Introduction
The incidence of papillary thyroid carcinoma (PTC)
is highest in patients with thyroid cancer, reaching 70–80%
(1). The main clinical
manifestation of PTC is a slowly growing thyroid mass with
multifocality and a propensity for regional lymph node metastasis
(2). Some PTCs are highly
aggressive; some tend to dedifferentiate and eventually develop
into poorly differentiated or undifferentiated carcinomas, leading
to a decreased survival rate and compromised quality of life
(2). Therefore, it is important to
further improve the cure rate and survival rate of patients with
PTC to study the growth, invasion and metastasis of PTC cells and
explore the molecular mechanism, on the basis of which it would be
possible to find biomarkers with differential expression and
metastatic prediction in PTC, as well as molecules that contribute
to therapy. An increasing number of studies have revealed that
aberrant expression of microRNAs (miRNAs) plays an important role
in regulating the growth and metastasis of PTC (3,4).
Therefore, by studying the biological function and mechanism of
miRNAs in PTC, new insights can be provided for the diagnosis and
treatment of PTC.
It is widely accepted that miR-122-5p has a tumour
suppressive function in a variety of carcinomas such as
hepatocellular carcinoma, bile duct carcinoma and gastric cancer
(5–7). However, the role of miR-122-5p in PTC
has not been clarified. Dual specificity phosphatase 4 (DUSP4) is a
member of the dual specificity phosphatase family and could
negatively regulate the activity of the MAP kinases (MAPK)
(8). DUSP4 has been widely
recognized as a biological marker of multiple malignant tumours.
Alterations in DUSP4 expression are involved in the oncogenesis of
a variety of tumours. DUSP4 acts as a tumour suppressor in most
cancers (8–10) but contributes to the occurrence and
development of a small portion of tumours, including PTC (7,11,12). A
previous study revealed that DUSP4 expression in PTC was
significantly higher than that in adjacent normal tissues (12). Moreover, the high expression of
DUSP4 was not only related to lymph node metastasis and
extrathyroidal extension of PTC but also an independent risk factor
for lymph node metastasis. In addition, DUSP4 expression was
associated with BRAF mutations in PTC cancer tissues and cell lines
(12). Therefore, DUSP4 is
considered a potential biomarker for the pathogenesis of PTC.
Through a bioinformatics study, it was revealed that the 3′-UTR of
DUSP4 mRNA has a specific binding sequence for miR-122-5p (7). Xu et al (7) revealed for the first time that
miR-122-5p inhibited the migration, invasion and metastasis of
gastric cancer (GC) cells by downregulating DUSP4. In summary, it
is hypothesized that miR-122-5p inhibits tumour development by
inhibiting DUSP4 in PTC cells.
The present study aimed to explore the role of
miR-122-5p in PTC oncogenesis. The expression pattern of miR-122-5p
in PTC cancer tissues and PTC cell lines was investigated, and the
roles of miR-122-5p in PTC were explored.
Materials and methods
Clinical tissue specimens
A total of 45 pairs of PTC tissues and adjacent
non-cancerous tissues were obtained with written informed consent
via surgical resection at the Second Hospital of Hebei Medical
University (Shujiazhuang, China) between January 2016 and February
2019. The age range of the patients was 25–77 years, and the median
age was 52 years. There were 16 male patients and 29 female
patients. The distance range between the tumour samples and
adjacent non-cancerous tissues was between 1.3 and 2.3 cm. All
tissue specimens were histopathologically examined by three
independent pathologists. Thus, all subjects were confirmed as PTC.
No chemotherapy or radiotherapy was performed for these patients
before the surgery. Fresh tissue specimens were frozen in liquid
nitrogen and stored at −80°C until use. All the clinical samples
were acquired with written informed consent from the participants.
The Institutional Review Board of the Second Hospital of Hebei
Medical University reviewed and approved the present study
(approval no. 2016-R269). The clinical studies were conducted
according to the principles expressed in the Declaration of
Helsinki.
Cell lines and culture
The human normal thyroid epithelial cell line
FRTL-5, thyroid cancer cell line BCPAP and PTC cell lines TPC-1,
and K1 were obtained from the American Type Culture Collection.
FRTL-5 was incubated in Keratinocyte Serum Free Medium (KSFM;
Thermo Fisher Scientific, Inc.). TPC-1, K1 and BCPAP were incubated
in RPMI-1640 medium (Thermo Fisher Scientific, Inc.) along with 10%
fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.). The whole
cells were maintained in humidified atmosphere at 37°C with 5%
CO2. BCPAP and K1 were authenticated by STR
profiling.
Reverse transcription-quantitative PCR
(RT-qPCR) assays
The total RNA from PTC tissues, adjacent
non-cancerous tissues and all cell lines was extracted and purified
using TRIzol reagent (Thermo Fisher Scientific, Inc.). Synthesis of
cDNA and RT-qPCR measurements were carried out as previously
described (13,14). The designed primer sequences for
RT-qPCR were as follows: miR-122-5p,
5′-TATTCGCACTGGATACGACACAAAC-3′ (sense) and
5′-GCCCGTGGAGTGTGACAATGGT-3′ (anti-sense); DUSP4,
5′-CTACATCCTAGGTTCGGTCAAC-3′ (sense) and 5′-TAGACGATGACCGCCGAGTA-3′
(anti-sense); GAPDH, 5′-CCTGCCTCTACTGGCGCTGC-3′ (sense) and
5′-GCAGTGGGGACACGGAAGGC-3′ (anti-sense). Relative expression was
calculated using the 2−ΔΔCq method (15).
GAPDH was used as the internal reference. RT-qPCR
was performed using SYBR Premix Ex Taq™ kit (Takara Bio, Inc.) and
ABI7500 PCR system (Applied Biosystems; Thermo Fisher Scientific,
Inc.).
Plasmid construction and
transfection
miR-122-5p full-length sequences were PCR amplified
using Phusion HSII Flash High-Fidelity PCR Master Mix (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
miR-122-5p mimic (5′-UGGAGUGUGACAAUGGUGUUUG-3′), miR-mimic control
(5′-GUGCACGAAGGCUCAUCAUU-3′), miR-122-5p inhibitor
(5′-AACACCAUUGUCACACUCCAUU-3′) and miR-inhibitor control
(5′-UUCUCCGAACGUGUCACGUTT-3′) were obtained from Shanghai
GenePharma Co., Ltd. miRNA mimics, miRNA inhibitors, miRNA mimics
control and miRNA-inhibitors control in serum-free Accell™ medium
(Waters Corporation) were transfected into K1 cells at a final
concentration of 50 nM using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols. Incubation was performed for 24 h at
37°C, and then transfected cells were transferred to RPMI-1640
medium. All tests were performed 72 h after transfection. The
transfection efficiency of plasmids was identified by qPCR.
Assessment of cell viability
To assess cell proliferation, Cell Counting Kit-8
(CCK-8) assays were performed. For CCK-8 assays, the K1 cells in
each group were plated into 96-well plates with 2,000 cells/well.
At the indicated time-points, 10 µl CCK-8 reagent (Dojindo
Molecular Technologies, Inc.) was added into the treated cells.
Following incubation for 2 h at 37°C, the optical density at 450 nm
(OD450) was measured using Varioskan Flash reader (Thermo Fisher
Scientific, Inc.).
Cell invasion and migration
assays
Cell invasion was assessed using a Transwell assay
and cell migration was measured using a scratch/wound healing
assay. For the Transwell assay, cells (1×105 cells/well)
suspended in serum-free DMEM supplemented with 1 mg/ml mitomycin C
(inhibitor of cell proliferation) were seeded onto the upper
chamber of the Transwell inserts (24-well insert, pore size 8 mm).
The filter membranes were pre-coated with Matrigel at 37°C for 30
min. DMEM containing 20% FBS was added to lower chamber. After 36 h
of incubation, the cells migrating into lower surface of the
inserts were fixed for 30 min, stained with 1% crystal violet at
37°C for 30 min, and photographed under an inverted light
microscope (Olympus Corporation; scale bar, 100 µm). Invasion was
measured by counting the number of stained cells.
For the scratch/wound healing assay, cells
(1×105 cells/well) were seeded in 12-well plates and
cultured until 75% confluence. Then, a wound was created by
manually scratching the cell monolayer with a 200-µl pipette tip.
The cultures were washed to remove floating cells, and then the
adherent cells were incubated in serum-free DMEM. Cell migration
into the wound was observed under an inverted light microscope
(Olympus Corporation; scale bar, 200 µm) at 0 and 24 h for each
group. The scratch area was calculated using ImageJ software
(v1.8.0; National Institutes of Health).
Western blotting
Whole-cell lysate protein from cells with indicated
interventions in 6-well plates were extracted using ice-cold RIPA
lysis buffer (Beyotime Institute of Biotechnology). The protein
content was assessed using the BCA method. The concentration range
of protein loaded per well was 5.2–6.7 µg/µl. A total of 40 µg
protein of each sample was separated via SDS-PAGE on 10% gels, and
subsequently separated proteins were transferred onto
polyvinylidene fluoride (PVDF) membranes. After being blocked with
5% non-fat milk for 1 h at room temperature, PVDF membranes were
incubated with the antibodies against DUSP4 (1:10,000; cat. no.
5149) and β-tubulin (1:10,000; cat. no. 2146) (both from Cell
Signaling Technology, Inc.) overnight at 4°C. Then, a
HRP-conjugated secondary antibody (1:10,000; cat. no. SA00001-1;
Wuhan Sanying Biotechnology) was used at room temperature for 1 h.
The bands were visualized using enhanced chemiluminescent substrate
reagent kit (Amersham; Cytiva) and chemiluminescence system
(Amersham Image 600; General Electric; Cytiva). The signal
densitometry was semi-quantified using ImageJ software (v1.8.0;
National Institutes of Health).
Immunohistochemistry (IHC)
The tumor tissues were fixed in 4% paraformaldehyde
buffered with phosphate-buffered saline for 24 h at room
temperature, embedded in paraffin and serially sectioned (4 µm).
Next, the tissue sections were blocked in 5% goat serum (Wuhan
Servicebio Technology Co., Ltd.) for 15 min at room temperature,
and incubated with primary DUSP4 antibody (1:100; cat. no. ab72593;
Abcam) at 4°C overnight. The sections were incubated with
HRP-conjugated anti-rabbit (1:200; cat. no. GB23303; Wuhan
Servicebio Technology Co., Ltd.) at 37°C for 20 min, and then
visualized using a PV-9000 DAB detection kit according to the
manufacturer's protocol. The sections were counterstained with
hematoxylin for 3 min at room temperature and observed under a
light microscope (Olympus Corporation; scale bar, 100 µm). DUSP4
staining was graded semi-quantitatively as previously described
(16). Staining intensity was
graded as 1 (no staining), 2 (weak staining), 3 (clear staining),
or 4 (strong staining). The IHC images regarding Grade 1–4 tumour
tissues are presented in Fig. 1.
The intensity and abundance (expressed as a fraction) were
multiplied to obtain the total immunoreactivity score.
Animal experiments
A total of 30 male athymic BALB/c nude mice (6 weeks
old; 19–22 g) were obtained from the Shanghai SLAC Laboratory
Animal Co., Ltd. miR-122-5p mimic-transfected and control K1 cells
were inoculated subcutaneously on the ventral side of the right rib
at the density of 2×106 cells per mouse (10 mice per
group). All mice were divided into two groups: Control group and
the miR-122-5p mimics group. After 30 days, tumour-bearing mice
were anesthetized with ether (6 ml; inhalation anesthesia), and
then sacrificed via cervical dislocation. The sacrifice of mice was
confirmed when the heart and breathing stopped. Subsequently, all
tumours were removed and weighed. Tumour volumes were calculated
using the following formula: Volume (cm3)=length (L; cm)
× width (W; cm)2/2. The largest tumour volume was ~1.5
cm3. Athymic nude mice assays were conducted in
accordance with the Institutional principles for the concern and
use of animals, and the corresponding protocols (including
euthanasia protocol) were approved by the Institutional Animal Care
and Use Committee of the Second hospital of Hebei Medical
University. The mice were housed in a common environment in which
the room temperature was ~20-30°C and the humidity ~60–80% and fed
a general laboratory diet.
Luciferase reporter assays
TargetScan (http://www.targetscan.org) was used to predict the
presence of complementary binding sites between miR-122-5p and the
sequence 3′-UTR of DUSP4. To establish the mutated DUSP4 reporter
vector, the site-specific mutagenesis system (Thermo Fisher
Scientific, Inc.) was used to mutate the complementary binding
site. The wild-type (WT) 3′-UTR luciferase reporter plasmid
(pMIR-DUSP4-WT) and mutant (MUT) reporter plasmid (pMIR-DUSP4-MUT)
were subsequently constructed. The 3′-UTRs (including WT and MUT)
of DUSP4 was chemically synthesized and cloned into pMIR expression
vectors including luciferase genes (pmiR-GLO Dual-Luciferase
reporter vector; Promega Corporation) to construct pMIR-DUSP4-WT or
pMIR-DUSP4-MUT plasmid as previously described (7). The sequences of miR-122-5p mimic and
miR-mimic control were described above (Shanghai GenePharma Co.,
Ltd.). miR-122-5p mimic or miR-mimic control, and pMIR-DUSP4-WT or
pMIR-DUSP4-MUT were co-transfected into K1 cells using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) for
48 h. After transfection, luciferase activity in cell lysates was
measured using a Dual-luciferase reporter system (Promega
Corporation) according to the manufacturer's protocol. Relative
luciferase intensity was normalized to Renilla luciferase
activity.
Statistical analysis
All data are presented as the mean ± SEM. The
experiments were repeated at least three times. All statistical
analyses were performed using the GraphPad Prism Software 6
(GraphPad Software, Inc.). For comparisons, Wilcoxon signed-rank
test, Wilcoxon rank sum test, Pearson, one-way ANOVA or paired
Student's t-test were performed as indicated. Pearson's correlation
analysis was performed for correlation analysis. Pearson's
chi-square test was performed to determine the association between
miR-122-5p expression and clinicopathological characteristics.
Bonferroni post hoc test was performed following one-way ANOVA.
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-122-5p is downregulated in PTC
specimens
miR-122-5p expression levels in 45 pairs of PTC
tissues and adjacent non-cancerous tissues were detected by
RT-qPCR. As revealed in Fig. 2A,
miR-122-5p was significantly downregulated in PTC tissue specimens
compared with adjacent non-cancerous tissues. The Pearson's
chi-square test revealed that high miR-122-5p expression was
negatively associated with higher CDFI classification (17), stronger invasion, larger PTC tumours
and advanced pathological T/N stage (Table I). Moreover, 21 PTC specimens with
metastasis had lower miR-122-5p expression than 24 non-metastatic
PTC specimens (Fig. 2B).
Accordingly, compared with 18 PTC specimens with diameters <3
cm, 27 PTC specimens with diameters >3 cm exhibited lower
expression of miR-122-5p (Fig. 2C).
In addition, miR-122-5p expression levels in the normal thyroid
epithelial cell line FRTL-5, thyroid cancer cell line BCPAP and PTC
cell lines TPC-1, K1 were detected by RT-qPCR. As presented in
Fig. 2D, miR-122-5p was also
significantly downregulated in PTC cell lines.
 | Table I.Association between miR-122-5p
expression levels and clinicopathological characteristics of
patients with papillary thyroid carcinoma. |
Table I.
Association between miR-122-5p
expression levels and clinicopathological characteristics of
patients with papillary thyroid carcinoma.
|
|
| miR-122-5p
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristics | Cases | Low | High | P-value |
|---|
| Total | 45 | 23 | 22 |
|
| Sex |
|
|
| 0.5589 |
| Male | 12 | 7 | 5 |
|
|
Female | 33 | 16 | 17 |
|
| Age, years |
|
|
| 0.3666 |
|
≥65 | 20 | 13 | 7 |
|
|
<65 | 25 | 12 | 13 |
|
| CDFI classification
grade |
|
|
| 0.0080a |
| I | 28 | 10 | 18 |
|
|
II–III | 17 | 13 | 4 |
|
| Degree of
invasion |
|
|
| 0.0018a |
|
Intrathyroid | 22 | 6 | 16 |
|
|
Extrathyroid | 23 | 17 | 6 |
|
| PTC diameter,
cm |
|
|
| 0.0001a |
|
≥3.0 | 27 | 21 | 6 |
|
|
<3.0 | 18 | 2 | 16 |
|
| Pathological stage
(T) |
|
|
| 0.0027a |
| T2 | 24 | 7 | 17 |
|
|
T3-T4 | 21 | 16 | 5 |
|
| Pathological stage
(N) |
|
|
| 0.0305a |
| N0 | 32 | 12 | 20 |
|
| N1 | 13 | 11 | 2 |
|
| Outcome |
|
|
| 0.2150 |
|
Persistent/recurrent | 2 | 2 | 0 |
|
|
Death | 1 | 1 | 0 |
|
|
Cured | 42 | 20 | 22 |
|
miR-122-5p inhibits the proliferation,
invasion and migration of PTC
To explore the roles of miR-122-5p in PTC,
miR-122-5p was overexpressed or silenced by transducing miR-122-5p
mimics or miR-122-5p inhibitors into K1 cells. The transfection
efficiency of plasmids was determined through the detection of mRNA
levels (Fig. 3A). CCK-8 assays
revealed that miR-122-5p overexpression reduced the proliferation
of K1 cells (Fig. 3B). In addition,
Transwell assays revealed that miR-122-5p overexpression inhibited
cell invasion, and scratch assays revealed that miR-122-5p
overexpression decreased the number of migratory cells (Fig. 3C-F). By contrast, miR-122-5p
knockdown generated the opposite results in all the aforementioned
parameters (Fig. 3B-F).
 | Figure 3.miR-122-5p promotes the proliferation,
invasion and migration of papillary thyroid carcinoma. (A)
miR-122-5p expression in K1 cells transfected with miR inhibitor
control, miR-122-5p inhibitors, miR mimics control and miR-122-5p
mimics. (B) Cell proliferation of treated K1 cells was evaluated
using Cell Counting Kit-8 assays. (C and D) The invasion of treated
K1 cells was evaluated by Transwell assays. Scale bar, 100 µm. (E)
The migration of treated K1 cells was assessed by scratch/wound
healing assays, and images were acquired under a microscope at 0
and 24 h. Scale bar, 200 µm. (F) Cell migration ratios (24 h/0 h)
are presented in the histogram. Results are presented as the mean ±
SEM from three independent experiments. ***P<0.001 by one-way
ANOVA. (G and H) miR-122-5p-overexpressing or control K1 cells was
inoculated into nude mice. Then, 30 days later, tumour-bearing mice
were sacrificed, and tumours were removed and weighed. The
statistical graph indicates the quantitative results of the tumour
weights (n=10). Results are presented as the mean ± SEM.
***P<0.001 by Student's t-test. miR, microRNA. |
To explore the significance in vivo of
miR-122-5p in PTC, miR-122-5p-overexpressing and control K1 cells
were injected subcutaneously into nude mice. The weight of the
xenograft tumours was measured 30 days after injection. As revealed
in Fig. 3G and H, miR-122-5p
overexpression significantly reduced the weights of PTC xenograft
tumours.
miR-122-5p suppresses DUSP4
expression
Next, it was investigated whether the negative
regulation of miR-122-5p on DUSP4 exists in vivo and in
vitro. DUSP4 expression levels in the same 45 pairs of PTC
tissues and adjacent non-cancerous tissues used in Fig. 2A were observed using IHC. As
revealed in Fig. 4A and B, DUSP4
protein levels were significantly upregulated in PTC tissue
specimens compared with adjacent non-cancerous tissues. The
correlation analyses between DUSP4 expression and miR-122-5p
expression in PTC tissues revealed that DUSP4 expression levels
were negatively correlated with that of miR-122-5p in PTC tissues
(r=−0.9028, P<0.0001; Fig. 4C).
Accordingly, the regulatory effects of miR-122-5p on DUSP4 in
vitro were further explored. The mRNA and protein expression
levels of DUSP4 in miR-122-5p-overexpressed and silenced K1 cells
were observed by RT-qPCR and western blotting. As revealed in
Fig. 4D-F, both the mRNA and
protein levels of DUSP4 were decreased in the
miR-122-5p-overexpressed cells and increased in the
miR-122-5p-silenced cells. Next, the significance of miR-122-5p in
DUSP4 luciferase activity was further clarified. The luciferase
reporter assay revealed that miR-122-5p overexpression reduced the
luciferase activity of K1 cells, which did not occur in the
pMIR-DUSP4-MUT plasmid-transfected cells that lacked the effective
binding region of DUSP4 (Fig.
4G).
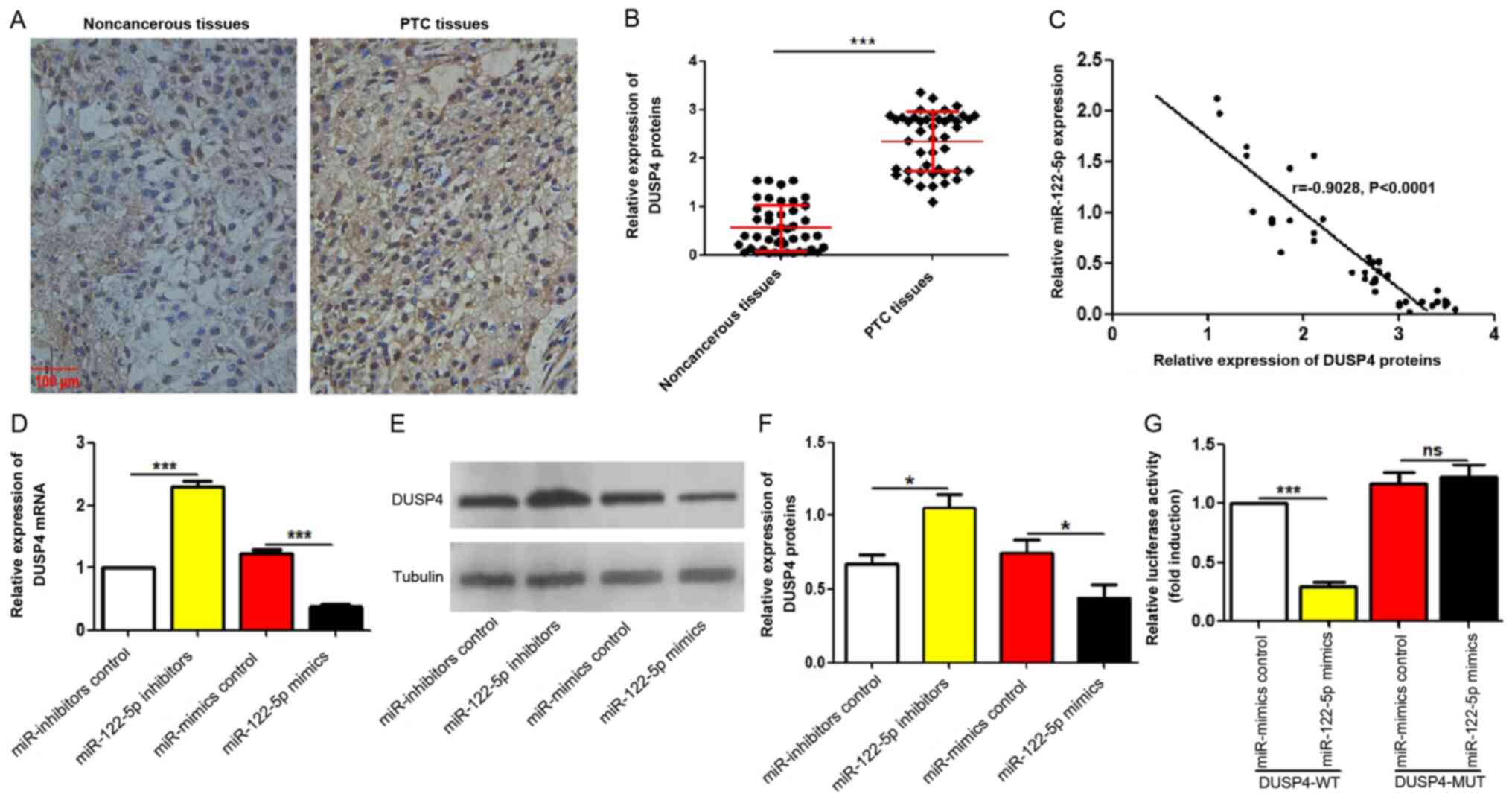 | Figure 4.miR-122-5p suppresses DUSP4
expression. (A and B) DUSP4 protein expression in 45 pairs of PTC
tissues and adjacent non-cancerous tissues were evaluated by
immunohistochemical assay. Scale bar, 100 µm. The quantitative
results are presented as the median with interquartile range.
***P<0.001 by Wilcoxon signed-rank test. (C) The correlation
between DUSP4 protein levels and miR-122-5p mRNA levels in PTC
tissues was analyzed. n=45, r=−0.9028, P<0.0001 by Pearson's
correlation analysis. (D-F) After transfection with corresponding
plasmids for 72 h, DUSP4 levels in treated K1 cells were evaluated
by reverse transcription-quantitative PCR or western blotting. The
results in F represent the protein expression of DUSP4. The
relative expression refers to the ratio of DUSP4 to tubulin.
*P<0.05 and ***P<0.001 by one-way ANOVA. (G) After
transfection with constructed pMIR-DUSP4-WT or pMIR-DUSP4-MUT
plasmid (including miR-122-5p mimics and miR mimics control) for 48
h, the luciferase activity of each group was analyzed by
dual-luciferase reporter assays. Results are presented as the mean
± SEM from three independent experiments. ***P<0.001 by one-way
ANOVA. miR, microRNA; DUSP4, dual specificity phosphatase 4; PTC,
papillary thyroid carcinoma; WT, wild-type; MUT, mutant; ns, no
significance. |
Discussion
The key role of miRNAs in tumorigenesis has
attracted great attention. Notably, an increasing number of miRNAs
have been confirmed to be implicated in PTC. Some miRNAs have
negative effects in PTC, for example, miR-222 can promote the
invasion and metastasis of PTC (18), and miR-146b-5p can enhance the
migration and invasion of PTC cells (19). By contrast, some miRNAs have
positive effects in PTC, for example, miR-599 has been found to
promote apoptosis and inhibit proliferation and the
epithelial-mesenchymal transition in PTC cells (3). In addition, miR-215 can suppress the
proliferation, migration and invasion of PTC cells (4). Similar reports also confirmed the
inhibitory effect of miR-let-7e, miR-200b, miR-188-5p and
miR-524-5p on PTC oncogenesis (20–23).
In the present study, miR-122-5p was focused on, which is widely
reported to suppress the growth and function of tumours (5–7) and
specifically bind to the 3′-UTR of PTC promoters (7). However, there is a lack of studies
regarding the role of miR-122-5p in PTC. As anticipated, miR-122-5p
was decreased in PTC tissues and cell lines compared with
non-cancerous tissues and a normal thyroid epithelial cell line,
respectively. Furthermore, with the growth and metastasis of PTC,
miR-122-5p expression in cancer tissues was further reduced. The
association analyses between miR-122-5p expression and
clinicopathological characteristics revealed that the reduced
expression of miR-122-5p indicated poor overall survival. Previous
studies have revealed several miRNAs that can predict the prognosis
of PTC, including miR-599 and miR-215 (3,4,24,25).
Thus, the aforementioned data suggested that miR-122-5p may be a
novel promising prognostic biomarker for PTC.
Functional assays revealed that overexpression of
miR-122-5p inhibited the proliferation, invasion and migration of
PTC in vitro. Conversely, knockdown of miR-122-5p promoted
the proliferation, invasion and migration of PTC in vitro.
Nude mouse xenograft experiments revealed that miR-122-5p
overexpression suppressed PTC growth in vivo. Several miRNAs
have been revealed to inhibit the carcinogenesis of PTC, such as
miR-let-7e and miR-200b (20–23).
These data indicated that miR-122-5p may have a potential
therapeutic effect on PTC.
A previous bioinformatics study revealed that the
3′-UTR of DUSP4 mRNA has a specific binding sequence for miR-122-5p
(7), deducing that DUSP4 may be a
downstream target of miR-122-5p. The expression of miR-122-5p was
negatively associated with DUSP4 expression in PTC tissues, which
confirmed the negative regulation of DUSP4 expression by
miR-122-5p. Furthermore, cytology experiments revealed that
miR-122-5p overexpression inhibited DUSP4 expression, while
miR-122-5p knockdown promoted DUSP4 expression. Notably, a
luciferase reporter assay demonstrated that miR-122-5p can
negatively regulate the luciferase activity of the DUSP4 reported
in PTC cells by binding to the 3′-UTR, which explains the
inhibitory effect of miR-122-5p on DUSP4 expression in PTC cells.
DUSP4 has been demonstrated to contribute to the occurrence and
development of PTC (12). The
aforementioned results indicated that the negative regulation of
DUSP4 at least partially accounts for the roles of miR-122-5p in
PTC. The working model of the study is presented in Fig. 5. However, the intrinsic mechanism
underlying DUSP4-regulated PTC carcinogenesis requires further
investigation.
In conclusion, the present study confirmed that
miR-122-5p is a tumour-suppressor miRNA in PTC and a promising
prognostic biomarker of PTC. Moreover, miR-122-5p mimics may be a
potential treatment strategy for PTC. Based on these results,
further improvement in the diagnosis and treatment of PTC may be
achieved in the future.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Natural
Science Foundation of Hebei Province (grant no. H2018206180).
Availability of data and materials
The data generated or analysed during this study are
included in this published article.
Authors' contributions
LZ and NH conceived and designed the experiments. NH
and YT performed the experiments, analyzed the data, and prepared
the figures. YS helped with the analysis of the data. LZ and NH
wrote the manuscript. LZ and NH confirm the authenticity of all the
raw data. All the authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study protocol was approved by the
Research Ethics Committee of the Second Hospital of Hebei Medical
University (Shujiazhuang, China) and written informed consent was
obtained from all the participants for their tissues to be used for
the purposes of this research. The clinical studies were conducted
according to the principles expressed in the Declaration of
Helsinki. Athymic nude mice assays were conducted in accordance
with the Institutional principles for the concern and use of
animals, and the corresponding protocols (including euthanasia
protocol) was approved by the Institutional Animal Care and Use
Committee of the Second Hospital of Hebei Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
PTC
|
papillary thyroid carcinoma
|
|
CD
|
cluster of differentiation
|
|
IL
|
interleukin
|
|
FBS
|
fetal bovine serum
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
CCK-8
|
Cell Counting Kit-8
|
|
IHC
|
immunohistochemistry
|
References
|
1
|
Yang Z, Li G, Ding C, Sun W and Zhang J:
Long non-coding RNA HULC exerts oncogenic activity on papillary
thyroid cancer in vitro and in vivo. Artif Cells Nanomed
Biotechnol. 48:326–335. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wiltshire JJ, Drake TM, Uttley L and
Balasubramanian SP: Systematic review of trends in the incidence
rates of thyroid cancer. Thyroid. 26:1541–1552. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang DP, Tang XZ, Liang QK, Zeng XJ, Yang
JB and Xu J: microRNA-599 promotes apoptosis and represses
proliferation and epithelial-mesenchymal transition of papillary
thyroid carcinoma cells via downregulation of Hey2-depentent Notch
signaling pathway. J Cell Physiol. 235:2492–2505. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han J, Zhang M, Nie C, Jia J, Wang F, Yu
J, Bi W, Liu B, Sheng R, He G, et al: miR-215 suppresses papillary
thyroid cancer proliferation, migration, and invasion through the
AKT/GSK-3β/Snail signaling by targeting ARFGEF1. Cell Death Dis.
10:1952019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ma J, Li T, Han X and Yuan H: Knockdown of
LncRNA ANRIL suppresses cell proliferation, metastasis, and
invasion via regulating miR-122-5p expression in hepatocellular
carcinoma. J Cancer Res Clin Oncol. 144:205–214. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xu Z, Liu G, Zhang M, Zhang Z, Jia Y, Peng
L, Zhu Y, Hu J, Huang R and Sun X: miR-122-5p inhibits the
proliferation, invasion and growth of bile duct carcinoma cells by
targeting ALDOA. Cell Physiol Biochem. 48:2596–2606. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu X, Gao F, Wang J, Tao L, Ye J, Ding L,
Ji W and Chen X: MiR-122-5p inhibits cell migration and invasion in
gastric cancer by down-regulating DUSP4. Cancer Biol Ther.
19:427–435. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Balko JM, Schwarz LJ, Bhola NE, Kurupi R,
Owens P, Miller TW, Gómez H, Cook RS and Arteaga CL: Activation of
MAPK pathways due to DUSP4 loss promotes cancer stem cell-like
phenotypes in basal-like breast cancer. Cancer Res. 73:6346–6358.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hijiya N, Tsukamoto Y, Nakada C, Tung
Nguyen L, Kai T, Matsuura K, Shibata K, Inomata M, Uchida T,
Tokunaga A, et al: Genomic loss of DUSP4 contributes to the
progression of intraepithelial neoplasm of pancreas to invasive
carcinoma. Cancer Res. 76:2612–2625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen M, Zhang J, Berger AH, Diolombi MS,
Ng C, Fung J, Bronson RT, Castillo-Martin M, Thin TH, Cordon-Cardo
C, et al: Compound haploinsufficiency of Dok2 and Dusp4 promotes
lung tumorigenesis. J Clin Invest. 129:215–222. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gröschl B, Bettstetter M, Giedl C,
Woenckhaus M, Edmonston T, Hofstädter F and Dietmaier W: Expression
of the MAP kinase phosphatase DUSP4 is associated with
microsatellite instability in colorectal cancer (CRC) and causes
increased cell proliferation. Int J Cancer. 132:1537–1546. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ma B, Shi R, Yang S, Zhou L, Qu N, Liao T,
Wang Y, Wang Y and Ji Q: DUSP4/MKP2 overexpression is associated
with BRAF(V600E) mutation and aggressive behavior of papillary
thyroid cancer. Onco Targets Ther. 9:2255–2263. 2016.PubMed/NCBI
|
|
13
|
Heinemann FG, Tolkach Y, Deng M, Schmidt
D, Perner S, Kristiansen G, Müller SC and Ellinger J: Serum
miR-122-5p and miR-206 expression: Non-invasive prognostic
biomarkers for renal cell carcinoma. Clin Epigenetics. 10:112018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheng G, Li Y, Liu Z and Song X: The
microRNA-429/DUSP4 axis regulates the sensitivity of colorectal
cancer cells to nintedanib. Mol Med Rep. 23:12012.
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Masson D, Rioux-Leclercq N, Fergelot P,
Jouan F, Mottier S, Théoleyre S, Bach-Ngohou K, Patard JJ and Denis
MG: Loss of expression of TIMP3 in clear cell renal cell carcinoma.
Eur J Cancer. 46:1430–1437. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kong QF, Lv B, Wang B, Zhang XP, Sun HJ
and Liu J: Association of von Willebrand factor (vWF) expression
with lymph node metastasis and hemodynamics in papillary thyroid
carcinoma. Eur Rev Med Pharmacol Sci. 24:2564–2571. 2020.PubMed/NCBI
|
|
18
|
Huang Y, Yu S, Cao S, Yin Y, Hong S, Guan
H, Li Y and Xiao H: MicroRNA-222 promotes invasion and metastasis
of papillary thyroid cancer through targeting protein phosphatase 2
regulatory subunit B alpha expression. Thyroid. 28:1162–1173. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lima CR, Geraldo MV, Fuziwara CS, Kimura
ET and Santos MF: MiRNA-146b-5p upregulates migration and invasion
of different papillary thyroid carcinoma cells. BMC Cancer.
16:1082016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ding C, Yu H, Shi C, Shi T, Qin H and Cui
Y: MiR-let-7e inhibits invasion and magration and regulates HMGB1
expression in papillary thyroid carcinoma. Biomed Pharmacother.
110:528–536. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou B, Xu J, Chen Y, Gao S, Feng X and Lu
X: miR-200b/c-RAP1B axis represses tumorigenesis and malignant
progression of papillary thyroid carcinoma through inhibiting the
NF-κB/Twist1 pathway. Exp Cell Res. 387:1117852020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou P, Irving A, Wu H, Luo J, Aguirre J,
Costa M, Khamsuree M, Gerads N and Liu W: Validation of
MicroRNA-188-5p inhibition power on tumor cell proliferation in
papillary thyroid carcinoma. Cell Transplant.
29:9636897209183002020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu H, Chen X, Lin T, Chen X, Yan J and
Jiang S: MicroRNA-524-5p suppresses the progression of papillary
thyroid carcinoma cells via targeting on FOXE1 and ITGA3 in cell
autophagy and cycling pathways. J Cell Physiol. 234:18382–18391.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Santiago K, Chen Wongworawat Y and Khan S:
Differential MicroRNA-signatures in thyroid cancer subtypes. J
Oncol. 2020:20523962020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jiang K, Li G, Chen W, Song L, Wei T, Li
Z, Gong R, Lei J, Shi H and Zhu J: Plasma exosomal miR-146b-5p and
miR-222-3p are potential biomarkers for lymph node metastasis in
papillary thyroid carcinomas. Onco Targets Ther. 13:1311–1319.
2020. View Article : Google Scholar : PubMed/NCBI
|



















