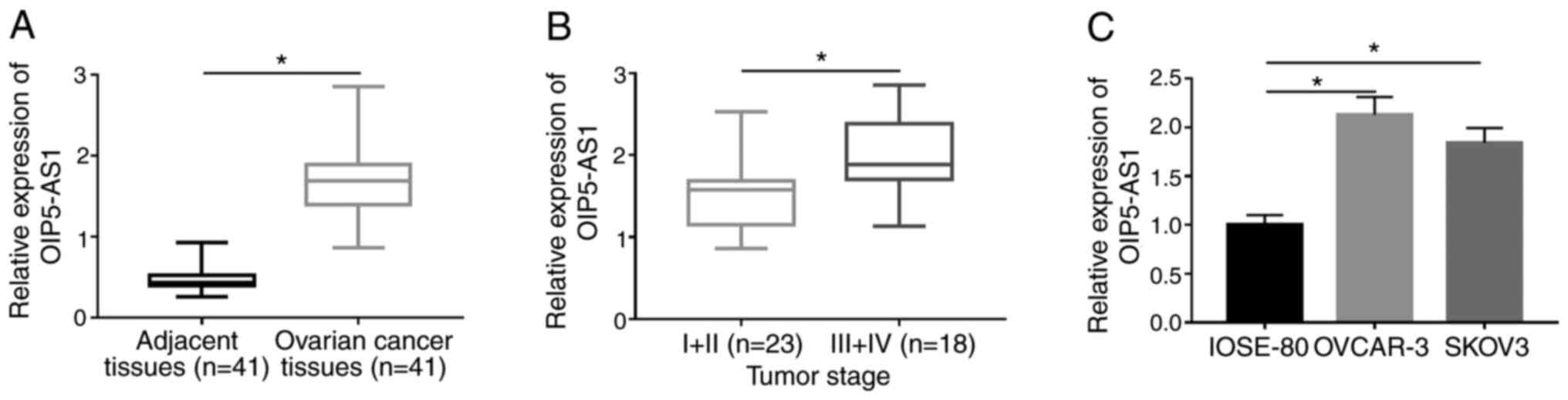
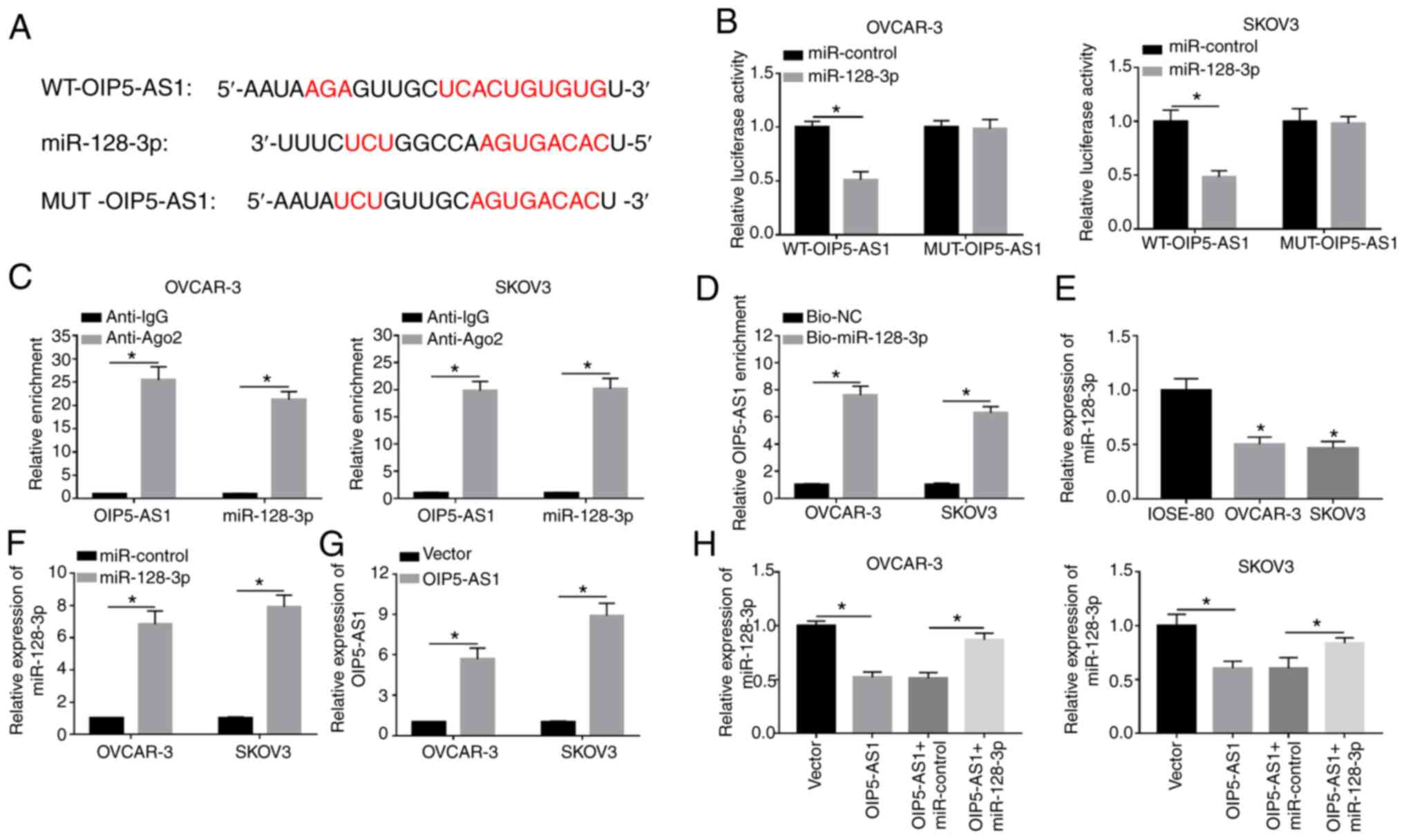
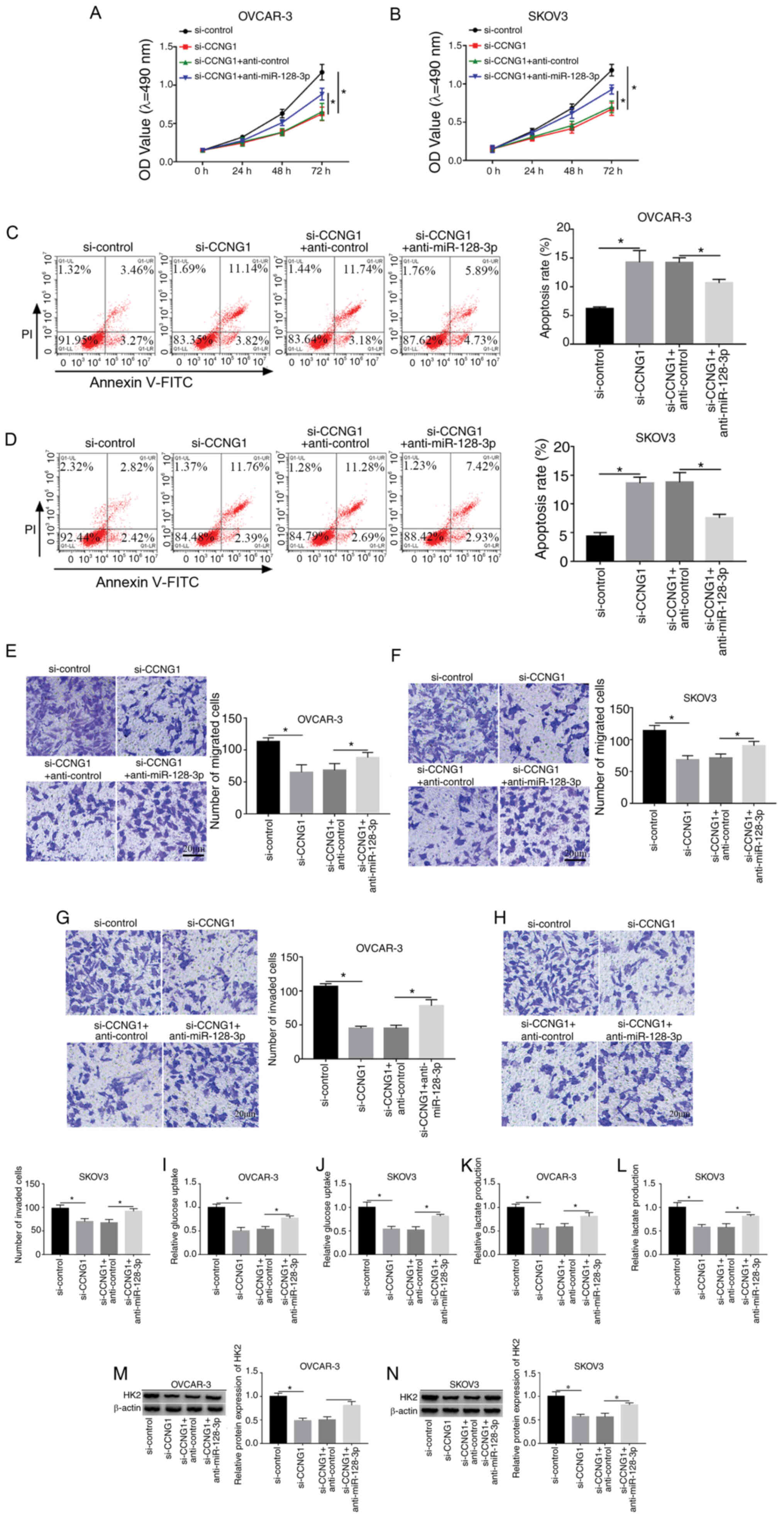
|
1
|
Reid BM, Permuth JB and Sellers TA:
Epidemiology of ovarian cancer: A review. Cancer Biol Med. 14:9–32.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et
al: MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu E, Liu Z and Zhou Y:
Carboplatin-docetaxel-induced activity against ovarian cancer is
dependent on up-regulated lncRNA PVT1. Int J Clin Exp Pathol.
8:3803–3810. 2015.PubMed/NCBI
|
|
4
|
Gao Y, Meng H, Liu S, Hu J, Zhang Y, Jiao
T, Liu Y, Ou J, Wang D, Yao L, et al: LncRNA-HOST2 regulates cell
biological behaviors in epithelial ovarian cancer through a
mechanism involving microRNA let-7b. Hum Mol Genet. 24:841–852.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chai Y, Liu J, Zhang Z and Liu L:
HuR-regulated lncRNA NEAT1 stability in tumorigenesis and
progression of ovarian cancer. Cancer Med. 5:1588–1598. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ganapathy-Kanniappan S: Molecular
intricacies of aerobic glycolysis in cancer: Current insights into
the classic metabolic phenotype. Crit Rev Biochem Mol Biol.
53:667–682. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vaupel P, Schmidberger H and Mayer A: The
warburg effect: Essential part of metabolic reprogramming and
central contributor to cancer progression. Int J Radiat Biol.
95:912–919. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang B, Zhang L, Cao Y, Chen S, Cao J, Wu
D, Chen J, Xiong H, Pan Z, Qiu F, et al: Overexpression of lncRNA
IGFBP4-1 reprograms energy metabolism to promote lung cancer
progression. Mol Cancer. 16:1542017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hu Y, Sun H, Hu J and Zhang X: LncRNA
DLX6-AS1 promotes the progression of neuroblastoma by activating
STAT2 via targeting miR-506-3p. Cancer Manag Res. 12:7451–7463.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li N and Zhan X and Zhan X: The lncRNA
SNHG3 regulates energy metabolism of ovarian cancer by an analysis
of mitochondrial proteomes. Gynecol Oncol. 150:343–354. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Peng WX, Koirala P and Mo YY:
LncRNA-mediated regulation of cell signaling in cancer. Oncogene.
36:5661–5667. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gutschner T and Diederichs S: The
hallmarks of cancer: A long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang M, Sun X, Yang Y and Jiao W: Long
non-coding RNA OIP5-AS1 promotes proliferation of lung cancer cells
and leads to poor prognosis by targeting miR-378a-3p. Thorac
Cancer. 9:939–949. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu X, Zheng J, Xue Y, Yu H, Gong W, Wang
P, Li Z and Liu Y: PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop
regulates the biological behavior of glioma cells. Theranostics.
8:1084–1105. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo L, Chen J, Liu D and Liu L:
OIP5-AS1/miR-137/ZNF217 axis promotes malignant behaviors in
epithelial ovarian cancer. Cancer Manag Res. 12:6707–6717. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu QY, Jiang XX, Tian HN, Guo HL, Guo H
and Guo Y: Long non-coding RNA OIP5-AS1 plays an oncogenic role in
ovarian cancer through targeting miR-324-3p/NFIB axis. Eur Rev Med
Pharmacol Sci. 24:7266–7275. 2020.PubMed/NCBI
|
|
18
|
Cho WC: OncomiRs: The discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen J, Zhao D and Meng Q: Knockdown of
HCP5 exerts tumor-suppressive functions by up-regulating tumor
suppressor miR-128-3p in anaplastic thyroid cancer. Biomed
Pharmacother. 116:1089662019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhao J, Li D and Fang L: MiR-128-3p
suppresses breast cancer cellular progression via targeting LIMK1.
Biomed Pharmacother. 115:1089472019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang C, Xie L, Liang H and Cui Y: LncRNA
MIAT facilitates osteosarcoma progression by regulating
mir-128-3p/VEGFC axis. IUBMB Life. 71:845–853. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu M, Zhou K, Wu Y, Wang L and Lu S:
Linc0a0161 regulated the drug resistance of ovarian cancer by
sponging microRNA-128 and modulating MAPK1. Mol Carcinog.
58:577–587. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li R, Gong L, Li P, Wang J and Bi L:
MicroRNA-128/homeobox B8 axis regulates ovarian cancer cell
progression. Basic Clin Pharmacol Toxicol. 125:499–507. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Baek WK, Kim D, Jung N, Yi YW, Kim JM, Cha
SD, Bae I and Cho CH: Increased expression of cyclin G1 in
leiomyoma compared with normal myometrium. Am J Obstet Gynecol.
188:634–639. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Perez R, Wu N, Klipfel AA and Beart RW Jr:
A better cell cycle target for gene therapy of colorectal cancer:
Cyclin G. Gastroenterology. 7:884–889. 2003.
|
|
27
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu X, Ma L, Rao Q, Mao Y, Xin Y, Xu H, Li
C and Wang X: MiR-1271 inhibits ovarian cancer growth by targeting
cyclin G1. Med Sci Monit. 21:3152–3158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yan J, Jiang JY, Meng XN, Xiu YL and Zong
ZH: MiR-23b targets cyclin G1 and suppresses ovarian cancer
tumorigenesis and progression. J Exp Clin Cancer Res. 35:312016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Albright JL: Dairy animal welfare: Current
and needed research. J Dairy Sci. 70:2711–2731. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Swearengen JR: Common challenges in
safety: A review and analysis of AAALAC findings. ILAR J.
59:127–133. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lou W, Ding B and Fu P: Pseudogene-derived
lncRNAs and their miRNA sponging mechanism in human cancer. Front
Cell Dev Biol. 8:852020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu DD, Chen X, Sun KX, Wang LL, Chen S and
Zhao Y: Role of the lncRNA ABHD11-AS1 in the tumorigenesis and
progression of epithelial ovarian cancer through targeted
regulation of RhoC. Mol Cancer. 16:1382017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhu FF, Zheng FY, Wang HO, Zheng JJ and
Zhang Q: Downregulation of lncRNA TUBA4B is associated with poor
prognosis for epithelial ovarian cancer. Pathol Oncol Res.
24:419–425. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Arunkumar G, Anand S, Raksha P,
Dhamodharan S, Prasanna Srinivasa Rao H, Subbiah S, Murugan AK and
Munirajan AK: LncRNA OIP5-AS1 is overexpressed in undifferentiated
oral tumors and integrated analysis identifies as a downstream
effector of stemness-associated transcription factors. Sci Rep.
8:70182018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hu GW, Wu L, Kuang W, Chen Y, Zhu XG, Guo
H and Lang HL: Knockdown of linc-OIP5 inhibits proliferation and
migration of glioma cells through down-regulation of YAP-NOTCH
signaling pathway. Gene. 610:24–31. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Naemura M, Kuroki M, Tsunoda T, Arikawa N,
Sawata Y, Shirasawa S and Kotake Y: The long noncoding RNA OIP5-AS1
is involved in the regulation of cell proliferation. Anticancer
Res. 38:77–81. 2018.PubMed/NCBI
|
|
39
|
Bai Y and Li S: Long noncoding RNA
OIP5-AS1 aggravates cell proliferation, migration in gastric cancer
by epigenetically silencing NLRP6 expression via binding EZH2. J
Cell Biochem. 121:353–362. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sen R, Ghosal S, Das S, Balti S and
Chakrabarti J: Competing endogenous RNA: The key to
posttranscriptional regulation. Sci World J. 2014:8962062014.
View Article : Google Scholar
|
|
41
|
Tan JY, Sirey T, Honti F, Graham B,
Piovesan A, Merkenschlager M, Webber C, Ponting CP and Marques AC:
Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in
mouse embryonic stem cells. Genome Res. 25:655–666. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao
WZ, Peng H, Hong WW, Yin LH, Pu YP and Liang GY: Integrated
analysis of long non-coding RNA-associated ceRNA network reveals
potential lncRNA biomarkers in human lung adenocarcinoma. Int J
Oncol. 49:2023–2036. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun WL, Kang T, Wang YY, Sun JP, Li C, Liu
HJ, Yang Y and Jiao BH: Long noncoding RNA OIP5-AS1 targets Wnt-7b
to affect glioma progression via modulation of miR-410. Biosci Rep.
39:BSR201803952019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Escoté X and Fajas L: Metabolic adaptation
to cancer growth: From the cell to the organism. Cancer Lett.
356:171–175. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Icard P, Fournel L, Wu Z, Alifano M and
Lincet H: Interconnection between metabolism and cell cycle in
cancer. Trends Biochem Sci. 44:490–501. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Horne MC, Goolsby GL, Donaldson KL, Tran
D, Neubauer M and Wahl AF: Cyclin G1 and cyclin G2 comprise a new
family of cyclins with contrasting tissue-specific and cell
cycle-regulated expression. J Biol Chem. 271:6050–6061. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jensen MR, Factor VM, Fantozzi A, Helin K,
Huh CG and Thorgeirsson SS: Reduced hepatic tumor incidence in
cyclin G1-deficient mice. Hepatology. 37:862–870. 2003. View Article : Google Scholar : PubMed/NCBI
|