Introduction
The basic functions of the skeletal musculature
include facilitating locomotor activity, postural behavior and
breathing (1). In addition, the
skeletal musculature is a key metabolic organ in the human body,
which is involved in regulating metabolite homeostasis and
maintaining metabolic health via processes such as mediating
glucose uptake and insulin sensitivity (2). Skeletal muscle has been identified as
a secretory organ, which may exert autocrine, paracrine or
endocrine effects to affect the function of adipose tissue or the
cardiovascular system (3).
Therefore, impaired or loss of function of skeletal muscle can
affect functional capacity and increase the risk of a number of
diseases, such as diabetes mellitus (4), muscle atrophy (5) and cancer (6). Skeletal myogenesis is a complex
biological process that involves myoblast proliferation, migration
and differentiation, as well as fusion of multicellular myotubes
into contractile skeletal muscle fibers (7). In addition, according to skeletal
muscle contractility and expression levels of myosin heavy chain
(MyHC) subtypes, skeletal muscle fibers are classified into two
major types: Fast- (MyHC2b and MyHC2×) and slow-twitch fibers
(MyHC1 and MyHC2a) (8–10). A number of transcription factors
mediate skeletal myogenesis (11–15),
including the myogenic regulatory family (MRF) and the myocyte
enhancer 2 family (MEF2), which are among the most studied gene
families involved in regulating skeletal myogenesis (14). The MRF family, also known as the
MyoD family, consists of MyoD, MyoG, Myf5 and MRF4
(14), and serves a key role in
regulating specific gene transcription in muscle cells, as well as
cell growth cycle and differentiation (16). For instance, MyoD and
Myf5 induce expression levels of myogenin and other MEF2
family transcription factors (17). Subsequently, myogenin and MEF2
family members cooperate in the activation of muscle structural
genes during differentiation and the formation of multinucleated
myotubes (17). Megeney et
al (18) demonstrated that
mice lacking both MyoD and dystrophin displayed a marked
increase in severity of myopathy leading to premature death,
suggesting a role for MyoD in muscle regeneration.
Evidence has demonstrated that microRNAs
(miRNAs/miRs) are a class of non-coding RNAs 17–24 nucleotides in
length (19,20), which serve key roles in numerous
physiological and pathological processes, such as cell
proliferation, cell differentiation, or tumorigenesis (21–23).
A number of studies have shown that miRNAs regulate the expression
levels of key functional genes and transcription factors at the
posttranscriptional level, and induce mRNA degradation or
translation inhibition by interacting with the 3′ untranslated
regions (UTRs) of their target mRNAs (20,24).
Involvement of miRNAs in skeletal muscle development and function
has previously been reported (25,26).
A number of studies have demonstrated that miRNAs function as key
regulators of myogenesis via regulating myoblast proliferation and
differentiation (25,27,28).
miR-1, for example, promotes myogenesis via targeting
histone deacetylase 4, a transcriptional repressor of the
myogenesis gene (29).
miRNA-133 enhances myoblast proliferation by repressing
serum response factor (29).
miRNA-206 regulates skeletal muscle satellite cell
proliferation and differentiation by repressing Pax7
(26), and miRNA-139-5p
regulates C2C12 cell myogenesis by blocking the Wnt/β-catenin
signaling pathway (30). In
addition, miRNA-27b regulates Pax3 protein levels, and
downregulation of Pax3 causes rapid entry into the myogenic
differentiation program (31).
Changes in miRNA expression levels have been reported to be
associated with skeletal myogenesis (32,33).
Long non-coding RNAs (lncRNAs), a class of
non-protein-coding RNAs >200 nucleotides in length, have been
identified as competing endogenous RNAs (ceRNAs) that sponge miRNAs
via complementary base pairing to regulate biological process
(34). To date, numerous lncRNAs
have been identified in mice and humans, among which some serve
important roles in muscle development and myogenesis (35,36).
A number of studies have indicated that expression levels of
miR-23a and miR-23b are increased in the early stage
of C2C12 differentiation, and that these miRNAs are involved in the
regulation of TrxR1 expression levels via direct binding to
the 3′ UTR of TrxR1 mRNA (37,38).
Wang et al (10) reported
that miR-23a-3p downregulates the expression levels of
Myh1, 2 and 4 via targeting the 3′ UTR of mRNAs, thereby
inhibiting myogenesis of C2C12 myoblasts and fast-twitch MyHC
isoforms. However, the role of miR-23a-5p in myoblast
proliferation and differentiation has not yet been fully
elucidated. The present study demonstrated that miR-23a-5p
affected myoblast proliferation and differentiation. It was
hypothesized that miR-23a-5p promoted C2C12 myoblast
proliferation and inhibited differentiation, which may be involved
in lncDum regulation.
Materials and methods
Animal experiments
A total of 21 female C57BL/6 mice (Dashuo Co., Ltd.)
had free access to food and water in natural light cycle and 37°C
temperature conditions. The muscle tissues of 2–6-week-old C57BL/6
mice were collected for RNA isolation. A total of three mice were
included each group (2-week-old, 3-week-old, 4-week-old,
5-week-old, 6-week-old C57B/L6 mice). Tissues (liver, spleen, lung,
heart, kidney, muscle, inguinal fat, perirenal fat, brain, shoulder
fat and gonads fat) of 6-week-old C57BL/6 mice were collected for
RNA isolation. In order to establish a muscle atrophy model, muscle
atrophy was induced by Dexamethasone (Dex; Beijing Solarbio Science
& Technology Co., Ltd.)-mediated injury as described by
Morimoto et al (39). In
brief, three 10-week-old mice were intramuscularly injected in the
left hind leg with 100 µl 10 µM Dex. As a control, the right legs
of the same mice were intramuscularly injected with PBS. The
injection was performed every 7 days to ensure success. At 14 days
after the first injection, muscle samples were collected for RNA
isolation.
All animal procedures were approved by the Animal
Care and Ethics Committee of Sichuan Agricultural University
[approval no. DKY-(S20176903)] and performed in accordance with the
institutional guidelines for the care and use of laboratory
animals.
Cell culture
C2C12 myoblasts and HeLa cells (Beijing Stem Cell
Bank; Chinese Academy of Science) were cultured in growth medium
(GM) containing DMEM and 10% FBS (both Gibco; Thermo Fisher
Scientific, Inc.). When C2C12 myoblasts reached 80% confluence, GM
was replaced with differentiation medium (DM) containing DMEM
supplemented with 2% horse serum (Gibco; Thermo Fisher Scientific,
Inc.). Cells were cultured at 37°C with 5% CO2.
Cell transfection
miR-23a-5p mimics (cat. no. miR10017019-1-5),
negative control (NC) mimics (cat. no. miR01201-1-5) and small
interfering (si)RNA molecules directed against lncDum
(si-lncDum) were designed and synthesized by Guangzhou RiboBio Co.,
Ltd. Overexpression of lncDum (p-lncDum) was achieved by
cloning lncDum complementary DNA into vector pcDNA3.1 (Beijing
Tsingke New Industry TsingKe Biotechnology Co., Ltd.). Briefly,
5×105 or 4×104 C2C12 myoblasts were seeded in
12- or 96-well plates and grown to 80% confluency, then transfected
with miRNA-23a-5p mimics, NC mimics, si-lncDum, p-lncDum, empty
pcDNA3.1+ vector or siRNA control (si-NC) at a
concentration of about 20 µM using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). The transfections
were carried out every 48 h. Cells were harvested for 6 days, and
bright-field microscope was used to observe the ability of C2C12
myoblasts to differentiate into myotubes 6 days post-transfection.
The sequences of synthesized RNA oligonucleotides are presented in
Table SI.
Reverse transcription-quantitative PCR
(RT-qPCR)
RNA from muscle tissue and C2C12 cells was obtained
using TRIzol® (Takara Biotechnology Co., Ltd.) according
to the manufacturer's instructions. Reverse transcription was
performed using a PrimeScript RT Master Mix kit for mRNA, or a
PrimeScript™ miRNA RTPCR kit (both Takara Biotechnology Co., Ltd.)
for miRNA, according to the manufacturer's instructions. qPCR was
performed using the SYBR Premix Ex Taq kit (Takara Biotechnology
Co., Ltd.) using a CFX96 system (Bio-Rad Laboratories, Inc.).
U6 and β-actin served as endogenous controls for
miRNA and mRNA, respectively. The expression levels of miRNA and
mRNA were quantified using the comparative threshold cycle
(2−ΔΔCq) method (40).
Primer sequences are presented in Table SII.
Cell proliferation analysis
In brief, C2C12 myoblasts were seeded in 96-well
plates, and transfected with miR-23a-5p mimics, NC mimics,
si-lncDum, p-lncDum, si-NC or pcDNA3.1+ empty vector,
after which cell proliferation was determined at 0, 12, 24, 48, 72
and 96 h using a Cell Counting Kit (CCK)-8 (Beyotime Institute of
Biotechnology) according to the manufacturer's instructions. For
5-ethynyl-2′-deoxyuridine (EdU) proliferation analysis, C2C12
myoblasts were treated with 10 µM EdU (Guangzhou RiboBio Co., Ltd.)
after 24 h post-transfection and incubated for 3 h at 37°C. EdU
staining was performed according to the manufacturer's protocol.
Cell nuclei were stained with DAPI (Beyotime Institute of
Biotechnology) for 10 min at room temperature. Images
(magnification, ×100) of the entire well were captured using an
IX53 fluorescent inverted microscope (Olympus Corporation).
Western blotting
Total cellular proteins were extracted using RIPA
lysis buffer (Wuhan Servicebio Technology Co., Ltd.) 96 h after
transfection. The native protein lysate was collected, and the
protein concentration was measured using the BCA protein
concentration assay kit (Wuhan Servicebio Technology Co., Ltd.)
according to the manufacturer's instructions. Proteins (30–50
µg/per well) were resolved by SDS-PAGE with 10% gels, then
transferred to PVDF membranes (EMD Millipore). The membranes were
blocked with 5% non-fat milk in 0.5% TBS-Tween-20 (TBST) for 1 h at
room temperature. The membranes were then incubated overnight at
4°C with primary antibodies against β-actin (dilution 1:1,000; cat.
no. GB12001, Wuhan Servicebio Technology Co., Ltd.); MyoG (dilution
ratio 1:1,000; cat. no. A6664, ABclonal), and MyHC1 (dilution ratio
1:1,000; cat. no. A7564, ABclonal). The membranes were washed with
TBST and then incubated with the secondary antibody (horseradish
peroxidase-conjugated goat anti-rabbit IgG; dilution 1:3,000; cat.
no. GB23303; Wuhan Servicebio Technology Co., Ltd.) for 30 min at
room temperature. The signals were visualized using enhanced
chemiluminescent reagent (Wuhan Servicebio Technology Co., Ltd.),
and the expression levels of β-actin were used for normalization.
The AlphaEaseFC™ software processing system (version 3.2β; Alpha
Innotech) was used to analyze the optical density value of the
target bands.
Immunofluorescence analysis
For immunofluorescence analysis, C2C12 myoblasts
were transfected with miR-23a-5p mimics, NC, si-lncDum, p-lncDum,
si-NC or pcDNA3.1+ empty vector during differentiation,
then washed three times with PBS, and fixed in 4% paraformaldehyde
for 30 min at room temperature. Next, cells were washed three times
with PBS, and blocked with 5% normal goat serum in PBS for 1 h at
room temperature. Then, cells were incubated with an anti-myosin
slow antibody (anti-MyHC1; dilution 1:300; cat. no. bs-9862R;
BIOSS) overnight at 4°C, washed three times with PBS, and incubated
with a fluorescent secondary antibody (dilution ratio 1:300; cat.
no. bs-0295G-FITC; BIOSS) at 37°C for 1 h. Cell nuclei were stained
with DAPI (Beyotime Institute of Biotechnology) for 20 min at room
temperature. Images (magnification, ×100) were captured using an
IX53 fluorescent inverted microscope (Olympus Corporation). The
fusion index of myotubes as calculated by Image J software (Image J
1.50i; National Institutes of Health).
Luciferase reporter assay
HeLa cells were seeded in 96-well plates and
cultured to 90% confluency prior to transfection. Wild-type
(WT-lncDum) and mutant lncDum (Mut-lncDum) were amplified using
primers containing XhoI and NotI restriction sites
and cloned into a psi-CHECK™-2 promoter vector at the 3′ end of the
Renilla gene Beijing Tsingke New Industry TsingKe
Biotechnology Co., Ltd.). For the luciferase reporter analysis,
4×104 HeLa cells were seeded in 96-well plates and grown
to 80% confluency, then co-transfected with either WT-lncDum or
Mut-lncDum in conjunction with either miR-23a-5p mimics or mimics
control (Guangzhou RiboBio Co., Ltd.) at a final concentration of
100 nM using Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Luciferase activity was measured using a
Dual-Glo Luciferase Assay System (Promega Corporation) 24 h after
transfection, according to the manufacturer's instructions.
Luciferase activity levels were normalized to those of firefly
luciferase.
Statistical analysis
Data are presented as the mean ± SEM of three
experiments. Statistical analysis was performed using SPSS software
(version 22.0; IBM Corp.). Differences between groups were
determined using an unpaired Student's t-test. Differences between
3 or more groups were determined by one-way ANOVA, followed by the
Least Significant Difference method. P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-23a-5p is associated with skeletal
myogenesis
miR-23a-5p exhibited a similar expression
profile to three myoblast-specific miRNAs [miR-206-3p
(41), miR-181a-5p
(42) and miR-221 (43); Fig.
1A] in C2C12 myoblasts. The expression levels of
miR-23a-5p in numerous types of tissue derived from
6-week-old WT mice were assessed, which demonstrated that
miR-23a-5p was highly expressed in muscle, compared with
adipose tissue and brain. However, miR-23a-5p was expressed
at lower levels in muscle tissue compared with major organs, such
as the liver, spleen, lung, heart and kidney (Fig. 1B). Furthermore, miR-23a-5p
was highly expressed in the muscle tissue of 2–3-week-old mice, and
notably decreased after 3 weeks and remained at low levels as mice
aged (Fig. 1C). In order to
confirm these results, a muscle atrophy model was established using
Dex injection, as previously described (5,44,45).
Compared with the control group, Dex treatment caused a significant
decrease in miR-23a-5p expression levels in muscle tissue
(Fig. 1D). Thus, the present
findings indicated that miR-23a-5p may be associated with
skeletal myogenesis.
miR-23a-5p promotes C2C12 myoblast
proliferation
miR-23a-5p exhibited a notably decreased
expression level during the proliferation and differentiation of
C2C12 myoblasts compared with myogenesis-specific miRNAs, including
miR-206-3p, miR-181a-5p, miR-221/222 and miR-29b-3p
(Fig. 2A). In order to identify
the role of miR-23a-5p in regulating skeletal myogenesis,
miR-23a-5p mimics were transfected into C2C12 myoblasts
using GM to evaluate whether miR-23a-5p mediates
proliferation of these cells. Transfection of miR-23a-5p
mimics increased the expression levels of miR-23a-5p by
1,327-fold compared with the NC group, indicating that
miR-23a-5p was successfully overexpressed in proliferating
C2C12 myoblasts (Fig. 2B).
Subsequently, an EdU proliferation assay was performed to evaluate
the effect of miR-23a-5p on C2C12 myoblast proliferation; the
results demonstrated that miR-23a-5p significantly increased the
proliferation of EdU-positive cells compared with NC cells
(Fig. 2C and D). These findings
were also validated by an CCK-8 assay: Following transfection for
72 h, a greater number of C2C12 myoblasts were observed in the
miR-23a-5p mimics group compared with the NC group (Fig. 2E). Furthermore, RT-qPCR analysis
demonstrated that the expression levels of Cyclin E and
Cyclin B were significantly upregulated in C2C12 myoblasts
transfected with miR-23a-5p mimics, compared with the NC group
(Fig. 2F). Although increasing
miR-23a-5p tended to cause a decrease in P21
expression levels compared with the NC group, the difference was
not significant. Taken together, these data indicated that
miR-23a-5p positively regulated C2C12 myoblast
proliferation.
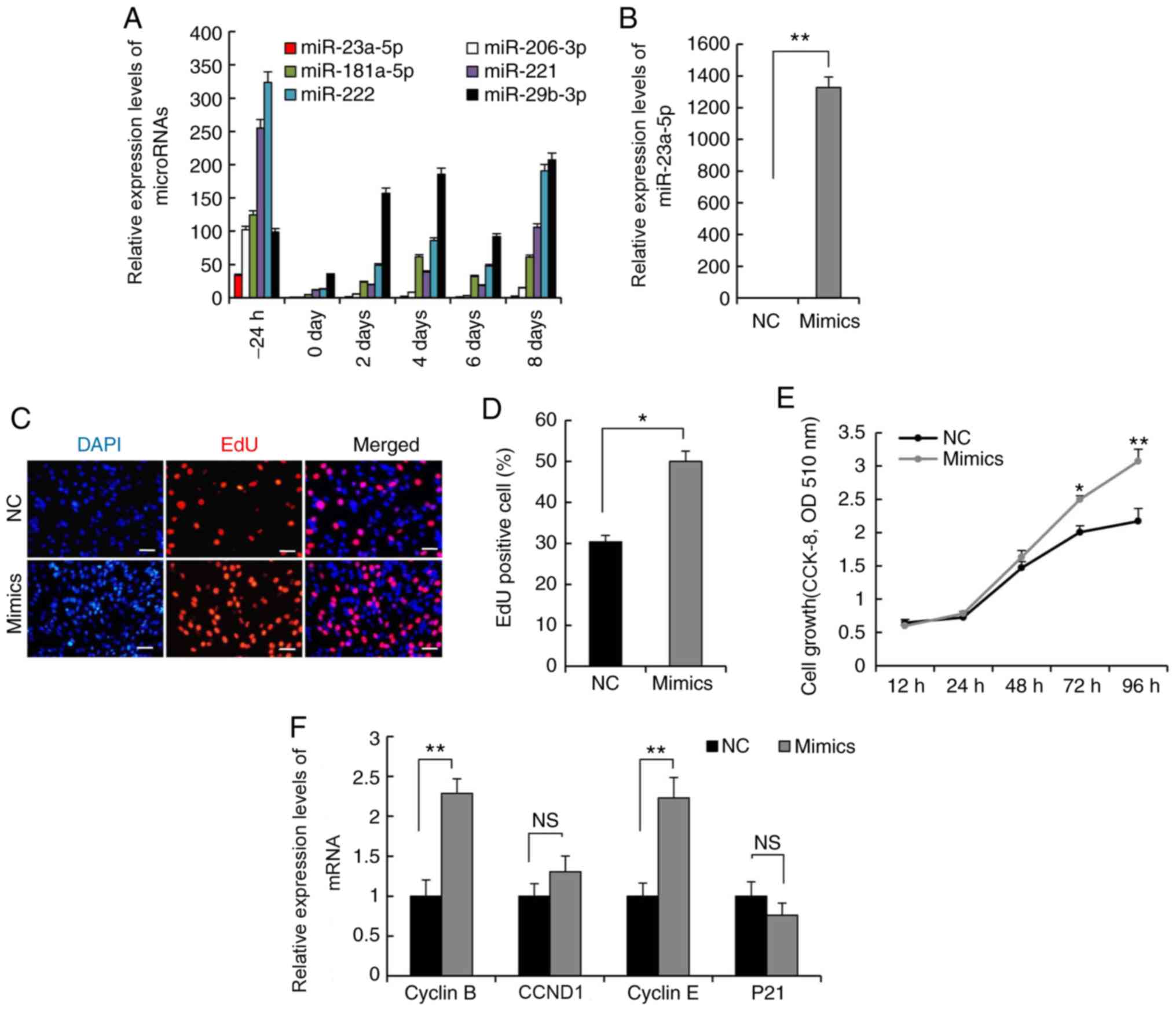 | Figure 2.miR-23a-5p positively modulates C2C12
myoblast proliferation. RT-qPCR was used to measure expression
levels of (A) miRNAs during C2C12 myoblast proliferation and
differentiation, and (B) miR-23a-5p in C2C12 myoblasts transfected
with miR-23a-5p mimics or NC during proliferation. (C) EdU assay
was performed following transfection for 24 h. Cells undergoing DNA
replication were stained with EdU (red) and cell nuclei were
stained with DAPI (blue). Scale bar, 100 µm. (D) Percentage of
EdU-positive cells/DAPI-positive cells was quantified. (E) Cell
count was measured by an CCK-8 assay. (F) RT-qPCR analysis of the
expression levels of Cyclin E, Cyclin B, CCND1 and
P21. Data are presented as the mean ± SEM (n=3). *P<0.05,
**P<0.01 vs. NC or as indicated. miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; NC, negative control; CCK-8, Cell
Counting Kit-8; EdU, 5-ethynyl-2′-deoxyuridine; OD, optical
density; NS, not significant. |
miR-23a-5p inhibits C2C12 myoblast
differentiation
In order to investigate the effect of
miR-23a-5p on myoblast differentiation, miR-23a-5p mimics
were transfected into C2C12 myoblasts using DM. At 6 days after
transfection of miR-23a-5p mimics (Fig. 3A), bright-field microscopy was
performed, which demonstrated that miR-23a-5p overexpression
markedly inhibited the fusion of myotubes compared with the NC
group (Fig. 3B). In order to
confirm these results, the expression levels of various myogenic
transcription factors, such as MyoD, Myf5, MyoG, MRF4 and
MyHC, were evaluated. Transfection with miR-23a-5p mimics
resulted in a significant decrease in the expression levels of
MyoD, MyoG and Myf5 compared with the NC group
(Fig. 3C). Furthermore, MyoG
protein expression levels in miR-23a-5p-overexpressing C2C12
cells were assessed. It was revealed that MyoG was downregulated
following miR-23a-5p overexpression, as determined via
western blotting analysis (Fig.
3D).
 | Figure 3.miR-23a-5p negatively modulates C2C12
myoblast differentiation and formation of muscle fiber type. (A)
RT-qPCR analysis of the expression levels of miR-23a-5p in
C2C12 myoblasts transfected with miR-23a-5p mimics or NC during
differentiation. (B) Bright-field microscopy was used to observe
the ability of C2C12 myoblasts to differentiate into myotubes at 6
days post-transfection. Scale bar, 100 µm. (C) RT-qPCR analysis of
the expression levels of MyoG, MyoD, MRF4, Myf5 and
MyHC. (D) Western blotting analysis of MyoG protein levels
in C2C12 myoblasts transfected with miR-23a-5p mimics or NC during
differentiation. RT-qPCR analysis of (E) expression levels of genes
associated with fast- and slow-twitch fibers, and (F) The
composition of MyHC (MyHC1, MyHC2a, MyHC2b and MyHC2×) in myoblasts
transfected with miR-23a-5p mimics or NC. (G) Western blotting
analysis of MyHC1 protein levels in C2C12 myoblasts transfected
with miR-23a-5p mimics or NC during differentiation. (H)
Immunofluorescence staining of MyHC1 was used to analyze
myosin-slow-positive myotubes. The fusion index was calculated as
MyHC-positive cells to total nuclei (total DAPI-positive cells).
Scale bar, 100 µm. Data are presented as the mean ± SEM of three
independent repeats. *P<0.05, **P<0.01. miR, microRNA;
RT-qPCR, reverse transcription-quantitative PCR; NC, negative
control; MyHC, myosin heavy chain; NS, not significant. |
As muscle fiber type composition is key for muscle
biological function and body metabolism, the present study aimed to
determine whether miR-23a-5p mediates muscle fiber type
composition. RT-qPCR demonstrated that overexpression of miR-23a-5p
notably inhibited the expression levels of several genes associated
with fast- (Tnni2, Tnnc2, MyHC2b and MyHC2×) and
slow-twitch fibers (Tnni1, Tnnc1, MyHC2a and MyHC1)
(46) compared with the NC group,
indicating that miR-23a-5p may be involved in regulating the
composition of skeletal muscle fiber type (Fig. 3E). The expression levels of
MyHC1, −2a, −2b and −2×, following transfection of
C2C12 myoblasts with miR-23a-5p mimics or NC were further analyzed
during differentiation. Overexpression of miR-23a-5p in C212
myotubes decreased the percentage of MyHC1 and increased the
percentage of MyHC2a by RT-qPCR (Fig. 3F). In addition, overexpression of
miR-23a-5p decreased MyHC1 protein level by western blotting
analysis (Fig. 3G), as
demonstrated by immunofluorescence analysis of MyHC1 (Fig. 3H). Taken together, these results
indicated that miR-23a-5p may affect C2C12 myoblast
differentiation and mediate skeletal muscle fiber type
composition.
miR-23a-5p regulates C2C12 myoblast
proliferation and differentiation by interacting with lncDum
A previous study have demonstrated that
lncDum serves a key role in regulating cell differentiation
and muscle regeneration (47). In
order to confirm the function of lncDum on C2C12 myoblast
proliferation and differentiation, si-lncDum, si-NC, p-lncDum or
empty pcDNA3.1+ vector were transfected into C2C12
myoblasts during proliferation (Fig.
4A). CCK-8 and EdU assays revealed that inhibition of
lncDum significantly promoted C2C12 myoblast proliferation
compared with the control (Fig.
4B-D). Consistent with these findings, Cyclin E, CCND1
and Cyclin B were upregulated, and P21 was
downregulated in C212 myoblasts that were transfected with
si-lncDum (Fig. 4E). Consistent
with previous findings (47) that
lncDum enhances C2C12 myoblast differentiation,
overexpression of lncDum promoted the fusion of myotubes and
increased the expression levels of MyoD, MRF4, MyoG, Myf5
and MyHC, whereas lncDum inhibition decreased the
fusion of myotubes and downregulated these genes (Fig. 4F-H). In addition, western blotting
analysis showed that the expression levels of MyoG were also
upregulated by lncDum overexpression and downregulated by
lncDum inhibition compared with the control (Fig. 4I), thereby indicating that
lncDum promoted myoblast differentiation.
 | Figure 4.miR-23a-5p regulates C2C12 myoblast
proliferation and differentiation by interacting with
lncDum. (A) RT-qPCR analysis of the expression levels of
lncDum in myoblasts transfected with si-lncDum, si-NC,
p-lncDum or pcDNA3.1+ empty vector during proliferation.
(B) EdU assay was performed following 24 h of transfection. Scale
bar, 100 µm. (C) Percentage of EdU-positive/DAPI-positive cells was
quantified. (D) Cell count was measured via a CCK-8 assay in C2C12
myoblasts transfected with si-lncDum, si-NC, pcDNA3.1+
empty vector or p-lncDum. RT-qPCR analysis of the expression levels
of (E) Cyclin E, Cyclin B, CCND1 and P21, and (F)
lncDum in myoblasts transfected with si-lncDum, si-NC,
pcDNA3.1+ empty vector or p-lncDum during
differentiation. (G) Bright-field microscopy was used to observe
the ability of C2C12 myoblasts to differentiate into myotubes at 6
days after transfecting with si-lncDum, si-NC, pcDNA3.1+
empty vector or p-lncDum. Scale bar, 100 µm. (H) RT-qPCR measured
the expression levels of MyoG, MyoD, MRF4, Myf5 and
MyHC. (I) Western blotting analysis of MyoG protein levels
in C2C12 myoblasts transfected with si-lncDum, si-NC, p-lncDum or
pcDNA3.1+ empty vector during differentiation. (J)
lncDum contained a binding site for miR-23a-5p. RT-qPCR
analysis of the expression levels of (K) lncDum and
miR-23a-5p during myoblast proliferation and differentiation, and
(L) lncDum in myoblasts transfected with miR-23a-5p mimics
and p-lncDum, NC or p-lncDum during proliferation. (M) Luciferase
assays assessed the effect of miR-23a-5p on the activity of
lncDum. (N) Cell count was measured via a CCK-8 assay in
C2C12 myoblasts transfected with p-lncDum, NC or p-lncDum and
miR-23a-5p mimics. (O) RT-qPCR analysis of the expression levels of
Cyclin E, Cyclin B, CCND1 and P21. (P) Bright-field
was used to microscopy observe the ability of C2C12 myoblasts to
differentiate into myotubes at 6 days after transfecting with
p-lncDum, NC or p-lncDum and miR-23a-5p mimics. Scale bar, 100 µm.
(Q) RT-qPCR analysis of the expression levels of MyoG, MyoD,
MRF4, Myf5 and MyHC. (R) Western blotting analysis of
MyoG protein levels in C2C12 myoblasts transfected with miR-23a-5p
mimics and p-lncDum, NC or p-lncDum during differentiation. Data
are presented as the mean ± SEM of three independent repeats.
*P<0.05, **P<0.01. miR, microRNA; lnc, long non-coding RNA;
RT-qPCR, reverse transcription-quantitative PCR; si, small
interfering; NC, negative control; CCK-8, Cell Counting Kit-8;
MyHC, myosin heavy chain; OD, optical density; NS, not
significant. |
A number of lncRNAs that function in skeletal
myogenesis have previously been identified as ceRNAs that sponge
miRNAs via complementary base pairing (48,49).
In the present study, it was observed that lncDum contained
a complementary sequence to that of miR-23a-5p, and that
lncDum and miR-23a-5p exhibited opposite mRNA
expression level patterns during C2C12 myoblast proliferation and
differentiation (Fig. 4J and K).
Moreover, RT-qPCR analysis demonstrated that overexpression of
miR-23a-5p significantly inhibited the increase of
lncDum induced by transfection of p-lncDum (Fig. 4L). A luciferase reporter assay
demonstrated that, compared with the control, overexpression of
miR-23a-5p suppressed the luciferase activity of WT-lncDum,
but had little effect on miR-23a-5p binding sites in
Mut-lncDum (Fig. 4M),
demonstrating that miR-23a-5p interacted with lncDum
via complementary base pairing. Moreover, rescue experiments were
performed to evaluate whether the effects of miR-23a-5p on
the proliferation and differentiation of C2C12 myoblasts were
regulated by lncDum. The results indicated that
miR-23a-5p significantly attenuated the effect of
lncDum overexpression on C2C12 myoblast proliferation and
differentiation (Fig. 4N-R). Taken
together, these data suggested that miR-23a-5p mediated
myogenic proliferation and differentiation via interactions with
lncDum.
Discussion
Skeletal myogenesis is a complex biological process
(7). In addition to the
involvement of myoblast proliferation and differentiation,
differentiated myoblasts fuse into multinucleate myotubes, which
give rise to diverse types of muscle fiber that build the complex
skeletal muscle architecture essential for body movement, postural
behavior and breathing (1,50). Previous studies have also
demonstrated the roles of miRNAs in skeletal myogenesis. For
example, Naguibneva et al (42) reported that miR-181 regulates
mammalian myoblast differentiation via targeting homeobox protein
Hox-A11. Mi et al (30)
reported that miR-139-5p regulates C2C12 cell myogenesis via
blocking the Wnt/β-catenin signaling pathway. Wang et al
(10) reported that
miR-23a-3p regulates myogenic differentiation by inhibiting
the expression levels of fast MyHC isoforms. To the best of our
knowledge, however, the role and molecular mechanism of
miR-23a-5p in myoblast proliferation and differentiation has
not previously been fully elucidated. The present study aimed to
determine whether miR-23a-5p was involved in skeletal
myogenesis. Previous studies have indicated that miR-206-3p
(41), miR-181a-5p
(42) and miR-221 (43) serve key roles in the regulation of
skeletal myogenesis. The results of the present study demonstrated
that miR-23a-5p exhibited a similar expression level pattern
to these three myoblast-specific miRNAs. The present study also
revealed that miR-23a-5p was ubiquitously expressed in
different types of tissue and moderately expressed in muscle
tissue. Furthermore, miR-23a-5p expression levels in muscle
tissue were significantly decreased following Dex treatment. Dex, a
potent synthetic glucocorticoid, has been widely used to induce
muscle atrophy due to its ability to stimulate protein degradation
(51). Muscle atrophy induced by
muscular dysfunction can affect functional capacity. The data
obtained in the present study supported the hypothesis that
miR-23a-5p is associated with skeletal myogenesis. As
miR-23a-5p exhibited low expression during the proliferation
and differentiation of C2C12 myoblasts compared with
myogenesis-specific miRNAs, including miR-206-3p, miR-181a-5p,
miR-221/222 and miR-29b-3p, it was hypothesized that
overexpression of miR-23a-5p could affect skeletal
myogenesis.
Myoblast proliferation and differentiation are key
processes in skeletal myogenesis (7). Cyclin E, Cyclin B and
CCND1, which bind cyclin-dependent protein kinases (Cdk) to
control cell cycle progression, such as G1-S and G2-M transition,
are expressed during the course of the cell cycle (52–54).
By contrast, as a Cdk inhibitor, p21 can bind and inactivate
Cdk-cyclin complexes to repress specific steps of cell cycle
progression (55–57). The results of the present study
demonstrated that miR-23a-5p positively regulate the
expression levels of these genes during C2C12 myoblast
proliferation. EdU and CCK-8 assays also showed that
miR-23a-5p significantly promoted C2C12 myoblast
proliferation.
Previous studies have shown that myogenic
differentiation is controlled by complex myogenic transcription
factors, such as MyoD, Myf5, MyoG, MRF4 and MyHC
(58,59). MyoD and Myf5
participate in controlling early differentiation, and MyoG
and MRF4 induce myoblast differentiation at later stages
(30,42). MyoG and MyoD are key
transcription factors in myogenesis and regulate transcription of
the majority of muscle-specific genes (60). MyoG and MyoD serve an
important role in the regulation of myoblast differentiation.
MyoD is considered to act as a myogenic determination gene
(61,62), whereas MyoG is essential for
terminal differentiation of committed myoblasts (59). The present results indicated that
miR-23a-5p decreased the expression levels of
myogenesis-specific factors and that miR-23a-5p inhibits
C2C12 myoblast differentiation. Previous studies have demonstrated
that regulators could modulate muscle fiber type transition via
altering the percentage of MyHC isoforms associated with slow- or
fast-twitch fibers (63–65). Fibers expressing MyHC2a and 2× have
intermediate characteristics between MyHC types 1 and 2b (66). In addition, muscle fiber type
composition is key for muscle biological function and body
metabolism; therefore, the present study aimed to determine whether
miR-23a-5p mediates muscle fiber type composition. The
results suggested that miR-23a-5p is involved in regulating
the composition of skeletal muscle fiber type in a more complex
manner. Taken together, these results indicated that
miR-23a-5p may affect C2C12 myoblast differentiation and
mediate skeletal muscle fiber-type composition.
A number of lncRNAs that are key regulators of
skeletal muscle physiology and are involved in skeletal myogenesis
have been identified as ceRNAs that sponge miRNAs via complementary
base pairing (34,67). Wang et al (47) indicated that lncDum serves
an important role in regulating cell differentiation and muscle
regeneration. These results suggest that miR-23a-5p
interacted with lncDum by complementary base pairing, and
that lncDum promotes C2C12 myoblast proliferation and
differentiation. Taken together, these data indicated that
miR-23a-5p mediates myogenic proliferation and
differentiation via interacting with lncDum.
In conclusion, the present study demonstrated that
the expression levels of miR-23a-5p were showed a dynamic
change, from decrease to increase, during myogenesis in mice.
Analysis revealed that miR-23a-5p overexpression promoted
C2C12 myoblast proliferation, inhibited C2C12 myoblast
differentiation and regulated muscle fiber type composition. The
present study indicated that lncDum affected C2C12 myoblast
proliferation and differentiation, which was contrary to the
effects of miR-23a-5p. Taken together, the present results
suggested that lncDum may be a target of miR-23a-5p
in the regulation of skeletal myogenesis. However, the association
between miR-23a-5p inhibition and the regulation of skeletal
myogenesis was not determined in the present study, and further
investigation is required to elucidate the regulatory mechanisms
underlying miR-23a-5p in skeletal muscle development.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Sichuan Sci
& Tech Support Program (grant nos. 2016NYZ0050 and SCCXTD-008),
the National Natural Science Foundation of China (grant no.
31530073) and the earmarked fund for China Agriculture Research
System (grant no. CARS-36-05B).
Availability of data and material
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
SZ, LZ and ML conceptualized and designed the
experiments. XZ, HG, LW and PZ performed the experiments. JD, LS
and DJ collected the animal samples. JW and XL analyzed the data.
XZ drafted the manuscript. All authors read and approved the final
version of the manuscript.
Ethics approval and consent to
participate
All animal procedures were conducted in accordance
with institutional guidelines for the care and use of laboratory
animals was approved by the Animal Care and Ethics Committee of
Sichuan Agricultural University, Sichuan, China [approval no.
DKY-(S20176903)].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chargé SBP and Rudnicki MA: Cellular and
molecular regulation of muscle regeneration. Physiol Rev.
84:209–238. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ijuin T, Hatano N, Hosooka T and Takenawa
T: Regulation of insulin signaling in skeletal muscle by PIP3
phosphatase, SKIP, and endoplasmic reticulum molecular chaperone
glucose-regulated protein 78. Biochim Biophys Acta. 1853:3192–3201.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pedersen BK and Febbraio MA: Muscles,
exercise and obesity: Skeletal muscle as a secretory organ. Nat Rev
Endocrinol. 8:457–465. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krook A, Björnholm M, Galuska D, Jiang XJ,
Fahlman R, Myers MG Jr, Wallberg-Henriksson H and Zierath JR:
Characterization of signal transduction and glucose transport in
skeletal muscle from type 2 diabetic patients. Diabetes.
49:284–292. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jackman RW and Kandarian SC: The molecular
basis of skeletal muscle atrophy. Am J Physiol Cell Physiol.
287:C834–C843. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tisdale MJ: Loss of skeletal muscle in
cancer: biochemical mechanisms. Front Biosci. 6:D164–D174. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Buckingham M, Bajard L, Chang T, Daubas P,
Hadchouel J, Meilhac S, Montarras D, Rocancourt D and Relaix F: The
formation of skeletal muscle: From somite to limb. J Anat.
202:59–68. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schiaffino S and Reggiani C: Fiber types
in mammalian skeletal muscles. Physiol Rev. 91:1447–1531. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Choe JH, Choi YM, Lee SH, Shin HG, Ryu YC,
Hong KC and Kim BC: The relation between glycogen, lactate content
and muscle fiber type composition, and their influence on
postmortem glycolytic rate and pork quality. Meat Sci. 80:355–362.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Chen X, Zheng Y, Li F, Lu Z, Chen
C, Liu J, Wang Y, Peng Y, Shen Z, et al: miR-23a inhibits myogenic
differentiation through down regulation of fast myosin heavy chain
isoforms. Exp Cell Res. 318:2324–2334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Buckingham M: Myogenic progenitor cells
and skeletal myogenesis in vertebrates. Curr Opin Genet Dev.
16:525–532. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rudnicki MA and Jaenisch R: The MyoD
family of transcription factors and skeletal myogenesis. BioEssays.
17:203–209. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu J, McKinsey TA, Zhang CL and Olson EN:
Regulation of skeletal myogenesis by association of the MEF2
transcription factor with class II histone deacetylases. Mol Cell.
6:233–244. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Talarico AP: Myf5 does not induce
apoptosis in skeletal myoblasts but is regulated by oncogenic ras
expression (unpublished PhD thesis). Cleveland State University;
2009
|
|
15
|
Estrella NL, Desjardins CA, Nocco SE,
Clark AL, Maksimenko Y and Naya FJ: MEF2 transcription factors
regulate distinct gene programs in mammalian skeletal muscle
differentiation. J Biol Chem. 290:1256–1268. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
te Pas MF, Soumillion A, Harders FL,
Verburg FJ, van den Bosch TJ, Galesloot P and Meuwissen TH:
Influences of myogenin genotypes on birth weight, growth rate,
carcass weight, backfat thickness, and lean weight of pigs. J Anim
Sci. 77:2352–2356. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Keren A, Tamir Y and Bengal E: The p38
MAPK signaling pathway: A major regulator of skeletal muscle
development. Mol Cell Endocrinol. 252:224–230. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Megeney LA, Kablar B, Garrett K, Anderson
JE and Rudnicki MA: MyoD is required for myogenic stem cell
function in adult skeletal muscle. Genes Dev. 10:1173–1183. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Song G, Xu G, Ji C, Shi C, Shen Y, Chen L,
Zhu L, Yang L, Zhao Y and Guo X: The role of microRNA-26b in human
adipocyte differentiation and proliferation. Gene. 533:481–487.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bian H, Zhou Y, Zhou D, Zhang Y, Shang D
and Qi J: The latest progress on miR-374 and its functional
implications in physiological and pathological processes. J Cell
Mol Med. 23:3063–3076. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao Y, Lin L, Li T, Yang J and Wei Y: The
role of miRNA-223 in cancer: Function, diagnosis and therapy. Gene.
616:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cheng Y, Liu X, Zhang S, Lin Y, Yang J and
Zhang C: MicroRNA-21 protects against the H(2)O(2)-induced injury
on cardiac myocytes via its target gene PDCD4. J Mol Cell Cardiol.
47:5–14. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Callis TE, Deng Z, Chen JF and Wang DZ:
Muscling through the microRNA world. Exp Biol Med (Maywood).
233:131–138. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen JF, Tao Y, Li J, Deng Z, Yan Z, Xiao
X and Wang DZ: microRNA-1 and microRNA-206 regulate skeletal muscle
satellite cell proliferation and differentiation by repressing
Pax7. J Cell Biol. 190:867–879. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luo W, Nie Q and Zhang X: MicroRNAs
involved in skeletal muscle differentiation. J Genet Genomics.
40:107–116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Russell AP, Lamon S, Boon H, Wada S,
Güller I, Brown EL, Chibalin AV, Zierath JR, Snow RJ, Stepto N, et
al: Regulation of miRNAs in human skeletal muscle following acute
endurance exercise and short-term endurance training. J Physiol.
591:4637–4653. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen JF, Mandel EM, Thomson JM, Wu Q,
Callis TE, Hammond SM, Conlon FL and Wang DZ: The role of
microRNA-1 and microRNA-133 in skeletal muscle proliferation and
differentiation. Nat Genet. 38:228–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mi L, Li Y, Zhang Q, Zhao C, Peng Y, Yang
G and Zheng X: MicroRNA-139-5p regulates C2C12 cell myogenesis
through blocking Wnt/β-catenin signaling pathway. Biochem Cell
Biol. 93:8–15. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Crist CG, Montarras D, Pallafacchina G,
Rocancourt D, Cumano A, Conway SJ and Buckingham M: Muscle stem
cell behavior is modified by microRNA-27 regulation of Pax3
expression. Proc Natl Acad Sci USA. 106:13383–13387. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ge Y, Sun Y and Chen J: IGF-II is
regulated by microRNA-125b in skeletal myogenesis. J Cell Biol.
192:69–81. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang MB, Xu H, Xie SJ, Zhou H and Qu LH:
Insulin-like growth factor-1 receptor is regulated by microRNA-133
during skeletal myogenesis. PLoS One. 6:e291732011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dong XZ, Hu Y, Liu P and Lu Y: The
research progress of lncRNA as CeRNA in gastric cancer. Chin
Pharmacol Bull. 32:1185–1188, 1189, 2016 (In Chinese).
|
|
35
|
Mueller AC, Cichewicz MA, Dey BK, Layer R,
Reon BJ, Gagan JR and Dutta A: MUNC, a long noncoding RNA that
facilitates the function of MyoD in skeletal myogenesis. Mol Cell
Biol. 35:498–513. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kang X, Zhao Y, Van Arsdell G, Nelson SF
and Touma M: Ppp1r1b-lncRNA inhibits PRC2 at myogenic regulatory
genes to promote cardiac and skeletal muscle development in mouse
and human. RNA. 26:481–491. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mercatelli N, Fittipaldi S, De Paola E,
Dimauro I, Paronetto MP, Jackson MJ and Caporossi D: miR-23-TrxR1
as a novel molecular axis in skeletal muscle differentiation. Sci
Rep. 7:72192017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mercatelli N, Fittipaldi S, Dimauro I,
Scalabrin M and Caporossi D: TrxR1/miR-23 as a novel molecular axis
acting on skeletal muscle differentation. Free Radic Biol Med.
96:S25–S26. 2016. View Article : Google Scholar
|
|
39
|
Morimoto Y, Kondo Y, Kataoka H, Honda Y,
Kozu R, Sakamoto J, Nakano J, Origuchi T, Yoshimura T and Okita M:
Heat treatment inhibits skeletal muscle atrophy of
glucocorticoid-induced myopathy in rats. Physiol Res. 64:897–905.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
McCarthy JJ: MicroRNA-206: The skeletal
muscle-specific myomiR. Biochim Biophys Acta. 1779:682–691. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Naguibneva I, Ameyar-Zazoua M, Polesskaya
A, Ait-Si-Ali S, Groisman R, Souidi M, Cuvellier S and Harel-Bellan
A: The microRNA miR-181 targets the homeobox protein Hox-A11 during
mammalian myoblast differentiation. Nat Cell Biol. 8:278–284. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cardinali B, Castellani L, Fasanaro P,
Basso A, Alemà S, Martelli F and Falcone G: Microrna-221 and
microrna-222 modulate differentiation and maturation of skeletal
muscle cells. PLoS One. 4:e76072009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Shen H, Liu T, Fu L, Zhao S, Fan B, Cao J
and Li X: Identification of microRNAs involved in
dexamethasone-induced muscle atrophy. Mol Cell Biochem.
381:105–113. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chromiak JA and Vandenburgh HH:
Glucocorticoid-induced skeletal muscle atrophy in vitro is
attenuated by mechanical stimulation. Am J Physiol.
262:C1471–C1477. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun Y, Wang G, Ji Z, Chao T, Liu Z, Wang
X, Liu G, Wu C and Wang J: Three slow skeletal muscle troponin
genes in small-tailed Han sheep (Ovis aries): Molecular cloning,
characterization and expression analysis. Mol Biol Rep.
43:999–1010. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang L, Zhao Y, Bao X, Zhu X, Kwok YK, Sun
K, Chen X, Huang Y, Jauch R, Esteban MA, et al: LncRNA Dum
interacts with Dnmts to regulate Dppa2 expression during myogenic
differentiation and muscle regeneration. Cell Res. 25:335–350.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chen Z, Liu H, Yang H, Gao Y, Zhang G and
Hu J: The long noncoding RNA, TINCR, functions as a competing
endogenous RNA to regulate PDK1 expression by sponging miR-375 in
gastric cancer. OncoTargets Ther. 10:3353–3362. 2017. View Article : Google Scholar
|
|
49
|
Liu W, Ma C, Yang B, Yin C, Zhang B and
Xiao Y: LncRNA Gm15290 sponges miR-27b to promote PPARγ-induced fat
deposition and contribute to body weight gain in mice. Biochem
Biophys Res Commun. 493:1168–1175. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Abmayr SM, Balagopalan L, Galletta BJ and
Hong SJ: Cell and molecular biology of myoblast fusion. Int Rev
Cytol. 225:33–89. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Mayer M, Shafrir E, Kaiser N, Milholland
RJ and Rosen F: Interaction of glucocorticoid hormones with rat
skeletal muscle: Catabolic effects and hormone binding. Metabolism.
25:157–167. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Schwartz GK and Shah MA: Targeting the
cell cycle: A new approach to cancer therapy. J Clin Oncol.
23:9408–9421. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bryant P, Zheng Q and Pumiglia K: Focal
adhesion kinase controls cellular levels of p27/Kip1 and p21/Cip1
through Skp2-dependent and -independent mechanisms. Mol Cell Biol.
26:4201–4213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Alt JR, Gladden AB and Diehl JA: p21(Cip1)
Promotes cyclin D1 nuclear accumulation via direct inhibition of
nuclear export. J Biol Chem. 277:8517–8523. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dupont J, Karas M and LeRoith D: The
cyclin dependent kinase inhibitor p21CIP/WAF is a positive
regulator of IGF-1-induced cell proliferation in MCF-7 human breast
cancer cells. J Biol Chem. 278:37256–37264. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang C, Chen P, Jin H, Yan X, Gan L, Li Y,
Zhou S, Chang J, Wang Y, Yang G, et al: Nidus vespae protein
inhibiting proliferation of HepG2 hepatoma cells through
extracellular signal-regulated kinase signaling pathways and
inducing G1 cell cycle arrest. Acta Biochim Biophys Sin (Shanghai).
40:970–978. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xiong Y, Hannon GJ, Zhang H, Casso D,
Kobayashi R and Beach D: p21 is a universal inhibitor of cyclin
kinases. Nature. 366:701–704. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zammit PS: Function of the myogenic
regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal
muscle, satellite cells and regenerative myogenesis. Semin Cell Dev
Biol. 72:19–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Montarras D, Chelly J, Bober E, Arnold H,
Ott MO, Gros F and Pinset C: Developmental patterns in the
expression of Myf5, MyoD, myogenin, and MRF4 during myogenesis. New
Biol. 3:592–600. 1991.PubMed/NCBI
|
|
60
|
Braun T and Gautel M: Transcriptional
mechanisms regulating skeletal muscle differentiation, growth and
homeostasis. Nat Rev Mol Cell Biol. 12:349–361. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Berkes CA and Tapscott SJ: MyoD and the
transcriptional control of myogenesis. Semin Cell Dev Biol.
16:585–595. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Edmondson DG and Olson EN: A gene with
homology to the myc similarity region of MyoD1 is expressed during
myogenesis and is sufficient to activate the muscle differentiation
program. Genes Dev. 4:1450. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Cheng X, Du J, Shen L, Tan Z, Jiang D,
Jiang A, Li Q, Tang G, Jiang Y, Wang J, et al: miR-204-5p regulates
C2C12 myoblast differentiation by targeting MEF2C and ERRγ. Biomed
Pharmacother. 101:528–535. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Du J, Zhang P, Zhao X, He J, Xu Y, Zou Q,
Luo J, Shen L, Gu H, Tang Q, et al: MicroRNA-351-5p mediates
skeletal myogenesis by directly targeting lactamase-β and is
regulated by lnc-mg. FASEB J. 3:1911–1926. 2019. View Article : Google Scholar
|
|
65
|
Shen L, Chen L, Zhang S, Du J, Bai L,
Zhang Y, Jiang Y, Li X, Wang J and Zhu L: MicroRNA-27b Regulates
Mitochondria Biogenesis in Myocytes. PLoS One. 11:e01485322016.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mizunoya W, Iwamoto Y, Sato Y, Tatsumi R
and Ikeuchi Y: Cold exposure increases slow-type myosin heavy chain
1 (MyHC1) composition of soleus muscle in rats. Anim Sci J.
85:293–304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Anderson DM, Anderson KM, Chang CL,
Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM,
Liou J, Bassel-Duby R, et al: A micropeptide encoded by a putative
long noncoding RNA regulates muscle performance. Cell. 160:595–606.
2015. View Article : Google Scholar : PubMed/NCBI
|


















