Introduction
The genes and mechanisms involved in complex
autoimmune diseases, which affect ~5% of the population, remain
obscure. Genome-wide association studies (GWAS) have identified
hundreds of autoimmune disease-associated loci without defining
causal variants (1). Understanding
the mechanisms that underlie these diseases may contribute to the
development of diseases-modifying therapeutic protocols.
Accumulating evidence has revealed that common genetic factors may
predispose individuals to multiple autoimmune diseases. In this
framework, several genetic polymorphisms have been associated with
systemic lupus erythematosus (SLE) and other autoimmune diseases
(2). In addition, it has been
reported that most of the identified systemic scleroderma (SSc)
susceptibility loci overlap with those of other autoimmune diseases
and in particular, disorders such as SLE and rheumatoid arthritis
(RA) (3). Thus, previous studies
have supported the hypothesis that common deregulated pathways
exist across multiple autoimmune diseases.
In an attempt to identify potential causal variants
that alter physiologic pathways and are involved in susceptibility
to autoimmune diseases, various studies have revealed that the
missense variant rs35677470 at the deoxyribonuclease I-like 3
(DNASE1L3) locus was associated with the development of SLE
(4,5), RA (6) and SSc (7), thus representing a shared risk factor
for these diseases. However, the exact functional consequence of
this polymorphism has not yet been thoroughly investigated.
Deoxyribonuclease I-like 3 (DNase 1L3) is a member of the human
DNase I family, representing a nuclease that cleaves
double-stranded DNA during apoptosis (8). The protein encoded by DNASE1L3 is one
of three human homologs of DNase I and functions as an endonuclease
capable of cleaving both single- and double-stranded DNA (9–11).
SLE is a chronic, severe, multiorgan systemic
autoimmune disease that predominantly affects women, with a complex
genetic inheritance and strong clustering in families (4). It is characterized by the production
of high titers of autoantibodies directed against native DNA, cell
surface and other cellular constituents that are associated with
high morbidity rates (12). RA is
a chronic, systemic multifactorial disease of unknown etiology,
characterized by progressive joint destruction resulting in severe
disability. It results from a complex interplay between genetic and
environmental factors (13,14).
SSc is a chronic, multisystem autoimmune disease clinically
characterized by progressive skin and internal organ fibrosis,
exhibiting one of the highest mortality rates among rheumatic
diseases, leading to premature death in affected individuals
(15,16). SSc affects connective tissue and
produces various heterogeneous clinical manifestations, including
inflammation, autoimmunity, vasculopathy and excessive
extracellular matrix production and deposition (17). SSc, RA and SLE are heterogeneous
diseases of the connective tissue that share clinical and
epidemiological manifestations as well as life-threatening
complications (18). Considering
that the production of autoantibodies represents a main feature of
these diseases, patients often exhibit an extensive deregulation of
the innate and adaptive immune response (19).
Given that autoimmune diseases may share common
susceptibility genes, potentially leading to the development of
shared therapeutic approaches, the present study aimed to
investigate the potential role of the rs35677470 variant at the
DNASE1L3 gene and the resultant R206C mutation in the
development of SLE, RA and SSc. The underlying mechanism
potentially affecting protein structure loss of function was also
assessed.
Materials and methods
Sequence retrieval
Protein sequences were retrieved from the UNIPROT
database (20) and the structural
information was obtained from the Protein Data Bank (21). To find homologs across species,
BLAST searches were performed with Mega BLAST (National Center for
Biotechnology Information) using the protein databases PDB and
SwissProt and Blastp (protein-protein BLAST) with default
parameters (22).
Sequence alignment
T-COFFEE, the multiple sequence alignment program
(23), was used to perform all
protein sequence alignments. Default parameters were used for the
alignment. ESPript was used to depict the sequence alignments and
to incorporate the available conservation or diversity information
and ENDscript was used for 3D homology representation (24,25).
Phylogenetic analysis
To test for phylogeny, trees were constructed using
maximum-likelihood analysis with 500 bootstrap replications
(26) implemented by MEGA7
(27). The evolutionary tree was
inferred from protein sequences using the Maximum Likelihood method
(28), based on the
Jones-Taylor-Thorntonmatrix model (28) and incorporated using the Gamma
distribution model. The tree with the highest log likelihood was
presented in all phylogeny figures. Initial trees for heuristic
searches were obtained automatically by applying Neighbor-Join
(29) and BioNJ (30) algorithms to a matrix of pairwise
distances, estimated using the Maximum Composite Likelihood
(31) approach and then by
selecting the topology with the superior log likelihood value.
Structural analysis
The structure of the DNASE1L3 protein
(Homo_sapiens_NP_004935.1) was built using homology modeling with
the SWISSMODEL program via the EXPASY server (32) and was based on the experimentally
determined crystal structures of homologous proteins retrieved from
the Protein Data Bank (IDs 1atn, 4awn and 3w3d) (33–35).
In silico mutagenesis of the Arg206Cys point mutation was created
in the homology model of the wild-type DNASE1L3 protein structure
using the PyMOL molecular-graphics system V.2.2 (Schrödinger, LLC)
(36). Multiple structure
alignments were performed using PyMOL (36) based on protein backbone RMSD (Root
Mean Square Deviation) optimization and sequence alignment.
ENDscript (25) was used to depict
weak and strong sequence conservations in structures across species
in order to pinpoint variable sites on the functional domain. All
figures depicting 3D models were generated using the PyMOL
molecular-graphics system V.2.2 (Schrödinger, LLC) (36).
Results and Discussion
Phylogenetics and sequence
analysis
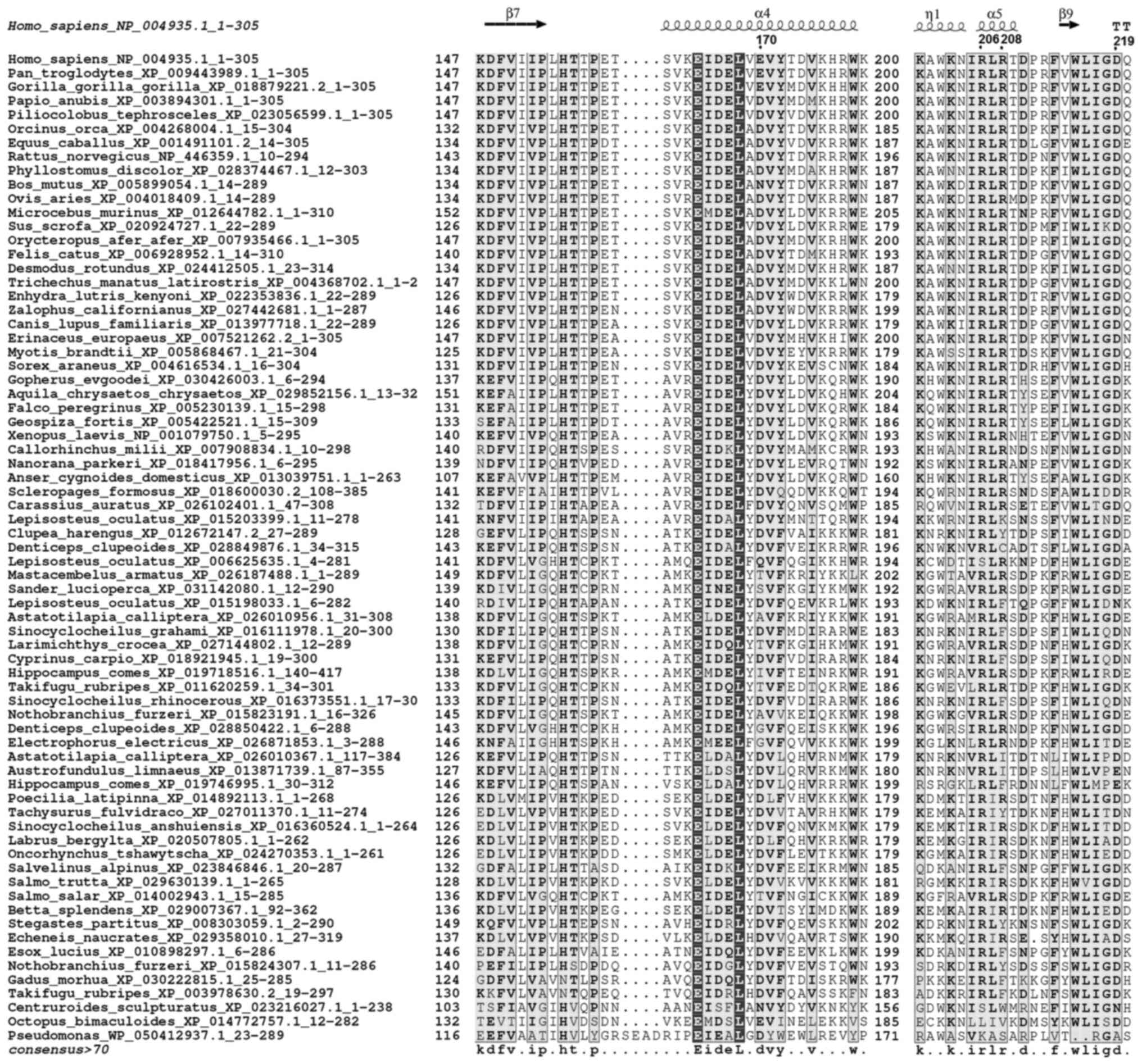
DNASE1L3 evolution was investigated to define
conservation elements in its protein sequence. Evolutionary
analysis is used to identify positions on the protein sequences
that are heavily conserved across species, indicating structural
importance (37). To determine the
evolutionary relationship among DNase1L3 members of the DNase I
family and to identify additional homologue DNAasesI in other
species, exhaustive BLAST searches were performed using a variety
of available protein sequence databases. Using multiple
phylogenetic approaches, the results revealed high sequence
conservation among species, with separation only occurring across
different animal taxa. Outgroups (mollusks and bacteria) were
aligned with high confidence, demonstrating good preservation of
important secondary structure elements (Figs. 1 and 2).
Structural analysis
The current study utilized 3D homology modeling to
localize polymorphisms in the DNASE1L3 protein. Structural analysis
revealed that the rs35677470 DNASE1L3 single nucleotide
polymorphism (SNP), which encodes the non-conservative amino acid
variation Arg206Cys, modified the conserved electrostatic network
that holds protein secondary structure elements in position
(Fig. 3A). Specifically, the
guanidinium group of the Arg206 side chain interacted with the
carboxylate group of Glu170, forming a strong salt bridge (Fig. 3B). Together with the Arg208 to
Asp219 charge interaction, an electrostatic salt bridge network was
formed, which stabilized two important scaffold α-helices. This
network was interrupted by the highly defective rs35677470 SNP
allele, in which arginine is replaced by a cysteine, affecting the
molecular architecture (Fig. 3C
and Table I). Previous studies on
the effect of this SNP in Caucasian populations, resulting in a
lower level of DNAse1L3 activity, are consistent with this
observation, demonstrating that the SNP affects position 206 in the
protein, thus producing a less active form of human DNAse1L3
(38,39). The introduction of the cysteine
residue at position 206 has no effect on disulphide bond formation,
since all four cystines forming disulphide bridges (24–52 and
194–231; Fig. 3) are distal to
position 206 and sterically inaccessible at this position. Herein,
the structural analysis showed that although position 206 is
distant to the nucleotide binding site forming residues, the effect
of this mutation destabilizes the nucleotide interacting loop
preceding that of position 206 (residue 193–196). The enhanced
mobility of this loop facing the nucleotide recognition site and
the subsequent direct interaction with the affected attacked
nucleotide may affect the enzymatic activity of
DNAse1L3.
 | Table I.Summary of salt bridge stabilizing
interactions. |
Table I.
Summary of salt bridge stabilizing
interactions.
| Salt bridge
interaction | Native | rs35677470
DNASE1L3 SNP (Arg206Cys) |
|---|
| Arg206→Glu170 | + | − |
| Arg208→Asp219 | + | + |
Arg206Cys mutation of DNASE1L3 in
patients with SLE, RA and SSc
The potentially causal missense variant rs35677470
localized to the DNASE1L3 gene (exon 8) at position 3p14.3
was identified following GWAS in patients with SLE, RA and SSc
(Table II). However, despite
exhibiting a pleiotropic effect in autoimmunity, this SNP was not
found to be associated with Type 1 Diabetes (6). A second SNP (rs7652027) of the
DNASE1L3 genomic region has also been recently associated
with SSc (40). However, the same
study revealed no association between the rs35677470 SNP with SSc
due to the absence of the SNP from the panel used. In other
studies, by Mayes et al (41) and Zochling et al (7), the association signal of the
rs35677470 SNP was also significantly associated with
anti-centromere-antibody-positive (ACA+) patients with
SSc but not with ACA− individuals. Therefore, it has
been hypothesized that the association between ACA and SSc in the
case of rs35677470 loss-of-function DNASE1L3 variants may
provide a link between defective apoptotic DNA breakdown and ACA
production (41).
 | Table II.Role overview of the rs35677470
(Arg206Cys) single nucleotide polymorphism in autoimmune
diseases. |
Table II.
Role overview of the rs35677470
(Arg206Cys) single nucleotide polymorphism in autoimmune
diseases.
| Disease | Effect | Author (Refs.) |
|---|
| SLE | Risk | Harley et al
2008, (4) |
|
|
| Gateva et al
2009, (5) |
| RA | Risk | Westra et al
2018, (6) |
| SSc | Risk | Martin et al
2012, (3) |
|
|
| Zochling et
al 2014, (7) |
|
|
| Mayes et al
2014, (41) |
| T1D | No genetic
association | Westra et al
2018, (6) |
In regards to the potential role of the rs35677470
SNP in SLE and taking into account that SLE pathogenesis is
associated with a reduced ability to clear DNA released from
apoptotic cells, Al-Mayouf et al (42) hypothesized that DNASE1L3
gene dysfunction may lead to impaired DNA breakdown and clearance
from apoptotic cells, thus resulting in the formation of antibodies
recognizing DNA and immune complexes. Furthermore, given that
similar types of DNA-driven immune complexes (such as anti-nuclear
and ACA antibodies) are also characteristic of SSc, this hypothesis
is also applicable in SLE (7).
Recently, decreased expression levels of DNASE1l3
have been detected in patients with SLE, resulting in the reduction
of nucleosome DNA digestion in serum, which is associated with the
development of anti-double stranded DNA antibodies. However, the
expression and activity of DNASE1l3 remained unchanged in patients
with RA, strengthening the hypothesis that the immunopathological
mechanism of RA differs from that of SLE (43). Furthermore, Serpas et al
(44) targeted the DNASE1L3 enzyme
as a potential therapeutic agent for SLE. To this end, a murine
model with DNASE1L3 deficiency was developed, in which mice exhibit
features of SLE and renal disease. Methods to treat this disease by
targeting the DNASE1L3 enzyme are currently being investigated.
Candidate gene studies and GWAS have attempted to
partially elucidate the complex genetic architecture of SLE through
the identification of >90 risk loci (45) and to determine the existing
differences in risk variants across different continents (46). These studies have established the
importance of several pathways in SLE, including innate immune
responses, the activation of lymphocytes and immune complex
clearing (45,47–49).
The present study comprehensively evaluated rs35677470 of the
shared autoimmune locus DNASE1L3, which has been reported to
produce an inactive form of DNaseIL3 (38,39).
Structural analysis performed in the current study elucidated the
potential role of mutations that modify the placement of structural
elements, consequently producing disordered protein folding and
thereby affecting its biological function. In the post-genomic era,
the identification of genes and molecules involved in the molecular
mechanisms of various diseases is of vital importance to elucidate
clinically relevant functional defects, thus contributing to drug
discovery and the production of novel therapies. Therefore,
determining the 3D structure and analyzing the role of any
causative mutation in the pathogenesis of disease, from the
structural/functional point of view, is necessary. As a
consequence, potential abnormalities in the conformation and
activity of proteins can be elucidated. Additionally, an efficient
integration of these data with other biological factors such as
antibodies, antagonists, inhibitors and binders (50) may identify novel targets for
pharmaceutical intervention. The results of the present study
further elucidated the biological significance of rs35677470 at the
DNASE1L3 locus in SLE, RA and SSc, and demonstrated the
value of pleiotropic gene studies for autoimmune diseases.
Acknowledgements
Not applicable.
Funding
This study was supported by the project ‘INSPIRED:
The National Research Infrastructures on Integrated Structural
Biology, Drug Screening Efforts and Drug Target Functional
Characterization’ (grant no. MIS 5002550), which is implemented
under the Action ‘Reinforcement of the Research and Innovation
Infrastructure’, funded by the Operational Program
‘Competitiveness, Entrepreneurship and Innovation’ (NSRF 2014–2020)
and co-financed by Greece and the European Union (European Regional
Development Fund).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
EEE, MIZ and GNG designed the current study and
drafted the manuscript. AA, GNG, MM, DAS and MIZ searched the
literature. EEE, MM, DAS, AA and MIZ analyzed and interpreted the
data. MIZ, AA and DAS critically revised the manuscript. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
DAS is the Managing Editor of the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
All other authors declare that they have no competing
interests.
References
|
1
|
Goulielmos GN, Zervou MI, Myrthianou E,
Burska A, Niewold TB and Ponchel F: Genetic data: The new challenge
of personalized medicine, insights for rheumatoid arthritis
patients. Gene. 583:90–101. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramos PS, Criswell LA, Moser KL, Comeau
ME, Williams AH, Pajewski NM, Chung SA, Graham RR, Zidovetzki R,
Kelly JA, et al: A comprehensive analysis of shared loci between
systemic lupus erythematosus (SLE) and sixteen autoimmune diseases
reveals limited genetic overlap. PLoS Genet. 7:e10024062011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Martín JE, Bossini-Castillo L and Martín
J: Unraveling the genetic component of systemic sclerosis. Hum
Genet. 131:1023–1037. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
International Consortium for Systemic
Lupus Erythematosus Genetics (SLEGEN), ; Harley JB,
Alarcón-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL,
Tsao BP, Vyse TJ, Langefeld CD, et al: Genomewide association scan
in women with systemic lupus erythematosus identifies
susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat
Genet. 40:204–210. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gateva V, Sandling JK, Hom G, Taylor KE,
Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, et
al: A large-scale replication study identifies TNIP1, PRDM1, JAZF1,
UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus.
Nat Genet. 41:1228–1233. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Westra HJ, Martínez-Bonet M,
Onengut-Gumuscu S, Lee A, Luo Y, Teslovich N, Worthington J, Martin
J, Huizinga T, Klareskog L, et al: Fine-mapping and functional
studies highlight potential causal variants for rheumatoid
arthritis and type 1 diabetes. Nat Genet. 50:1366–1374. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zochling J, Newell F, Charlesworth JC, Leo
P, Stankovich J, Cortes A, Zhou Y, Stevens W, Sahhar J, Roddy J, et
al: An ImmunoChip-based interrogation of scleroderma susceptibility
variants identifies a novel association at DNASE1L3. Arthritis Res
Ther. 16:4382014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sisirak VB, Sally B, D'Agati V,
Martinez-Ortiz W, Özçakar ZB, David J, Rashidfarrokhi A, Yeste A,
Panea C, Chida AS, et al: Digestion of chromatin in apoptotic cell
microparticles prevents autoimmunity. Cell. 166:88–101. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rodriguez AM, Rodin D, Nomura H, Morton
CC, Weremowicz S and Schneider MC: Identification, localization,
and expression of two novel human genes similar to
deoxyribonuclease I. Genomics. 42:507–513. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zeng Z, Parmelee D, Hyaw H, Coleman TA, Su
K, Zhang J, Gentz R, Ruben S, Rosen C and Li Y: Cloning and
characterization of a novel human DNase. Biochem Biophys Res
Commun. 23:499–504. 1997. View Article : Google Scholar
|
|
11
|
Chen WJ, Lee IS, Chen CY and Liao TH:
Biological functions of the disulfides in bovine pancreatic
deoxyribonuclease. Protein Sci. 13:875–883. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Helmick CG, Felson DT, Lawrence RC,
Gabriel S, Hirsch R, Kwoh CK, Liang MH, Kremers HM, Mayes MD,
Merkel PA, et al: Estimates of the prevalence of arthritis and
other rheumatic conditions in the United States. Part I. Arthritis
Rheum. 58:15–25. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Firestein GS: Evolving concepts of
rheumatoid arthritis. Nature. 423:356–361. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
McAllister KM, Eyre S and Orozco G:
Genetics of rheumatoid arthritis: GWAS and beyond. Open Access
Rheumatology Res Rev. 3:31–46. 2013.
|
|
15
|
Gabrielli A, Avvedimento EV and Krieg T:
Scleroderma. N Engl J Med. 360:1989–2003. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Barnes J and Mayes MD: Epidemiology of
systemic sclerosis: Incidence, prevalence, survival, risk factors,
malignancy, and environmental triggers. Curr Opin Rheumatol.
24:165–170. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chizzolini C: T cells, B cells, and
polarized immune response in the pathogenesisof fibrosis and
systemic sclerosis. Curr Opin Rheumatol. 20:707–712. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wallace B, Vummidi D and Khanna D:
Management of connective tissue diseases associated interstitial
lung disease: A review of the published literature. Curr Opin
Rheumatol. 28:236–245. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu Z and Davidson A: Taming lupus-a new
understanding of pathogenesis is leading to clinical advances. Nat
Med. 18:871–882. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
UniProt Consortium: UniProt: A worldwide
hub of protein knowledge. Nucleic Acids Res. 47:D506–D515. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The protein
data bank. Nucleic Acids Res. 28:235–242. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Altschul SF, Madden TL, Schäffer AA, Zhang
J, Zhang Z, Miller W and Lipman DJ: Gapped BLAST and PSI-BLAST: A
new generation of protein database search programs. Nucleic Acids
Res. 25:3389–3402. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Notredame C, Higgins DG and Heringa J: A
novel method for fast and accurate multiple sequence alignment. Mol
Biol. 302:205–217. 2000. View Article : Google Scholar
|
|
24
|
Robert X and Gouet P: Deciphering key
features in protein structures with the new ENDscript server.
Nucleic Acids Res. 42((Web Server Issue)): W320–W324. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gouet P, Robert X and Courcelle E:
ESPript/ENDscript: Extracting and rendering sequence and 3D
information from atomic structures of proteins. Nucleic Acids Res.
31:3320–3323. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Felsenstein J: Confidence limits on
phylogenies: An approach using the bootstrap. Evolution.
39:783–791. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kumar S, Stecher G and Tamur K: MEGA7:
Molecular evolutionary genetics analysis version 7.0 for bigger
datasets. Mol Biol Evol. 33:870–1874. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Felsenstein J: Evolutionary trees from DNA
sequences: A maximum likelihood approach. J Mol Evol. 17:368–376.
1981. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Saitou N and Nei M: The neighbor-joining
method: A new method for reconstructing phylogenetic trees. Mol
Biol Evol. 4:406–425. 1987.PubMed/NCBI
|
|
30
|
Gascuel O: BIONJ: An improved version of
the NJ algorithm based on a simple model of sequence data. Mol Biol
Evol. 14:685–695. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tamura K, Nei M and Kumar S: Prospects for
inferring very large phylogenies by using the neighbor-joining
method. Proc Natl Acad Sci USA. 101:11030–11035. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Waterhouse A, Bertoni M, Bienert S, Studer
G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C,
Bordoli L, et al: SWISS-MODEL: Homology modelling of protein
structures and complexes. Nucleic Acids Res. 46:W296–W303. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kabsch W, Mannherz HG, Suck D, Pai EF and
Holmes KC: Atomic structure of the actin: DNase I complex. Nature.
347:37–44. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Parsiegla G, Noguere C, Santell L, Lazarus
RA and Bourne Y: The structure of human DNase I bound to magnesium
and phosphate ions points to a catalytic mechanism common to
members of the DNase I-like superfamily. Biochemistry.
51:10250–10258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sasaki K, Sakabe K, Sakabe N, Kondo H and
Shimomura M: Refined structure and solvent network of chicken
gizzard G-actin DNase 1 complex at 1.8Å resolution. Acta Cryst.
A49:C111–C112. 1993. View Article : Google Scholar
|
|
36
|
The PyMOL molecular graphics system,
version 1.7.4. Schrödinger, LLC; simplehttp://www.pymol.org
|
|
37
|
Andreou A, Giastas P, Christoforides E and
Eliopoulos EE: Structural and evolutionary insights within the
polysaccharide deacetylase gene family of Bacillus anthracis and
Bacillus cereus. Genes (Basel). 9:3862018. View Article : Google Scholar
|
|
38
|
Ueki M, Takeshita H, Fujihara J, Iida R,
Yuasa I, Kato H, Panduro A, Nakajima T, Kominato Y and Yasuda T:
Caucasian-specific allele in non-synonymous single nucleotide
polymorphisms of the gene encoding deoxyribonuclease I-like 3,
potentially relevant to autoimmunity, produces an inactive enzyme.
Clin Chim Acta. 407:20–24. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ueki M, Fujihara J, Takeshita H,
Kimura-Kataoka K, Iida R, Yuasa I, Kato H and Yasuda T: Global
genetic analysis of all single nucleotide polymorphisms in exons of
the human deoxyribonuclease I-like 3 gene and their effect on its
catalytic activity. Electrophoresis. 32:1465–1472. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
López-Isac E, Acosta-Herrera M, Kerick M,
Assassi S, Satpathy AT, Granja J, Mumbach MR, Beretta L, Simeón CP,
Carreira P, et al: GWAS for systemic sclerosis identifies multiple
risk loci and highlights fibrotic and vasculopathy pathways. Nat
Commun. 10:49552019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mayes MD, Bossini-Castillo L, Gorlova O,
Martin JE, Zhou X, Chen WV, Assassi S, Ying J, Tan FK, Arnett FC,
et al: Immunochip analysis identifies multiple susceptibility loci
for systemic sclerosis. Am J Hum Genet. 94:47–61. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Al-Mayouf SM, Sunker A, Abdwani R, Abrawi
SA, Almurshedi F, Alhashmi N, Al Sonbul A, Sewairi W, Qari A,
Abdallah E, et al: Loss-of-function variant in DNASE1L3 causes a
familial form of systemic lupus erythematosus. Nat Genet.
43:1186–1188. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhao Q, Yang C, Wang J, Li Y and Yang P:
Serum level of DNase1l3 in patients with
dermatomyositis/polymyositis, systemic lupus erythematosus and
rheumatoid arthritis, and its association with disease activity.
Clin Exp Med. 17:459–465. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Serpas L, Chan RWY, Jiang P, Ni M, Sun K,
Rashidfarrokhi A, Soni C, Sisirak V, Lee WS, Cheng SH, et al:
Dnase1l3 deletion causes aberrations in length and end-motif
frequencies in plasma DNA. Proc Natl Acad Sci USA. 11:641–649.
2019. View Article : Google Scholar
|
|
45
|
Moser K, Kelly J, Lessard C and Harley JB:
Recent insights into the genetic basis of systemic lupus
erythematosus. Genes Immun. 10:373–379. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Goulielmos GN, Zervou MI, Vazgiourakis VM,
Ghodke-Puranik Y, Garyfallos A and Niewold TB: The genetics and
molecular pathogenesis of systemic lupus erythematosus (SLE) in
populations of different ancestry. Gene. 668:59–72. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Niewold TB: Interferon alpha-induced
lupus: Proof of principle. J Clin Rheumatol. 14:131–132. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ko K, Koldobskaya Y, Rosenzweig E and
Niewold TB: Activation of the interferon pathway is dependent upon
autoantibodies in African-American SLE patients, but not in
European-American SLE patients. Front Immunol. 4:3092013.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Rahman A and Isenberg DA: Systemic lupus
erythematosus. N Engl J Med. 358:929–939. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Márquez A, Kerick M, Zhernakova A,
Gutierrez-Achury J, Chen WM, Onengut-Gumuscu S, González-Álvaro I,
Rodriguez- Rodriguez L, Rios-Fernández R, González-Gay MA, et al:
Meta-analysis of Immunochip data of four autoimmune diseases
reveals novel single-disease and cross-phenotype associations.
Genome Med. 10:972018. View Article : Google Scholar : PubMed/NCBI
|