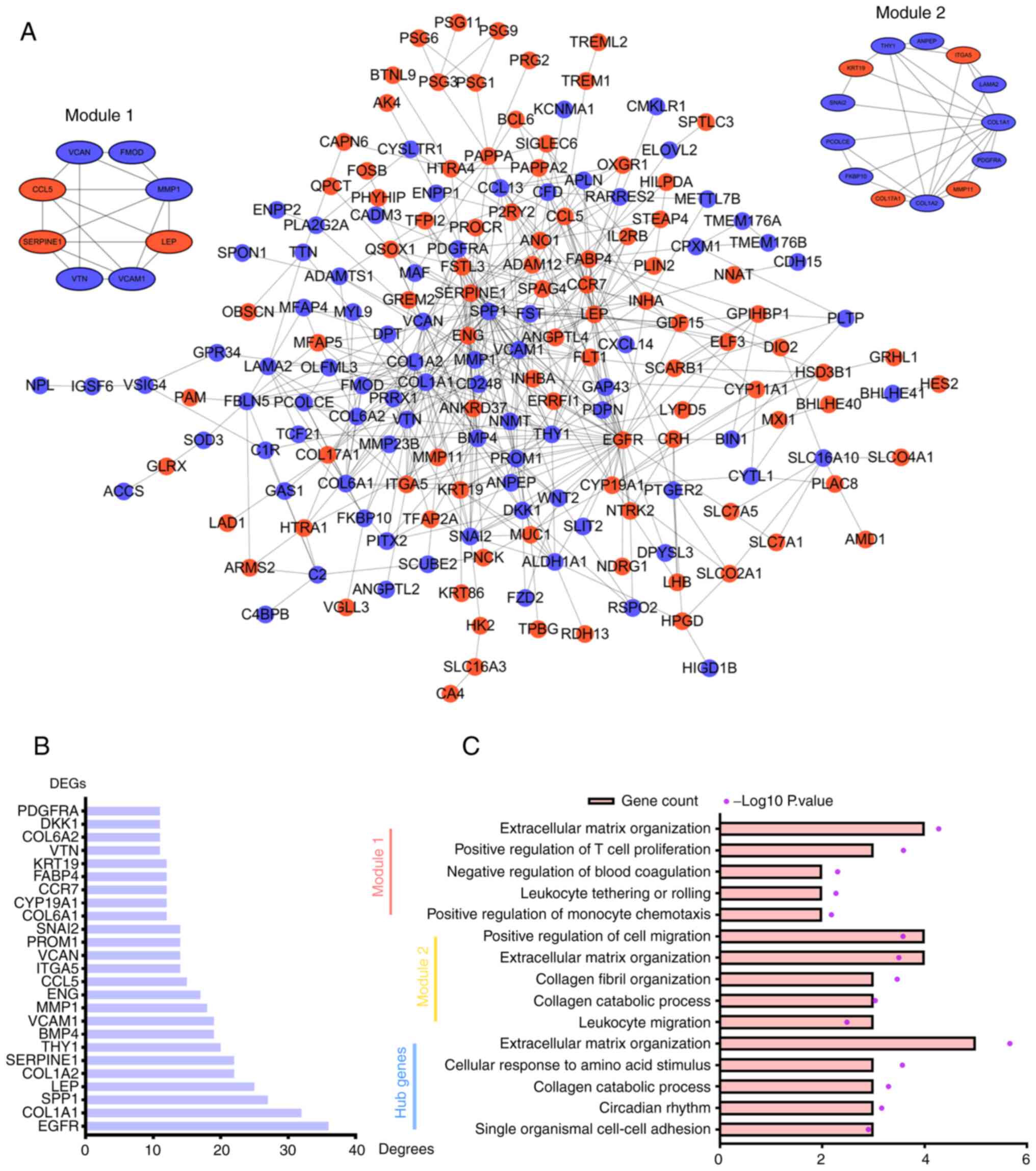
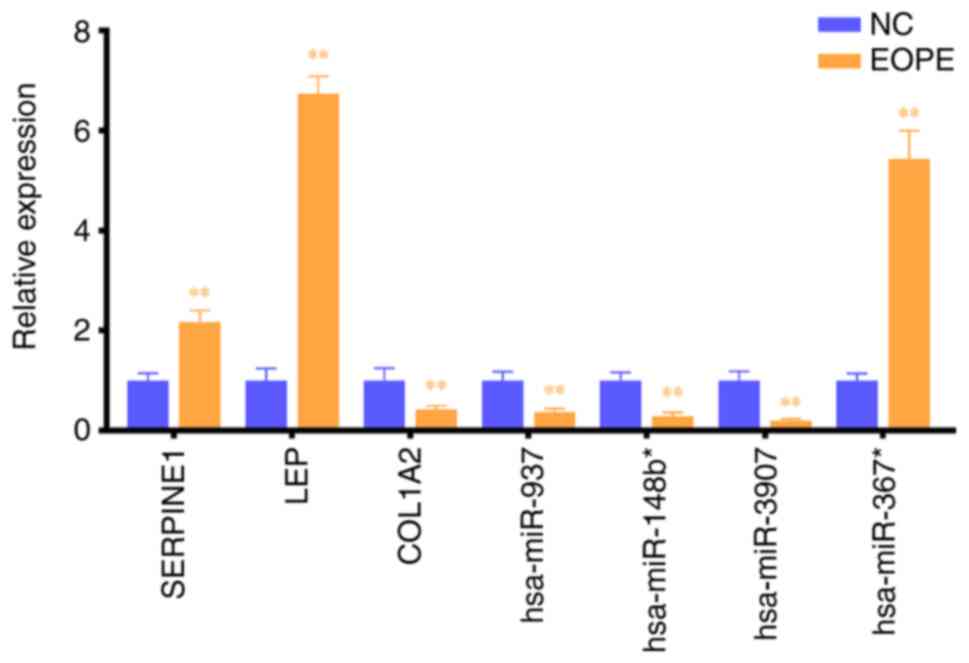
|
1
|
Steegers EA, von Dadelszen P, Duvekot JJ
and Pijnenborg R: Pre-eclampsia. Lancet. 376:631–644. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ghulmiyyah L and Sibai B: Maternal
mortality from preeclampsia/eclampsia. Semin Perinatol. 36:56–59.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Clark SL, Belfort MA, Dildy GA, Herbst MA,
Meyers JA and Hankins GD: Maternal death in the 21st century:
Causes, prevention, and relationship to cesarean delivery. Am J
Obstet Gynecol. 199:36.e1–e5, 91-2. e7-e11. 2008. View Article : Google Scholar
|
|
4
|
Roberts JM and Cooper DW: Pathogenesis and
genetics of pre-eclampsia. Lancet. 357:53–56. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
von Dadelszen P, Magee LA and Roberts JM:
Subclassification of preeclampsia. Hypertens Pregnancy. 22:143–148.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lisonkova S, Sabr Y, Mayer C, Young C,
Skoll A and Joseph KS: Maternal morbidity associated with
early-onset and late-onset preeclampsia. Obstet Gynecol.
124:771–781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brosens I, Pijnenborg R, Vercruysse L and
Romero R: The ‘Great Obstetrical Syndromes’ are associated with
disorders of deep placentation. Am J Obstet Gynecol. 204:193–201.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hata A: Functions of microRNAs in
cardiovascular biology and disease. Annu Rev Physiol. 75:69–93.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Jiao J, Ren P and Wu M:
Upregulation of miRNA-223-3p ameliorates RIP3-mediated necroptosis
and inflammatory responses via targeting RIP3 after spinal cord
injury. J Cell Biochem. Feb 28–2019.(Epub ahead of print).
|
|
11
|
Ranz JM and Machado CA: Uncovering
evolutionary patterns of gene expression using microarrays. Trends
Ecol Evol. 21:29–37. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang GH, Cao XY, Li YY, Zhou CC, Li L,
Wang K, Li H, Yu P, Jin Y and Gao L: Gene expression profile of the
hippocampus of rats subjected to traumatic brain injury. J Cell
Biochem. 120:15776–15789. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tan J, Hu L, Yang X, Zhang X, Wei C, Lu Q,
Chen Z and Li J: miRNA expression profiling uncovers a role of
miR-302b-3p in regulating skin fibroblasts senescence. J Cell
Biochem. 121:70–80. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Xie M, Yao Z, Niu Y, Bu Y and Gao
C: Three meta-analyses define a set of commonly overexpressed genes
from microarray datasets on astrocytomas. Mol Neurobiol.
47:325–336. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Del Carratore F, Jankevics A, Eisinga R,
Heskes T, Hong F and Breitling R: RankProd 2.0: A refactored
bioconductor package for detecting differentially expressed
features in molecular profiling datasets. Bioinformatics.
33:2774–2775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marot G, Foulley JL, Mayer CD and
Jaffrézic F: Moderated effect size and P-value combinations for
microarray meta-analyses. Bioinformatics. 25:2692–2699. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Heberle H, Meirelles GV, da Silva FR,
Telles GP and Minghim R: InteractiVenn: A web-based tool for the
analysis of sets through Venn diagrams. BMC Bioinformatics.
16:1692015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kolde R: Pheatmap: Pretty heatmaps: R
package version 1. 2012.
|
|
20
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
The Gene Ontology Consortium: The gene
ontology resource: 20 Years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43:D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wickham H: ggplot2: Elegant graphics for
data analysis. Springer International Publishing; 2016
|
|
30
|
Pajak M and Simpson TI: miRNAtap:
miRNAtap: microRNA Targets-Aggregated Predictions. R package
version 1.22.0. 2020.
|
|
31
|
Dhawan A, Scott JG, Harris AL and Buffa
FM: Pan-cancer characterisation of microRNA across cancer hallmarks
reveals microRNA-mediated downregulation of tumour suppressors. Nat
Commun. 9:52282018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Uzan J, Carbonnel M, Piconne O, Asmar R
and Ayoubi JM: Pre-eclampsia: Pathophysiology, diagnosis, and
management. Vasc Health Risk Manag. 7:467–474. 2011.PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Osungbade KO and Ige OK: Public health
perspectives of preeclampsia in developing countries: Implication
for health system strengthening. J Pregnancy. 2011:4810952011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen J and Khalil RA: Matrix
metalloproteinases in normal pregnancy and preeclampsia. Prog Mol
Biol Transl Sci. 148:87–165. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Karumanchi SA and Bdolah Y: Hypoxia and
sFlt-1 in preeclampsia: The ‘Chicken-and-Egg’ question.
Endocrinology. 145:4835–4837. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Janku F, Yap TA and Meric-Bernstam F:
Targeting the PI3K pathway in cancer: Are we making headway? Nat
Rev Clin Oncol. 15:273–291. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mayer IA and Arteaga CL: The PI3K/AKT
pathway as a target for cancer treatment. Annu Rev Med. 67:11–28.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cudmore MJ, Ahmad S, Sissaoui S, Ramma W,
Ma B, Fujisawa T, Al-Ani B, Wang K, Cai M, Crispi F, et al: Loss of
Akt activity increases circulating soluble endoglin release in
preeclampsia: Identification of inter-dependency between Akt-1 and
heme oxygenase-1. Eur Heart J. 33:1150–1158. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hastie R, Brownfoot FC, Pritchard N,
Hannan NJ, Cannon P, Nguyen V, Palmer K, Beard S, Tong S and
Kaitu'u-Lino TJ: EGFR (Epidermal Growth Factor Receptor) signaling
and the mitochondria regulate sFlt-1 (Soluble FMS-Like Tyrosine
Kinase-1) secretion. Hypertension. 73:659–670. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Goddard KA, Tromp G, Romero R, Olson JM,
Lu Q, Xu Z, Parimi N, Nien JK, Gomez R, Behnke E, et al:
Candidate-gene association study of mothers with pre-eclampsia, and
their infants, analyzing 775 SNPs in 190 genes. Hum Hered. 63:1–16.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gabinskaya T, Salafia CM, Gulle VE,
Holzman IR and Weintraub AS: Gestational age-dependent extravillous
cytotrophoblast osteopontin immunolocalization differentiates
between normal and preeclamptic pregnancies. Am J Reprod Immunol.
40:339–346. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xia J, Qiao F, Su F and Liu H: Implication
of expression of osteopontin and its receptor integrin alphanubeta3
in the placenta in the development of preeclampsia. J Huazhong Univ
Sci Technolog Med Sci. 29:755–760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Taylor BD, Ness RB, Olsen J, Hougaard DM,
Skogstrand K, Roberts JM and Haggerty CL: Serum leptin measured in
early pregnancy is higher in women with preeclampsia compared with
normotensive pregnant women. Hypertension. 65:594–599. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Salimi S, Farajian-Mashhadi F, Naghavi A,
Naghavi A, Mokhtari M, Shahrakipour M, Saravani M and Yaghmaei M:
Different profile of serum leptin between early onset and late
onset preeclampsia. Dis Markers. 2014:6284762014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Buurma AJ, Turner RJ, Driessen JH,
Mooyaart AL, Schoones JW, Bruijn JA, Bloemenkamp KW, Dekkers OM and
Baelde HJ: Genetic variants in pre-eclampsia: A meta-analysis. Hum
Reprod Update. 19:289–303. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Estrada-Gutierrez G, Cappello RE, Mishra
N, Romero R, Strauss JF III and Walsh SW: Increased expression of
matrix metalloproteinase-1 in systemic vessels of preeclamptic
women: A critical mediator of vascular dysfunction. Am J Pathol.
178:451–460. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang X and Meng T: MicroRNA-431 affects
trophoblast migration and invasion by targeting ZEB1 in
preeclampsia. Gene. 683:225–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lv Y, Lu X, Li C, Fan Y, Ji X, Long W,
Meng L, Wu L, Wang L, Lv M and Ding H: miR-145-5p promotes
trophoblast cell growth and invasion by targeting FLT1. Life Sci.
239:1170082019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Brkić J, Dunk C, O'Brien J, Fu G, Nadeem
L, Wang YL, Rosman D, Salem M, Shynlova O, Yougbaré I, et al:
MicroRNA-218-5p promotes endovascular trophoblast differentiation
and spiral artery remodeling. Mol Ther. 26:2189–2205. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hong F, Li Y and Xu Y: Decreased placental
miR-126 expression and vascular endothelial growth factor levels in
patients with pre-eclampsia. J Int Med Res. 42:1243–1251. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yan T, Liu Y, Cui K, Hu B, Wang F and Zou
L: MicroRNA-126 regulates EPCs function: Implications for a role of
miR-126 in preeclampsia. J Cell Biochem. 114:2148–2159. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
He P, Shao D, Ye M and Zhang G: Analysis
of gene expression identifies candidate markers and pathways in
pre-eclampsia. J Obstet Gynaecol. 35:578–584. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ma Y, Lin H, Zhang H, Song X and Yang H:
Identification of potential crucial genes associated with
early-onset pre-eclampsia via a microarray analysis. J Obstet
Gynaecol Res. 43:812–819. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Song J, Li Y and An RF: Identification of
early-onset preeclampsia-related genes and MicroRNAs by
bioinformatics approaches. Reprod Sci. 22:954–963. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gunel T, Hosseini MK, Gumusoglu E,
Kisakesen HI, Benian A and Aydinli K: Expression profiling of
maternal plasma and placenta microRNAs in preeclamptic pregnancies
by microarray technology. Placenta. 52:77–85. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Betoni JS, Derr K, Pahl MC, Rogers L,
Muller CL, Packard RE, Carey DJ, Kuivaniemi H and Tromp G: MicroRNA
analysis in placentas from patients with preeclampsia: Comparison
of new and published results. Hypertens Pregnancy. 32:321–339.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lykoudi A, Kolialexi A, Lambrou GI,
Braoudaki M, Siristatidis C, Papaioanou GK, Tzetis M, Mavrou A and
Papantoniou N: Dysregulated placental microRNAs in early and late
onset preeclampsia. Placenta. 61:24–32. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Yang-Dong O, Ya-Xian L and Xue-qiong Z:
Excavation and bioinformatics analysis of early-onset preeclampsia
related MicroRNA and target genes. J Nongken Med. 40:494–499.
2018.
|