp9X/Bag3
overcomes apoptotic resistance in glioblastoma in vitro and in
vivo. Oncotarget. 6:14507–14521. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu Z, Tang J, Wang J, Duan G, Zhou L and
Zhou X: miR-138 acts as a tumor suppressor by targeting EZH2 and
enhances cisplatin-induced apoptosis in osteosarcoma cells. PLoS
One. 11:e01500262016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ziyan W and Yang L: MicroRNA-21 regulates
the sensitivity to cisplatin in a human osteosarcoma cell line. Ir
J Med Sci. 185:85–91. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gai P, Sun H, Wang G, Xu Q and Jiang L:
miR-22 promotes apoptosis of osteosarcoma cells via inducing cell
cycle arrest. Oncol Lett. 13:2354–2358. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li X, Wang S, Chen Y, Liu G and Yang X:
miR-22 targets the 3′ UTR of HMGB1 and inhibits the
HMGB1-associated autophagy in osteosarcoma cells during
chemotherapy. Tumor Biol. 35:6021–6028. 2014. View Article : Google Scholar
|
|
38
|
Yan J, Zhang J, Zhang X, Li X, Li L, Li Z,
Chen R, Zhang L, Wu J, Wang X, et al: AEG-1 is involved in
hypoxia-induced autophagy and decreases chemosensitivity in T-cell
lymphoma. Mol Med. 24:352018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zou M, Zhu W, Wang L, Shi L and Hu G:
AEG-1/MTDH-activated autophagy enhances human malignant glioma
susceptibility to TGF-β1-triggered epithelial-mesenchymal
transition. Oncotarget. 7:13122–13138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pei G, Luo M, Ni X, Wu J, Wang S, Ma Y and
Yu J: Autophagy facilitates metadherin-induced chemotherapy
resistance through the AMPK/ATG5 pathway in gastric cancer. Cell
Physiol Biochem. 46:847–859. 2018. View Article : Google Scholar : PubMed/NCBI
|