Introduction
Colon cancer is the most frequent malignant tumor of
the digestive tract and has a high mortality due to its high
metastatic potential. It was responsible for ~1.4 million new cases
and ~700,000 deaths worldwide in 2012 (1). A previous study has reported that the
loss of CD82 expression and the increased expression of epidermal
growth factor receptor (EGFR) and/or its ligands are notable
biological characteristics of colon cancer, and these have been
associated with an increased risk of metastasis and hence poor
prognosis (2). CD82 is a member of
the tetraspanin superfamily of transmembrane proteins that are
ubiquitously expressed in normal tissues but expressed at low
levels in cancer cells and tissues. CD82 exerts its biological
effects by regulating cell migration, fusion, adhesion and
proliferation (3–5). An inverse relationship exists between
CD82 expression and the invasive and metastatic potentials of
cancer cells, and this relationship is frequently observed in a
wide range of malignant solid tumors, such as gastric, colon,
breast, skin, lung, pancreas and liver tumors (6–13).
Thus, CD82 is considered a wide-spectrum invasion- and
metastasis-suppressor.
CD82 suppresses cancer metastasis primarily by
inhibiting cancer cell migration and invasion. In the plasma
membrane, CD82 associates with other tetraspanin proteins to form
the tetraspanin web (14,15) consequently recruiting some membrane
components such as EGFR (2,16),
integrins (17,18) and gangliosides (19) into the tetraspanin web. The
formation of the tetraspanin web is a prerequisite for the proper
functioning of CD82. However, the mechanism through which CD82
redistributes the components of the tetraspanin web and the role of
CD82-associated proteins in the CD82-mediated suppression of cell
migration remains unclear.
Gangliosides are sialylated glycosphingolipids found
in the mammalian cell membrane that are involved in numerous
biological functions (20,21). It has been speculated that
gangliosides serve a role in a number of cell-surface events, such
as cell-to-cell adhesion, cell-extracellular matrix adhesion, and
plasma membrane receptor-mediated modulation of transmembrane
signaling (22–26). There is a growing body of evidence
that suggests that CD82 and gangliosides, particularly GM3
(NeuNAcα2, 3 Galβ1, 4Glc1, 1Cer) and GM2 (GalNacβ1, 4[NeuNAcα2, 3]
Galβ1, 4Glc1, 1Cer), which are characteristic components of
mammalian cell membranes, can synergistically inhibit cancer cell
migration (27–29). Previous studies have related the
low expression levels of GM3 to cancer development and
invasiveness, while GM2 are abundantly expressed on the cell
surface of certain types of human cancer. Although it is known that
quantitative and qualitative changes in the expression of
gangliosides are involved in oncogenic transformation of cells
(30–32), the exact mechanism has not been
fully elucidated. The present study aimed to investigate whether
EGFR signaling is involved in the mechanism through which CD82 and
gangliosides synergistically inhibit the motility and migration of
SW620 cells.
Materials and methods
Cell lines and cell culture
The human colorectal adenocarcinoma cell line SW620
was obtained from the Shanghai Institute of Biochemistry and Cell
Biology, Chinese Academy of Sciences. For all experiments, SW620
cells were grown in 10-cm cell culture dishes or in multiwell
plates in L15 medium supplemented with 10% heat-inactivated FBS
(both Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100
mg/ml streptomycin under a 5% CO2 atmosphere at
37°C.
Antibodies and reagents
Immunoglobulin G (IgG) antibodies against EGFR (cat.
no. sc-365829; 1:2,000), phosphorylated (p)-EGFR (Tyr-1173; cat.
no. sc-57542; 1:1,000), p-EGFR (Tyr-845) (cat. no. sc-57542;
1:1,000), p-EGFR (Tyr-1045; cat. no. sc-57541; 1:1,000), Akt1/2/3
(cat. no. sc-55523; 1:2,000), p-Akt1/2/3 (Ser473; cat. no.
sc-81433; 1:2,000), p-Akt1/2/3 (Thr308; cat. no. sc-16646-R;
1:2,000), MAPK (cat. no. sc-7383; 1:2,000) and p-p44/42 MAPK (cat.
no. sc-81492; 1:1,000) were obtained from Santa Cruz Biotechnology,
Inc. Anti-CD82 IgG (cat. no. ab59509; 1:5,000) and p-EGFR (Tyr-992;
cat. no. ab5638; 1:5,000) were obtained from Abcam, anti-GM3 IgM
(cat. no. 303-06541; 1:200) was obtained from Wako Pure Chemical
Industries Ltd., and anti-V5 IgG (cat. no. MA5-15253; 1:1,000) was
purchased from Invitrogen (Thermo Fisher Scientific, Inc.). Among
secondary antibodies, Goat anti-rabbit IgG-horseradish peroxidase
(cat. no. sc-2004; 1:2,000), rabbit anti-goat IgG-horseradish
peroxidase (cat. no. sc-2768; 1:2,000), goat anti-mouse
IgG-horseradish peroxidase (cat. no. sc-2005; 1:2,000) and Alexa
Fluor 488-conjugated anti-mouse IgG (cat. no. sc-362257; 1:200)
were obtained from Santa Cruz Biotechnology, Inc. FITC-conjugated
anti-mouse IgM was obtained from BD Biosciences Pharmingen (cat.
no. 555988; 1:500). Epidermal growth factor (EGF) was obtained from
ProSpec-Tany TechnoGene, Ltd., D-threo-1-phenyl-2-palmitoyl
amino-3-pyrrolidino-1-propanol (P4) was purchased from Matreya,
Inc., and GM3 and GM2 were purchased from Sigma-Aldrich (Merck
KGaA).
Glycosphingolipid extraction and
high-performance thin layer chromatography (HPTLC)
In order to decrease the ganglioside expression in
SW620 cells, the cells (5×106/dish) were grown in 10-cm
dishes at 37°C and treated with 1.0 µM P4, a specific inhibitor for
UDP-glucose ceramide glucosyltransferase and a key enzyme in the
synthesis of glucosylceramide. After 48 h, the cells were harvested
by trypsinization and washed three times with PBS. The gangliosides
were extracted and analyzed as previously described by Ladisch
et al (33). Briefly, the
cell pellet was extracted twice with chloroform:methanol (1:1,
v/v), and the extracts containing the total lipids (~15 ml) were
combined and dried under a stream of N2. The
gangliosides were purified by partitioning the dried samples in
diisopropyl ether: 1-butanol: 17 mM aqueous NaCl (6:4:5, v/v)
followed by Sephadex G-50 gel (Cytiva) filtration and
lyophilization. Samples (20 µl) were loaded and the individual
gangliosides were separated on silica gel 60 HPTLC plates (Merck
KGaA) with a solvent system consisting of chloroform:methanol:0.25%
aqueous CaCl2·2H2O (60:40:9, v/v/v). The
gangliosides were visualized as purple bands by spraying with
resorcinol-HCl reagent and heating at 120°C for 2 min, comparing
with the standards including GM3, GM2, GM1 GD1a, GD1b and GT1b.
To increase ganglioside expression in SW620 cells,
the cells (5×105/well) were treated with 50 µM GM3 for
48 h before harvesting. Cells were separated with trypsin + EDTA
and washed with PBS. Aliquots of cells (1×105) were
incubated with mouse anti-GM3 IgM (1:200; cat. no. 303-06541, Wako
Pure Chemical Industries Ltd.) for 1 h on ice. Subsequently, cells
were washed with PBS, incubated with FITC-conjugated anti-mouse IgM
(1:500; cat. no. 555988, BD Biosciences Pharmingen) for 40 min on
ice. The cells were fixed with 2% paraformaldehyde in PBS and
analyzed using a Coulter EPICS XL flow cytometer (Beckman Coulter,
Inc.).
Flow cytometry analysis
To identify the location of recombinant CD82, the
flow cytometry assay was performed in non-permeable and permeable
conditions. Cells were detached with trypsin/EDTA, and
5×104 cells were permeabilized by 0.1% TX-100 in TBS for
15 min, then washed with 1% BSA/0.1% NaN 3/PBS. Aliquots of the
cells (1×105) were incubated with primary antibodies
including anti-CD82 (1:1,000; cat. no. ab59509, Abcam) and anti-GM3
(1:200; cat. no. 303-06541, Wako Pure Chemical Industries Ltd.) in
5% goat serum (Thermo Fisher Scientific, Inc.), 1% BSA (Thermo
Fisher Scientific, Inc.) and 0.1% NaN3 in PBS for 1 h on ice, and
then with appropriate Alexa Fluor 488-conjugated secondary
antibodies for 30 min on ice. Cells were analyzed using a Coulter
EPICS XL flow cytometer and FlowJo software (version 7.6.1; FlowJo
LLC).
Transfections
The cDNA of CD82 was amplified from SW480
cells by PCR. CD82 gene was inserted into the pcDNA™4/V5
plasmid (Thermo Fisher Scientific, Inc.) at the restriction
endonuclease sites NheI and XhoI by the use of T4 DNA
ligase (Takara Biotechnology Co., Ltd.). The recombinant plasmid
was termed pcDNA™4/V5-CD82 and identified by sequencing
(Takara Biotechnology Co., Ltd.). The pcDNA™4/V5-CD82
plasmid was constructed and transfected into SW620 cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in Opti-MEM medium as OE (overexpression of
CD82), and the empty vector was also transfected as control.
The cells (1×105/well) were seeded in 12-well plate for
24 h with 1.6 µg of the recombinant plasmid and 2 µl Lipofectamine
2000 diluted with 100 µl Opti-MEM medium separately for 5 min at
room temperature. The mixture of diluted plasmid and Lipofectamine
2000 were incubated for 15 min at room temperature and then added
into the well. The transfect cells were analyzed after incubation
with the mixture at 37°C for 48 h following the manufacturer's
protocol.
Reverse transcription-quantitative PCR
(RT-qPCR)
The expression of CD82 was measured
using RT-qPCR after transfected into SW620 cells for 48 h. Total
RNA was extracted and purified using a MiniBEST Universal RNA
Extraction kit (Takara Biotechnology Co., Ltd.). The first strand
of cDNA was prepared from 2 µg RNA using TransScript First-Strand
cDNA Synthesis SuperMix (TransGen Biotech Co., Ltd.). qPCR was
performed using TransStart Top Green qPCR SuperMix (TransGen
Biotech Co., Ltd.) under StepOnePlus system (Applied Biosystems;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. The following thermocycling conditions were used for the
qPCR: 94°C for 30 sec; followed by 40 cycles of 94°C for 5 sec,
60°C for 15 sec and 72°C for 10 sec. The following primer pairs
were used for the qPCR: CD82 forward,
5′-GCCGACAAGAGCAGTTTCAT-3′ and reverse, 5′-GGAAGCCCATGAGCATAGTG-3′;
and GAPDH forward, 5′-AGTCCTACCACGATACCAAAGT-3′ and reverse,
5′-CATGAGAAGTACGACAACAGCCT-CAT. Relative mRNA expression levels
were determined by using the 2−ΔΔCq method. To ensure
test reliability, cDNA amounts were optimized, the Cq values were
kept below 35, and mRNA expression levels normalized to
GAPDH as the reference gene.
Western blotting
For western blot analysis, cells were harvested and
lysed in RIPA buffer (1% Triton X-100, 150 mM NaCl, 25 mM Tris, pH
7.5, 0.5% sodium deoxycholate, 0.1% SDS, 5 mM pyrophosphate, 50 mM
NaF) containing 1 mM Na3VO4, 1 mM DTT, 1%
protease inhibitor cocktail and 1% phosphatase inhibitor cocktail.
Protein concentration was determined by using a bicinchoninic acid
assay and 10 µg protein were separated by SDS-PAGE on 12%
polyacrylamide gels. The resolved proteins were transferred to PVDF
membranes (EMD Millipore) and incubated with primary antibodies
against EGFR (1:2,000; cat. no. sc-365829, Santa Cruz
Biotechnology, Inc.), p-EGFR (Tyr-1173; 1:1,000; cat. no. sc-57542,
Santa Cruz Biotechnology, Inc.), p-EGFR (Tyr-845; 1:1,000; cat. no.
sc-57542, Santa Cruz Biotechnology, Inc.), p-EGFR (Tyr-1045;
1:1,000; cat. no. sc-57541, Santa Cruz Biotechnology, Inc.),
Akt1/2/3 (1:2,000; cat. no. sc-55523, Santa Cruz Biotechnology,
Inc.), p-Akt1/2/3 (Ser473; 1:2,000; cat. no. sc-81433, Santa Cruz
Biotechnology, Inc.), p-Akt1/2/3 (Thr308; 1:2,000; cat. no.
sc-16646-R, Santa Cruz Biotechnology, Inc.), MAPK (1:2,000; cat.
no. sc-7383, Santa Cruz Biotechnology, Inc.), p-p44/42MAPK
(1:1,000; cat. no. sc-81492, Santa Cruz Biotechnology, Inc.),
p-EGFR (Tyr-992; 1:5,000; cat. no. ab5638, Abcam) and CD82 IgG
(1:5,000; cat. no. ab59509, Abcam). After washing with TBST (0.05%
Tween-20), the membranes were incubated with goat anti-rabbit
IgG-horseradish peroxidase (1:2,000; cat. no. sc-2004, Santa Cruz
Biotechnology, Inc.) and rabbit anti-goat IgG-horseradish
peroxidase (1:2,000; cat. no. sc-2768, Santa Cruz Biotechnology,
Inc.). Protein bands were visualized using the ECL kit (Cytiva) and
images captured using the VersaDoc™ Imaging system (Bio-Rad
Laboratories, Inc.). Densitometric semi-quantification of protein
expression levels was performed using the Gel-Pro analyzer v4.0
(Media Cybernetics, Inc.) and GraphPad Prism 5 (GraphPad Software,
Inc.) with β-actin as the loading control.
Wound healing assay
The invasive capacity of the SW620 cells was
verified through a wound healing assay. SW620 cells were seeded on
6-well plates at a cell density of 2×106 cells/well.
SW620 cells were treated with P4 and/or GM3 and/or transfected as
OE or empty vector as control, aforementioned. After incubating the
cells overnight in serum-free medium, the scratch was made when the
cells reached 85% confluence. Cells were washed three times with
PBS and incubated in serum-free medium containing 10 ng/ml EGF for
48 h at 37°C. Images of the migrated cells were captured using a
light microscope at ×200 magnification (Olympus Corporation) in
three randomly selected fields, wound closure was measured with
ImageJ software (version 1.47v, National Institutes of Health).
Transwell migration assay
For the Transwell migration assay, cells were
treated as aforementioned and subsequently incubated in serum-free
medium overnight at 37°C. Cells (1×105) were then
resuspended in 300 µl of serum-free medium containing 10 ng/ml EGF
and placed on the top compartment of a chamber with 8-µm pore
polycarbonate filters. The bottom chamber was filled with 250 µl
medium with 10% FBS. After 48 h, cells that migrated to the lower
surface of the membrane were fixed with methanol for 20 min at room
temperature and stained with 0.1% DAPI; images were captured from
five randomly chosen fields, and cells were counted under a light
microscope at ×100 magnification (Olympus Corporation). To block
MAPK and PI3K/Akt pathways with U0126 and LY294002
inhibitors, the SW620 cells were exposed to 10 µM of U0126 or 15 mM
of LY294002 for 4 h at 37°C. The cells were then harvested and used
for migration assay.
Statistical analysis
All experiments were repeated at least three times
and they consistently yielded similar results. The data were
analyzed using GraphPad Prism 5 (GraphPad Software, Inc.). Unpaired
t-test or one-way ANOVA followed by Dunnett's post hoc test were
used for statistical comparisons between groups. The results are
expressed as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Alteration of ganglioside content and
overexpression of CD82 in SW620 Cells
To investigate the role of gangliosides in SW620
cells, which may be related to a metastatic ability (34), the expression of gangliosides in
SW620 cells was altered by inhibiting their synthesis through
treatment with P4 and by the addition of exogenous GM3. The cells
were treated with 1 µM P4 for 48 h, and the gangliosides were then
extracted and monitored using HPTLC. The results showed that SW620
cells predominantly contained GM2 and that the gangliosides found
in SW620 cells were nearly depleted with 1 µM P4 (Fig. 1A). To increase the GM3 content,
cells were treated with 50 µM exogenous GM3 for 48 h, and the
expression of GM3 in the cells was determined using flow cytometry
(Fig. 1B).
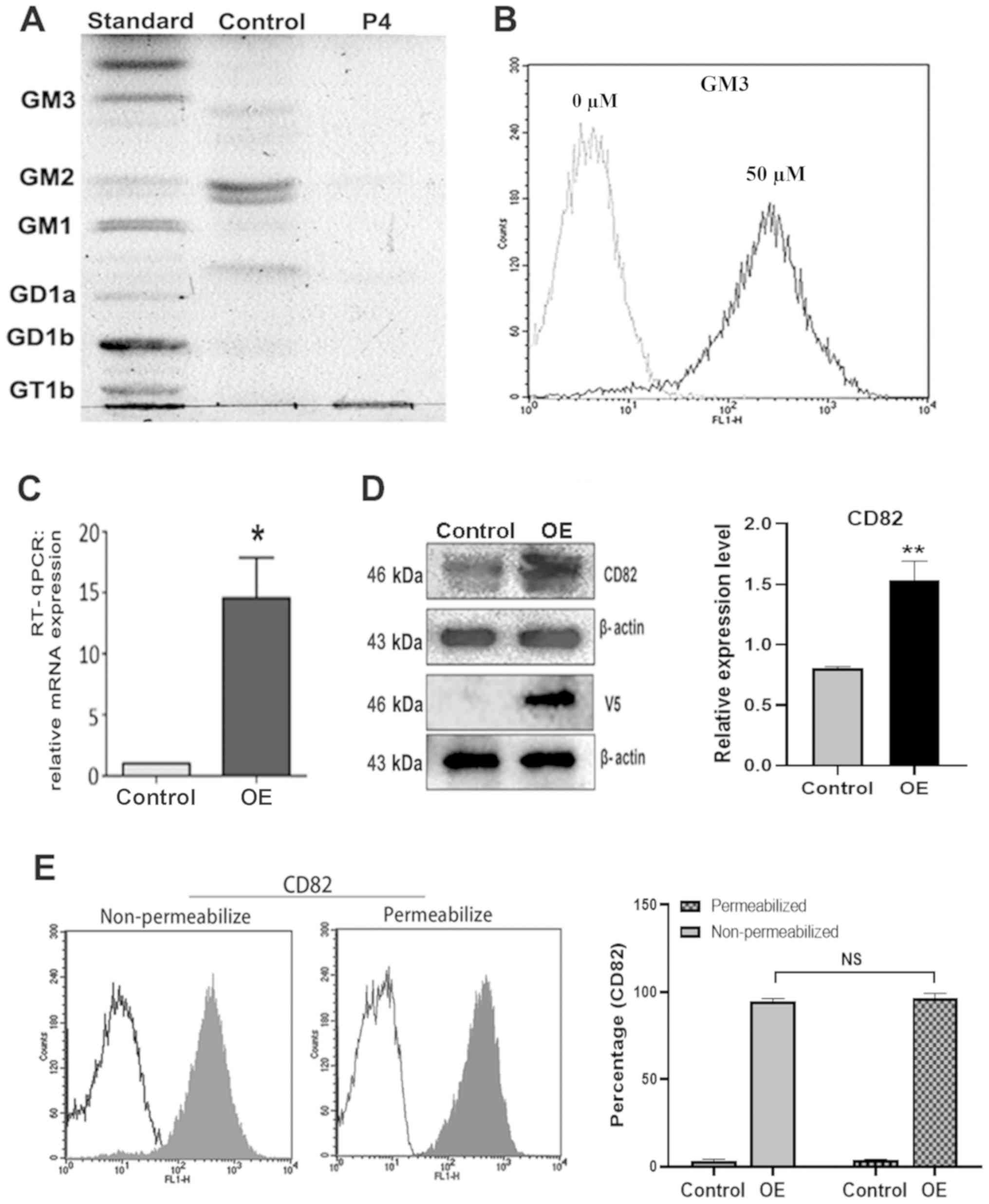 | Figure 1.Expression of gangliosides and CD82
in cells. (A and B) Cells were treated with either 1.0 µM P4 or 50
µM GM3. After 48 h of incubation, the gangliosides were extracted
and assayed by (A) HPTLC and (B) flow cytometry. (C and D)
pcDNA4/V5-CD82 or pcDNA4.0 (control) plasmids were constructed and
transfected into SW620 cells, and CD82 mRNA and protein expression
levels were measured by (C) RT-qPCR and (D) western blot,
respectively. Expression of V5- tagged CD82 protein was also
detected by western blot; β-actin was used as a loading control.
(E) Flow cytometric analysis of CD82 expression after cell
transfection. Shaded areas, SW620 cells transfected with
pcDNA4/V5-CD82; non-shaded areas, SW620 cells transfected with
pcDNA4.0 control. All the results shown are representative of three
independent experiments. Data are presented as the mean ± standard
deviation from three independent experiments. *P<0.05 and
**P<0.01 vs. Control group. CD82, CD82 antigen; GD1a and GD1b,
disialoganglioside; GM, monosialodihexosylganglioside; Ganglioside
GT1b; P4, D-threo-1-phenyl-2-palmitoyl
amino-3-pyrrolidino-1-propanol; RT-qPCR, reverse
transcription-quantitative PCR; OE, overexpression of wild-type
CD82. |
To investigate the influence of CD82 on SW620 cell
motility and migration, the cells were transfected with
pcDNA4/V5-CD82 as OE or pcDNA4.0 empty vector as control, and the
mRNA and protein expression levels of CD82 was measured using
RT-qPCR and western blotting, respectively (Fig. 1C and D). The pcDNA4/V5-CD82
plasmid also contained the fusion proteins including CD82 and V5
epitopes. To confirm the overexpression of CD82, the fusion protein
was detected by western blot with V5 antibody (Fig. 1D), and the CD82 expression profiles
were also compared using flow cytometry (Fig. 1E). The results demonstrated that
CD82 and V5 expression were significantly higher in OE of SW620
cells compared with the control. To identify the location of
recombinant CD82, the flow cytometry assay was performed in
non-permeable and permeable conditions. CD82 expression was
observed in 94.9 and 96.6% of CD82 transfected cells, but only in
3.4 and 3.6% of control cells, without or with permeabilization,
respectively (Fig. 1E). No notable
difference in the expression of CD82 was observed between
non-permeabilized and permeabilized cells transfected with
pcDNA4/V5-CD82 plasmid, which indicated that the
overexpression of CD82 was mainly on the cell surface.
Effect of gangliosides and CD82 on the
motility and migration of EGF-stimulated SW620 cells in vitro
To understand the influence of gangliosides and CD82
on the metastatic ability of SW620 cells, in vitro cell
motility and migration assays were performed using both wound
healing analysis and Transwell migration assay. Each group,
stimulated with EGF, was compared with the P4 treatment control
cells transfected with an empty vector, which had low expression of
gangliosides and CD82 (Fig. 2A1 and
B1). GM2 downregulation in SW620 cells had no significant
effect on cell motility and migration in vitro compared with
the control cells (Fig. 2A1, A2, B1
and B2). However, increasing GM3 inhibited cell motility and
migration in vitro (Fig. 2A3
and B3). CD82 alone or in combination with GM2 significantly
inhibited cell motility and migration in vitro (Fig. 2A4, B4, A5 and B5). GM3
overexpression further enhanced the inhibitory effect of CD82
(Fig. 2A6 and B6). The combination
of GM3 overexpression and uninhibited GM2 expression (that is,
without P4 treatment) markedly enhanced the inhibitory effect of
CD82 compared with either GM3 or GM2 alone (Fig. 2A7 and B7). These results suggested
that single factor of GM3 or CD82 and combination of CD82 with GM2
or GM3 can inhibit the EGF-stimulated SW620 cell motility and that
GM3, GM2 and CD82 in triple combination can show the most effect of
inhibition of the EGF-stimulated SW620 cell motility in
vitro.
 | Figure 2.Effect of gangliosides and CD82 on
the epidermal growth factor-induced migration of SW620 cells in
vitro. (A and B) SW620 cells were cultured and treated and/or
transfected with the indicated agents and plasmids for 48 h. Cell
migration assays were performed using (A) wound healing analysis
and (B) chemotaxis migration assays. For wound healing assay, the
widths of wound closure were measured (magnification, ×200). For
Transwell assay, the average number of cells that migrated through
the filter were counted (magnification, ×100). Data are presented
as the mean ± SD from three independent experiments. ***P<0.001
vs. P4+Control group. The expression of ganglioside and CD82 in
each group are shown as following, 1. No expression of GM2/GM3 and
CD82, 2. Only expression of GM2, 3. Only expression of GM3, 4. Only
expression of CD82, 5. Expression of GM2 and CD82, 6. Expression of
GM3 and CD82, 7. Expression of GM2, GM3 and CD82. GM3,
monosialodihexosylganglioside GM3; P4, D-threo-1-phenyl-2-palmitoyl
amino-3-pyrrolidino-1-propanol; OE, overexpression of wild-type
CD82 |
Effect of gangliosides and CD82 on the
phosphorylation of EGFR and EGFR-mediated intracellular signaling
pathways in EGF-stimulated SW620 cells
To investigate the effects of gangliosides and CD82
on the EGF-stimulated cell motility and migration in vitro,
the influence of gangliosides and CD82 on the activity of EGFR was
investigated by detecting the phosphorylation levels of three of
its tyrosine residues, all the experimental groups were compared
with the first group, which had low expression of gangliosides and
CD82. Whereas GM2 could not inhibit the phosphorylation at the
Tyr1173, Tyr1045 and Tyr845 residues (Fig. 3A; lane 2), GM3 could significantly
inhibit the EGF-stimulated phosphorylation levels at the Tyr1173
and Tyr1045 residues (Fig. 3A;
lane 3), and CD82 could significantly inhibit the phosphorylation
levels at the Tyr1045 residue (Fig.
3A; lane 4). The combination of GM3 and CD82 significantly
inhibited the phosphorylation of EGFR at both the Tyr1173 and
Tyr1045 residues (Fig. 3A; lane
6), and the combination of GM2 and CD82 notably inhibited the
phosphorylation of EGFR at the Tyr1045 residue (Fig. 3A; lane 5). The combination of GM3,
GM2 and CD82 resulted in the most effective inhibition of
phosphorylation levels at Tyr1173, Tyr1045 and Tyr845 residues
compared with the control group (Fig.
3A; lane 7). In addition, EGFR phosphorylation levels at the
Tyr992 residue was also measured and no obvious difference among
the groups was detected (data not shown). These results indicated
that gangliosides and CD82 downregulated the activity of EGFR by
inhibiting the phosphorylation of different tyrosine residues of
EGFR and that the effects of different gangliosides in combination
with CD82 are also different.
The EGFR-mediated intracellular signaling pathways
that are involved in the regulation of metastasis were also
studied. A previous study indicated that MAPK and PI3K/Akt
signaling pathways are associated with extracellular and
intracellular cues to control cell motility (34). To detect the activity of the MAPK
signaling pathway, the phosphorylation levels of MAPK were
determined. The results show that neither GM2 nor GM3 influenced
the EGF-stimulated phosphorylation of MAPK (Fig. 3B; lanes 2 and 3, respectively),
whereas CD82 significantly inhibited the EGF-stimulated
phosphorylation of MAPK (Fig. 3B;
lane 4). CD82 with GM2 or GM3 (Fig.
3B; lanes 5, and 6) or the mixture of GM2 and GM3 (Fig. 3B; lane 7) also showed notable
inhibitory effect on the phosphorylation of MAPK.
To detect the activity of the PI3K/Akt signaling
pathways, the phosphorylation levels of Akt were determined. The
results showed that GM3, but not GM2, can inhibit the
EGF-stimulated phosphorylation of Akt at Ser473 (Fig. 3C; lanes 3 and 2, respectively) and
that CD82 alone cannot inhibit the phosphorylation of Akt at Ser473
or T308 (Fig. 3C, lane 4). In
addition, CD82 with GM3 but not with GM2 can exert a synergistic
inhibitory effect on the EGF-stimulated phosphorylation of Akt at
Ser473 (Fig. 3C; lanes 6 and 5,
respectively). Addition of exogenous GM3 with overexpression of
CD82 notably reduced the EGF-induced phosphorylation of Akt at both
Ser473 comparing to the group of exogenous GM3 alone (Fig. 3C; lanes 3 and 6). The mixture of
GM3 and GM3 with CD82 also presented the notable inhibitory action
on the EGF-stimulated phosphorylation of Akt at Ser473 (Fig. 3C; lane 7).
Effect of blocking the MAPK and
PI3K/Akt pathways on cell motility and migration
To validate whether EGF-stimulated cell motility and
migration are mainly regulated by the MAPK and PI3K/Akt signaling
pathways, the activities of MAPK and PI3K/Akt in the cells were
blocked with U0126 and LY294002 inhibitors (Fig. 4). Blocking of the MAPK signaling
pathway with U0126 inhibited the phosphorylation of MAPK (Fig. 4A) and inhibited cell motility and
migration (Fig. 4C). Blocking of
the PI3K/Akt signaling pathway with LY294002 inhibited the
phosphorylation of Akt (Fig. 4B)
and inhibited the EGF-stimulated cell motility and migration
(Fig. 4C). These results show that
both the MAPK and PI3K/Akt pathways might be involved in the
modulation of SW620 cell motility and migration in
vitro.
Discussion
Colorectal cancer is a highly malignant tumor of the
digestive tract, and its incidence and mortality rates are
gradually increasing; colorectal cancer has a high mortality rate
owing to its high metastatic ability (1). Therefore, the elucidation of the
mechanisms involved in metastasis will allow for the selection of
anti-metastasis drugs contributing to a lower mortality rate. SW620
cells were originally isolated from the lymph node metastatic foci
of a patient with rectal cancer with high lymphatic metastasis
ability (35). The loss of CD82
expression and the increased expression of EGFR and/or its ligands
are the most notable biological characteristics of colon cancer
(7). These features have been
associated with an increased risk of metastasis, but their roles in
the lymph node metastatic processes have not been completely
elucidated.
CD82 is considered a wide-spectrum invasion- and
metastasis-suppressor. A number of studies have reported that the
combination of CD82 with gangliosides can exert a synergistic
inhibitory effect on cell motility and migration (25,29);
CD82 and gangliosides can inhibit the phosphorylation and activity
of EGFR. The present study aimed to understand the precise
mechanism by which CD82 and gangliosides influence EGF-stimulated
motility of SW620 cells with potential lymph node metastasis
(35) by investigating whether
EGFR signaling is involved in the mechanism by which CD82 and
gangliosides synergistically inhibit the motility and migration of
SW620 cells. The cells were starved overnight and then stimulated
with EGF instead of using siEGFR or EGFR selective inhibitors. The
results demonstrated that SW620 cells mainly express ganglioside
GM2 and low levels of CD82. Moreover, the present study
investigated the effects of gangliosides and CD82 on the migration
of SW620 cells in vitro by altering the contents of GM2 and
GM3 and the expression of CD82 in these cells. The results showed
that CD82 can suppress EGF-stimulated SW620 cell motility and
migration and that GM3, but not GM2, can suppress the
EGF-stimulated SW620 cell motility and migration. The results of
wound healing and migration assays in vitro indicated that
CD82 with either GM3 or GM2 can exert a synergistic inhibitory
effect on cell migration, and the mixture of CD82 with both GM2 and
GM3 presented the most notable synergistic inhibitory action.
Both GM3 and CD82 can inhibit the phosphorylation
and activity of EGFR. In order to understand whether CD82 and
gangliosides synergistically inhibit the motility and migration of
SW620 cells and whether they are associated with EGFR activity, the
present study investigated the effects of CD82 and the gangliosides
GM2 and GM3 on the phosphorylation of EGFR and found that
gangliosides and CD82 downregulated the activity of EGFR through
different mechanisms. GM3 had an inhibitory effect on EGFR
activation by inhibiting the phosphorylation of EGFR at the Tyr1045
and Tyr1173 residues, whereas CD82 downregulated the activity of
EGFR by inhibiting its phosphorylation at the Tyr1045 residue. The
current study also found that CD82 with either GM2 or GM3 can exert
a synergistic inhibitory effect on the activity of EGFR, but the
mechanisms responsible for the observed synergistic effects are
different. GM3 enhanced the inhibitory effect of CD82 on the
phosphorylation of EGFR at both the Tyr1045 and Tyr1173 residues,
whereas GM2 enhanced the inhibitory effect of CD82 on the
phosphorylation of EGFR only at the Tyr1045 residue.
Two intracellular signaling pathways, considered as
the important pathways involved in modulation of tumor metastasis
(36,37), which can be stimulated by EGFR were
analyzed. The overexpression of CD82 inhibited the phosphorylation
of MAPK. An increase in the level of GM3 inhibited the
phosphorylation of Akt. GM2 cannot influence the phosphorylation of
MAPK and Akt but can promote the inhibitory effect of CD82 on the
phosphorylation of MAPK. These results indicate that a single
factor of GM3 or CD82 inhibits SW620 cell migration only by
downregulating either of PI3K or MAPK signaling pathway, whereas
the combination of GM3 and CD82 can affect both PI3K and MAPK
signaling pathways to decrease cell migration. The most prominent
inhibitory effect shows when accumulation of PI3K and MAPK
signaling pathways with mixture of GM2, GM3 and CD82. In
conclusion, the gangliosides and CD82 inhibited SW620 cell motility
and migration through different mechanisms. GM3 suppressed SW620
cell motility and migration by inhibiting the EGF-stimulated
phosphorylation of EGFR at the Tyr1045 and Tyr1173 residues and the
downregulation of the MAPK signaling pathway, whereas CD82
suppressed SW620 cell motility and migration by inhibiting the
EGF-stimulated phosphorylation of EGFR at the Tyr1045 residue and
downregulation of the MAPK signaling pathway. The combination of
GM3, GM2 and CD82 also resulted in inhibition of phosphorylation
levels at Tyr845 residues, which is not an autophosphorylation
site. Phosphorylation of EGFR tyrosine 845 is a direct substrate of
Src, which is reported as synergism with EGFR through MAPK
signaling pathway (38). Thereby,
it was hypothesized that Tyr845 may be also important in activation
of MAPK pathway and cell migration.
CD82 with either of the gangliosides (GM3 and GM2)
exerts a synergistic inhibitory effect; however, the mechanisms are
different. GM3 enhanced the inhibitory effect of CD82 on the
phosphorylation of EGFR at both the Tyr1045 and Tyr1173 residues
and downregulated the PI3K and MAPK signaling pathways, whereas GM2
enhanced the inhibitory effect of CD82 on the phosphorylation of
EGFR at only the Tyr1045 residue and downregulated the MAPK
signaling pathway. CD82 inhibition of tumor cell movement might
depend on gangliosides by promoting internalization of EGFR, which
results from phosphorylation of EGFR at specific tyrosine residues
to activation of EGFR (39). The
internalized EGFR redistributing into distinct cellular
compartments causes attenuation of EGFR signaling. Signaling by
receptor tyrosine kinases (RTK) is mediated by alternative
interactions between specific phosphotyrosine residues in the
activated EGFR and individual Src homology 2 (SH2) domains of
cytoplasmic effectors. The multiple SH2-containing signal
transducers include phosphatidylinositol 3-kinase and Grb2, leading
to activation of downstream kinases including PI3K and MAPK
signaling pathways. Other studies have revealed that association of
a particular receptor with a tetraspanin-enriched microdomains may
lead either to the enhancement or attenuation of its activity, and
ganglioside expression levels clearly effect the ability of
tetraspanins to regulate RTK signaling. In addition, it was also
observed that formation of heterotypic GSL-GSL and GSL binding to a
GSL-binding protein could affect cellular phenotype by activation
of a signal transducer (26).
Hakomori (26) reported that
non-malignant HCV29 cells with GM2, GM3, and CD82 expression show
less motility and invasiveness than invasive YTS1 cells without
CD82 expression by regulating c-Met kinase activity. The present
study and these previous studies indicated that tetraspanin and
specific ganliosides in membrane microdomains may influence
signaling activation or block signaling responses by target growth
factor receptors. Taken together, the gangliosides and CD82
inhibited SW620 cell motility and migration by different
mechanisms. Future investigation of this possibility will help
clarify the synergistic role of gangliosides and CD82 on EGFR to
inhibit cell motility and invasion.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant no. 81602546 to X.H.
Huang and grant no. 81472716 to K.L. Ma).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KM and XH contributed to the conception and design
of the study. XH, YL and KM drafted the manuscript. YC and WW
performed the experiments. YL and SY analyzed and interpreted the
data. KM and XH were involved in revising the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kolligs FT: Diagnostics and Epidemiology
of Colorectal Cancer. Visc Med. 32:158–164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Odintsova E, Voortman J, Gilbert E and
Berditchevski F: Tetraspanin CD82 regulates compartmentalisation
and ligand-induced dimerization of EGFR. J Cell Sci. 116:4557–4566.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maecker HT, Todd SC and Levy S: The
tetraspanin superfamily: Molecular facilitators. FASEB J.
11:428–442. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pique C, Lagaudrière-Gesbert C, Delamarre
L, Rosenberg AR, Conjeaud H and Dokhélar MC: Interaction of CD82
tetraspanin proteins with HTLV-1 envelope glycoproteins inhibits
cell-to-cell fusion and virus transmission. Virology. 276:455–465.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sigala S, Faraoni I, Botticini D,
Paez-Pereda M, Missale C, Bonmassar E and Spanol P: Suppression of
telomerase, reexpression of KAI1, and abrogation of tumorigenicity
by nerve growth factor in prostate cancer cell lines. Clin Cancer
Res. 5:1211–1218. 1999.PubMed/NCBI
|
|
6
|
Hinoda Y, Adachi Y, Takaoka A, Mitsuuchi
H, Satoh Y, Itoh F, Kondoh Y and Imai K: Decreased expression of
the metastasis suppressor gene KAI1 in gastric cancer. Cancer Lett.
129:229–234. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lombardi DP, Geradts J, Foley JF, Chiao C,
Lamb PW and Barrett JC: Loss of KAI1 expression in the progression
of colorectal cancer. Cancer Res. 59:5724–5731. 1999.PubMed/NCBI
|
|
8
|
Yang X, Welch DR, Phillips KK, Weissman BE
and Wei LL: KAI1, a putative marker for metastatic potential in
human breast cancer. Cancer Lett. 119:149–155. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Geradts J, Maynard R, Birrer MJ, Hendricks
D, Abbondanzo SL, Fong KM, Barrett JC and Lombardi DP: Frequent
loss of KAI1 expression in squamous and lymphoid neoplasms. An
immunohistochemical study of archival tissues. Am J Pathol.
154:1665–1671. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Adachi M, Taki T, Ieki Y, Huang CL,
Higashiyama M and Miyake M: Correlation of KAI1/CD82 gene
expression with good prognosis in patients with non-small cell lung
cancer. Cancer Res. 56:1751–1755. 1996.PubMed/NCBI
|
|
11
|
Friess H, Guo XZ, Tempia-Caliera AA,
Fukuda A, Martignoni ME, Zimmermann A, Korc M and Büchler MW:
Differential expression of metastasis-associated genes in papilla
of vater and pancreatic cancer correlates with disease stage. J
Clin Oncol. 19:2422–2432. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sun HC, Tang ZY, Zhou G and Li XM: KAI1
gene expression in hepatocellular carcinoma and its relationship
with intrahepatic metastases. J Exp Clin Cancer Res. 17:307–311.
1998.PubMed/NCBI
|
|
13
|
Guo XZ, Friess H, Di Mola FF, Heinicke JM,
Abou-Shady M, Graber HU, Baer HU, Zimmermann A, Korc M and Büchler
MW: KAI1, a new metastasis suppressor gene, is reduced in
metastatic hepatocellular carcinoma. Hepatology. 28:1481–1488.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hemler ME: Tetraspanin proteins mediate
cellular penetration, invasion, and fusion events and define a
novel type of membrane microdomain. Annu Rev Cell Dev Biol.
19:397–422. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Claas C, Stipp CS and Hemler ME:
Evaluation of prototype transmembrane 4 superfamily protein
complexes and their relation to lipid rafts. J Biol Chem.
276:7974–7984. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yarden Y and Sliwkowski MX: Untangling the
ErbB signalling network. Nat Rev Mol Cell Biol. 2:127–137. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Iwata S, Kobayashi H, Miyake-Nishijima R,
Sasaki T, Souta-Kuribara A, Nori M, Hosono O, Kawasaki H, Tanaka H
and Morimoto C: Distinctive signaling pathways through CD82 and
beta1 integrins in human T cells. Eur J Immunol. 32:1328–1337.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sugiura T and Berditchevski F: Function of
alpha3beta1-tetraspanin protein complexes in tumor cell invasion.
Evidence for the role of the complexes in production of matrix
metalloproteinase 2 (MMP-2). J Cell Biol. 146:1375–1389. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Odintsova E, Sugiura T and Berditchevski
F: Attenuation of EGF receptor signaling by a metastasis
suppressor, the tetraspanin CD82/KAI-1. Curr Biol. 10:1009–1012.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
van Echten G and Sandhoff K: Ganglioside
metabolism. Enzymology, Topology, and regulation. J Biol Chem.
268:5341–5344. 1993.PubMed/NCBI
|
|
21
|
Huwiler A, Kolter T, Pfeilschifter J and
Sandhoff K: Physiology and pathophysiology of sphingolipid
metabolism and signaling. Biochim Biophys Acta. 1485:63–99. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hakomori S: Glycosphingolipids in cellular
interaction, differentiation, and oncogenesis. Annu Rev Biochem.
50:733–764. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hakomori S: Tumor malignancy defined by
aberrant glycosylation and sphingo(glyco)lipid metabolism. Cancer
Res. 56:5309–5318. 1996.PubMed/NCBI
|
|
24
|
Birklé S, Zeng G, Gao L, Yu RK and Aubry
J: Role of tumor-associated gangliosides in cancer progression.
Biochimie. 85:455–463. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Regina Todeschini A and Hakomori SI:
Functional role of glycosphingolipids and gangliosides in control
of cell adhesion, motility, and growth, through glycosynaptic
microdomains. Biochim Biophys Acta. 1780:421–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hakomori SI: Glycosynaptic microdomains
controlling tumor cell phenotype through alteration of cell growth,
adhesion, and motility. FEBS Lett. 584:1901–1906. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
von Lindern JJ, Rojo D, Grovit-Ferbas K,
Yeramian C, Deng C, Herbein G, Ferguson MR, Pappas TC, Decker JM,
Singh A, et al: Potential role for CD63 in CCR5-mediated human
immunodeficiency virus type 1 infection of macrophages. J Virol.
77:3624–3633. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park S-Y, Yoon S-J, Freire-de-Lima L, Kim
J-H and Hakomori SI: Control of cell motility by interaction of
gangliosides, tetraspanins, and epidermal growth factor receptor in
A431 versus KB epidermoid tumor cells. Carbohydr Res.
344:1479–1486. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Todeschini AR, Dos Santos JN, Handa K and
Hakomori SI: Ganglioside GM2/GM3 complex affixed on silica
nanospheres strongly inhibits cell motility through
CD82/cMet-mediated pathway. Proc Natl Acad Sci USA. 105:1925–1930.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Birchmeier C, Birchmeier W, Gherardi E and
Vande Woude GF: Met, metastasis, motility and more. Nat Rev Mol
Cell Biol. 4:915–925. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tringali C, Silvestri I, Testa F,
Baldassari P, Anastasia L, Mortarini R, Anichini A, López-Requena
A, Tettamanti G and Venerando B: Molecular subtyping of metastatic
melanoma based on cell ganglioside metabolism profiles. BMC Cancer.
14:5602014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tanaka K, Miyazawa M, Mikami M, Aoki D,
Kiguchi K and Iwamori M: Enhanced expression of unique gangliosides
with GM2-determinant in human uterine cervical carcinoma-derived
cell lines. Glycoconj J. 33:745–754. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ladisch S and Gillard B: A solvent
partition method for microscale ganglioside purification. Anal
Biochem. 146:220–231. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang X, Li Y, Zhang J, Xu Y, Tian Y and
Ma K: Ganglioside GM3 inhibits hepatoma cell motility via
down-regulating activity of EGFR and PI3K/AKT signaling pathway. J
Cell Biochem. 114:1616–1624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Leibovitz A, Stinson JC, McCombs WB III,
McCoy CE, Mazur KC and Mabry ND: Classification of human colorectal
adenocarcinoma cell lines. Cancer Res. 36:4562–4569.
1976.PubMed/NCBI
|
|
36
|
Davies MA: The role of the PI3K-AKT
pathway in melanoma. Cancer J. 18:142–147. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mook OR, Frederiks WM and Van Noorden CJ:
The role of gelatinases in colorectal cancer progression and
metastasis. Biochim Biophys Acta. 1705:69–89. 2004.PubMed/NCBI
|
|
38
|
Mueller KL, Powell K, Madden JM, Eblen ST
and Boerner JL: EGFR tyrosine 845 phosphorylation-dependent
proliferation and transformation of breast cancer cells require
activation of p38 MAPK. Transl Oncol. 5:327–334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Li Y, Huang X, Zhang J, Li Y and Ma K:
Synergistic inhibition of cell migration by tetraspanin CD82 and
gangliosides occurs via the EGFR or cMet-activated Pl3K/Akt
signalling pathway. Int J Biochem Cell Biol. 45:2349–2358. 2013.
View Article : Google Scholar : PubMed/NCBI
|


















