Introduction
Skeletal muscle atrophy is caused by various chronic
diseases, including obesity, cancer, diabetes, heart failure, AIDS,
sepsis, and rheumatoid arthritis (1). Atrophy also occurs in aging,
starvation, and muscle denervation, as well as in genetic
myopathies, such as muscular dystrophies, leading to a decrease in
muscle strength and mass (2).
Muscle atrophy is caused by decreased protein synthesis and
increased proteolysis, due primarily to hyperactivation of major
cellular degradation pathways, including the ubiquitin-proteasome
and autophagy-lysosomal systems (3–5).
Proinflammatory cytokines, such as tumor necrosis factor-α (TNF-α),
interleukin (IL)-1β, and IL-6, have been shown to promote the
breakdown of myofibrillar proteins and reduce protein synthesis,
leading directly to muscle loss (3–5). Among
the cytokines, TNF-α induces ubiquitin-dependent proteolysis by
activating various intracellular factors, including reactive oxygen
species (ROS), nuclear factor-κB (NF-κB), atrogin-1/muscle atrophy
F-box (MAFbx), and muscle RING-finger protein-1 (MuRF1), as well as
apoptosis (3–5).
TNF-α, a proinflammatory cytokine, has been
implicated in muscle catabolic conditions and has an important role
in a variety of biological processes, including cell growth and
differentiation (6), inflammation
(7), apoptosis, and necrosis
(8). Among its various effects on
skeletal muscle protein, TNF-α induces anabolic and catabolic
effects regulated by different signaling pathways. TNF-α acts by
binding to TNF receptor-1 (TNF-R1) and TNF-R2 and acts as a potent
activator of the NF-κB pathway by promoting endogenous production
of ROS via mitochondrial electron transport in various cell types,
including muscle (9).
Phosphorylation and proteasomal degradation of the NF-κB inhibitory
protein, inhibitor of κB (IκB), lead to the activation and nuclear
translocation of NF-κB (10).
Activated NF-κB causes skeletal muscle atrophy through three
potential mechanisms. First, NF-κB can increase the protein
expression of components of the ubiquitin-proteasome system, which
is involved in the degradation of specific muscle proteins during
muscle atrophy (11,12). Second, NF-κB can increase the
expression of inflammation-related molecules, such as IL-1β and
IL-6, which directly or indirectly activate muscle wasting
(13). Finally, NF-κB can inhibit
the expression of muscle differentiation-related genes, such as
myoblast determination protein 1 (MyoD) and myogenin, which are
involved in the regeneration of atrophied skeletal muscle (14,15).
MyoD and myogenin are myogenic transcription factors that have an
important role in regulating muscle differentiation, and activation
of NF-κB is known to reduce the cellular MyoD and myogenin protein
levels through post-transcriptional mechanisms (16). In summary, NF-κB appears to increase
the ubiquitin-proteasome pathway activity, increase IL-6
expression, and inhibit myogenesis, promoting muscle protein
breakdown, and leading to muscle atrophy.
Panduratin A has been shown to increase the
TNF-α-induced decrease in MyoD and myogenin mRNA expression and
attenuate the TNF-α-induced increase in atrogin-1/MAFbx and MAFbx
mRNA expression, and significantly inhibit the production of ROS by
increasing the mRNA expression of catalase and superoxide dismutase
in L6 myotubes (5). Furthermore,
S-allyl cysteine inhibits TNF-α-induced muscle atrophy by
suppressing endogenous pro-inflammatory molecules TNF-α, IL-6,
TNF-like weak inducer of apoptosis (TWEAK), TWEAK receptor
fibroblast growth factor-inducible receptor 14, IL-1β, NF-κB, and
E3 ubiquitin ligases, and inhibiting the degradation of
muscle-specific structural protein, fast myosin heavy chain (MHCf)
(7). Therefore, regulating both
TNF-α-induced oxidative stress and the NF-κB pathway is an
important strategy for improving muscle disorders, such as muscle
atrophy.
Pyropia yezoensis is a commercially important
edible red alga in Southeast Asia, including Korea, China, Japan,
and Taiwan (17). P.
yezoensis contains biologically active phytochemicals, such as
proteins, polysaccharides, essential fatty acids, vitamins,
minerals, tocopherols, carotenoids, and phytocyanins (17). In particular, its high protein
content (25-50%, dry matter basis) serves as a source of bioactive
peptides with antimuscular atrophy (18), antioxidant (19), anticancer (20), anti-inflammatory (21), collagen synthesis (22), and hepatoprotective effects
(23). A previous study on the
muscle atrophy protection mechanism of P. yezoensis protein
(PYCP) showed that PYCP prevented dexamethasone-induced muscle
atrophy in mice by increasing protein synthesis and decreasing
protein degradation via inhibiting the ubiquitin-proteasome and
autophagy-lysosomal pathways (18).
However, the efficacy of PYCP in the prevention of muscle atrophy
due to chronic low-grade inflammation has not yet been
demonstrated. Therefore, the present study investigated the
protective mechanisms and therapeutic potential of PYCP on
TNF-α-induced myotube atrophy in C2C12 myotubes.
Materials and methods
Preparation of PYCP
P. yezoensis was purchased from Suhyup, the
National Federation of Fisheries Cooperatives (Busan, Korea),
washed several times with tap water to remove salt and visible
epiphytes and stored at −50°C until use. P. yezoensis powder
(8 g) was diluted with 300 ml Tris-HCl buffer (50 mM, pH 7.8) and
stirred at 4°C for 1 h. The solution was centrifuged at 3,134 × g
at 4°C for 20 min and vacuum-filtered through a crucible. The
supernatant was added to 80%
(NH4)2SO4 and stirred at 4°C for
24 h, then centrifuged at 173,502 × g at 4°C for 30 min. After
dissolving the precipitate in Tris-HCl buffer, the salt was removed
by dialysis at 4°C for 72 h using a Spectra/Por membrane (Spectrum
Laboratories, Ltd.) with a molecular weight cutoff of 3.5 kDa. The
dialyzed solution was distributed into 1.5-ml tubes and
freeze-dried. The lyophilized powder (termed PYCP) was stored at
−70°C until use. PYCP was solubilized in phosphate-buffered saline
(PBS) for use in assays.
C2C12 cell culture and
differentiation
Myoblasts derived from mouse skeletal muscle (cat.
no. CRL-1772; American Type Culture Collection) were cultured in
6-well plates containing Dulbecco's modified Eagle's medium (DMEM;
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml
penicillin, and 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a humidified atmosphere of 5%
CO2. When myoblasts were approximately 80–90% confluent,
myotube differentiation was initiated by replacing the growth
medium with differentiation medium: DMEM supplemented with 2% FBS.
Differentiation was allowed to proceed for 6 days, with the medium
changed every 48 h, at which stage 90–100% of the cells had fused
into myotubes.
Treatment of cells with recombinant
TNF-α and PYCP
TNF-α (PeproTech, Inc.) was dissolved in PBS to a
final concentration of 10 µg/ml for the stock solution and then
diluted to the indicated concentrations. Differentiated myotubes
were treated with 20 ng/ml TNF-α, various concentrations of PYCP
(25, 50 or 100 µg/ml), or both factors in combination for 48 h. For
the control group, the myotubes were incubated with differentiation
medium and PBS only.
Cell viability assay
Cell viability was determined using the CellTiter 96
Aqueous Cell Proliferation Assay (Promega Corporation), which is
based on the formation of a formazan product from the tetrazolium
compound (3-4,5-dimethythiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-
(4-sulfonyl)-2H-tetrazolium (MTS). Cells (1.5×104
cells/well) were seeded into 96-well plates in 100 µl DMEM
supplemented with 10% FBS and were allowed to attach at 37°C for 24
h. Following differentiation as aforementioned, the cells were
incubated with 20 ng/ml TNF-α and PYCP (25, 50, or 100 µg/ml) at
37°C for 48 h. MTS solution (10 µl) was added, and the cells were
incubated at 37°C for 30 min. The absorbance at 490 nm was measured
using a Gen5 microplate reader (BioTek Instruments, Inc.).
Experiments were performed in triplicate.
Measurement of myotube diameter
Myotube cultures were photographed under a
phase-contrast microscope at ×20 magnification following treatment
with 20 ng/ml TNF-α and 25, 50, or 100 µg/ml PYCP for 48 h. Myotube
diameters were determined based on the method described by
Castillero et al and Menconi et al (24,25).
The average diameter of each myotube was calculated as the mean of
five different myotube measurements in 10 different fields (n=50)
using ImageJ software (version 4.16; National Institutes of
Health). The measurements were conducted by a researcher blinded to
the origin of the experimental groups.
Measurement of intracellular ROS
production
Intracellular ROS were detected using the
redox-sensitive fluorescent dye 2′-7′-dichlorofluorescein diacetate
[DCF-DA (C24H14C12O7);
Sigma-Aldrich; Merck KGaA). Cells were plated at 1.5×104
cells/well in 96-well plates overnight and differentiated into
myotubes for 6 days. Following differentiation, the cells were
incubated with 20 ng/ml TNF-α and PYCP (25, 50, or 100 µg/ml) at
37°C for 48 h. At the end of the treatment, cells were washed with
cold PBS and incubated with 20 µM DCF-DA at 37°C for 30 min.
Following incubation, the cells were washed with cold PBS.
Fluorescence intensities of the stained cells were determined at
excitation and emission wavelengths of 485 and 535 nm,
respectively, using a Gen5 microplate fluorescence reader (BioTek
Instruments, Inc.).
Measurement of IL-6 production
IL-6 was measured in the supernatant using an ELISA
kit (cat. no. M6000B; R&D Systems, Inc.), according to the
manufacturer's instructions. Briefly, a monoclonal antibody
specific for mouse IL-6 was coated onto the microplates. After
adding 50 µl of standard or sample, 50 µl of assay diluent (RD1-14)
was added to the center of each well. Wells were incubated at room
temperature for 2 h, then washed five times. Afterward, 100 µl of
mouse IL-6 conjugate was added to each well, and the plate was
incubated for another 2 h, followed by repeated washing. Lastly,
wells were incubated in 100 µl of substrate solution for 30 min and
stopped with stop solution. The optical density of each well was
determined at 450 nm and corrected at 570 nm.
Measurement of 20S proteasome
activity
The chymotrypsin-like activity of the 20S proteasome
was measured based on changes in the fluorescence of
7-amino-4-methylcoumarin (AMC) conjugated to the chymotrypsin
peptide substrate LLVY using a 20S proteasome activity kit
(Chemicon; Thermo Fisher Scientific, Inc.). In brief, cells were
suspended in RIPA lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM
NaCl, 0.5% sodium deoxycholate, 1% Triton X-100, 0.1% SDS, and 2 mM
EDTA) containing protease inhibitors (1 mg/ml aprotinin, 1 mg/ml
leupeptin, 1 mg/ml pepstatin A, 200 mM
Na3VO4, 500 mM NaF, and 100 mM PMSF) and
centrifuged at 16,000 × g at 4°C for 10 min. The protein
concentration of supernatants was determined with a bicinchoninic
acid (BCA) protein assay (Pierce; Thermo Fisher Scientific, Inc.).
The cell lysates were incubated with a labeled substrate, LLVY-AMC,
at 37°C for 90 min. The cleavage activity was monitored by
detecting the free fluorophore AMC using a Gen5 microplate reader
(BioTek Instruments, Inc.) (18).
Preparation of total cell lysate
After C2C12 myoblasts were differentiated for 6
days, myotubes were treated with 20 ng/ml TNF-α and PYCP (25, 50,
or 100 µg/ml) at 37°C for 48 h. Cells were washed twice with PBS
(Gibco; Thermo Fisher Scientific, Inc.) and lysed with extraction
buffer [1% NP-40, 0.25% sodium deoxycholate, 1 mM ethylene
glycol-bis(β-aminoethyl ether)-N,N,N′N′-tetra-acetic acid, 150 mM
NaCl, and 50 mM Tris-HCl, pH 7.5] containing protease inhibitors (1
mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin A, 200 mM
Na3VO4, 500 mM NaF, and 100 mM PMSF) on ice.
After incubation at 4°C for 30 min, the extracts were centrifuged
at 16,000 × g at 4°C for 10 min. The protein levels in the
supernatants were quantified using a BCA protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. The supernatant was then used in western blot
analysis (18).
Preparation of cytosolic and nuclear
extracts
Cells were treated and harvested as described for
the total cell lysate extraction. Cells were then lysed with
hypotonic lysis buffer (25 mM HEPES pH 7.5, 5 mM EDTA, 5 mM
MgCl2, and 5 mM DTT) containing protease inhibitors (1
mg/ml aprotinin, 1 mg/ml leupeptin, 1 mg/ml pepstatin A, 200 mM
Na3VO4, 500 mM NaF, and 100 mM PMSF), and
incubated on ice for 10 min. Cells were further lysed by adding
ice-cold NP-40 to a final concentration of 2.5%. The tubes were
vortexed on the highest setting for 5 sec. After incubation on ice
for another 10 min, the tubes were vortexed again on the highest
setting for 5 sec, then centrifuged at 16,000 × g at 4°C for 5 min.
The supernatant (cytoplasmic extract) was immediately transferred
to clean, pre-chilled tubes. The insoluble (pellet) fraction
containing nuclei was resuspended in ice-cold cell extraction
buffer (10 mM HEPES pH 7.9, 100 mM NaCl, 1.5 mM MgCl2,
0.1 mM EDTA, and 0.2 mM DTT) containing protease inhibitor. The
samples were placed on ice and vortexed for 15 sec every 10 min for
a total of 40 min. The tubes were centrifuged at 16,000 × g at 4°C
for 10 min. The supernatant (nuclear extract) was immediately
transferred to clean, pre-chilled tubes. Protein levels were
determined using a BCA protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
Both extracts were then used in western blot analysis (18).
Western blot analysis
Equal amounts of protein (30 µg) were separated
using 7–12.5% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and transferred to polyvinylidene fluoride
membranes (EMD Millipore). The membranes were blocked by incubation
with 1% bovine serum albumin (BSA) in TBS-T (10 mM Tris-HCl, 150 mM
NaCl, and 0.1% Tween-20) at room temperature for 90 min, and then
incubated with specific primary antibodies at room temperature for
3 h (Table I). The membranes were
washed three times with TBS-T and incubated for 90 min at room
temperature with the appropriate horseradish peroxidase-conjugated
secondary anti-rabbit IgG (cat. no. 7074S; Cell Signaling
Technology, Inc.), anti-mouse IgG (cat. no. 7076S; Cell Signaling
Technology, Inc.), or donkey anti-goat IgG antibodies (cat. no.
A50-101P; Bethyl Laboratories, Inc.), diluted at 1:10,000-1:20,000
in TBS-T containing 1% BSA. Signals were detected using an enhanced
chemiluminescence western blot analysis kit (Thermo Fisher
Scientific, Inc.). Experiments were performed in triplicate, and
densitometry analysis was performed using Multi-Gauge software
version 3.0 (Fujifilm Life Sciences). GAPDH, β-actin, or lamin B1
were used as internal controls for equal protein loading in total
cell, cytoplasmic or nuclear extracts, respectively (18).
 | Table I.List of primary antibodies used in
western blot analysis. |
Table I.
List of primary antibodies used in
western blot analysis.
| Antibody | Catalog number | Species of
origin | Dilution |
|---|
|
Atrogin-1/MAFbx | sc-27645 | Goat | 1:1,000 |
| β-actin | sc-47778 | Mouse | 1:1,000 |
| GAPDH | sc-25778 | Mouse | 1:1,000 |
| IκBα | sc-7218 | Rabbit | 1:1,000 |
| Lamin B1 | sc-377000 | Mouse | 1:1,000 |
| MuRF1 | sc-27642 | Goat | 1:1,000 |
| MyoD | sc-377186 | Mouse | 1:1,000 |
| Myogenin | sc-12732 | Mouse | 1:1,000 |
| NF-κB p65 | sc-8008 | Mouse | 1:500 |
| p-IκBα | sc-8404 | Mouse | 1:1,000 |
| TNF-R1 | sc-8436 | Mouse | 1:1,000 |
Statistical analysis
The results are presented as the mean ± standard
deviation of at least three independent experiments. Data were
analyzed by one-way analysis of variance followed by the Duncan
multiple-range test using SPSS software version 18.0 (SPSS, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Effects of PYCP on cell viability
To examine the cytotoxic effects of PYCP on mouse
skeletal muscle cells, cell viability was measured using the MTS
assay. C2C12 myotubes were incubated without or with PYCP at
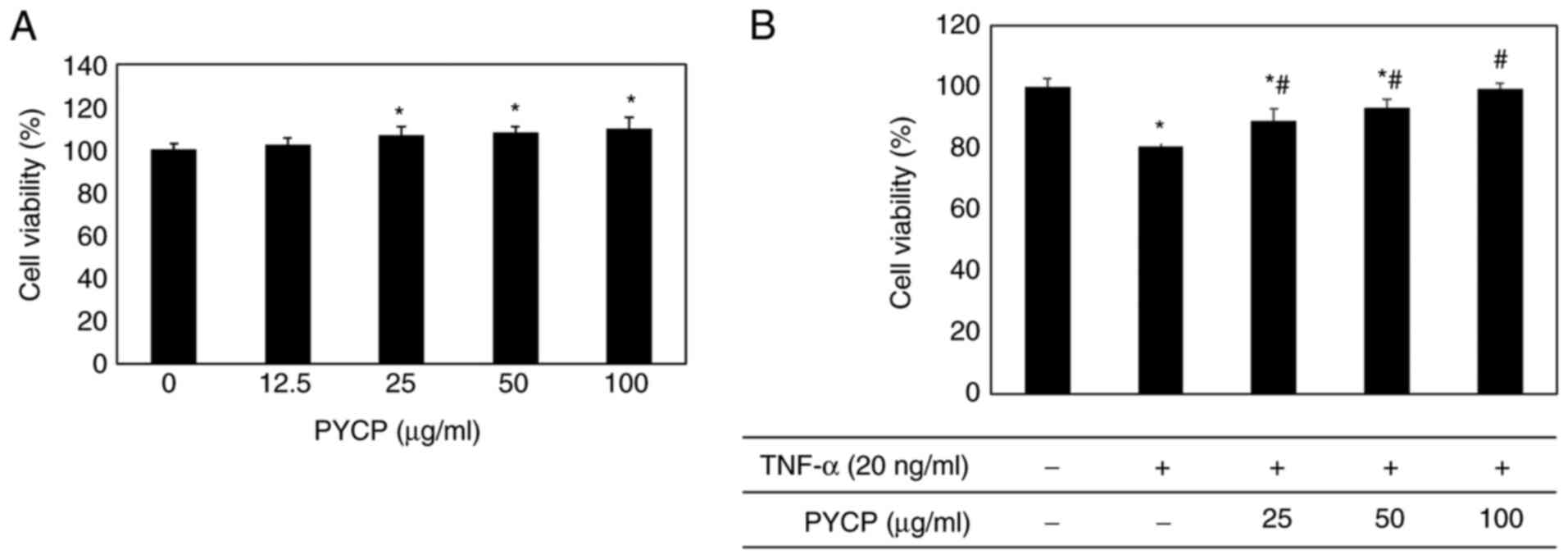
various concentrations (25-100 µg/ml). As shown in Fig. 1A, no significant toxicity was
observed in myotubes treated with PYCP for 48 h. To examine the
protective effects of PYCP against TNF-α-induced myotube damage,
mouse skeletal muscle C2C12 cells were incubated with various
concentrations of PYCP (25, 50, or 100 µg/ml). As shown in Fig. 1B, the cell viability of
TNF-α-treated myotubes was decreased compared with non-treated
myotubes. However, treatment with PYCP protected cells from the
TNF-α-induced decrease in cell viability (Fig. 1B). Therefore, 25, 50 and 100 µg/ml
PYCP were used for further experiments.
Effects of PYCP on myotube
diameter
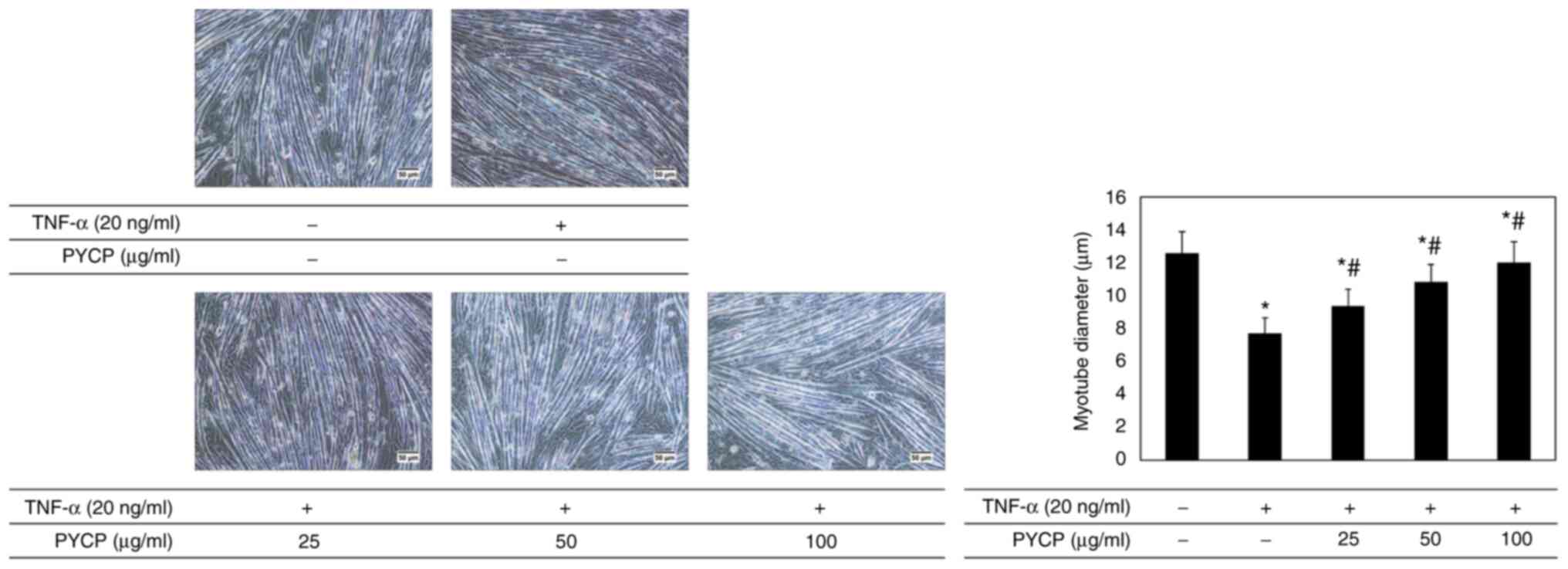
To investigate whether PYCP protects against myotube
atrophy in TNF-α-stimulated C2C12 myotubes, the myotube diameter
was measured. As shown in Fig. 2,
the myotube diameter in TNF-α-treated myotubes was decreased
compared with non-treated myotubes. However, TNF-α-treated myotubes
showed a significant increase in myotube diameter in the presence
of PYCP (Fig. 2).
Effects of PYCP on TNF-α-induced
intracellular ROS production
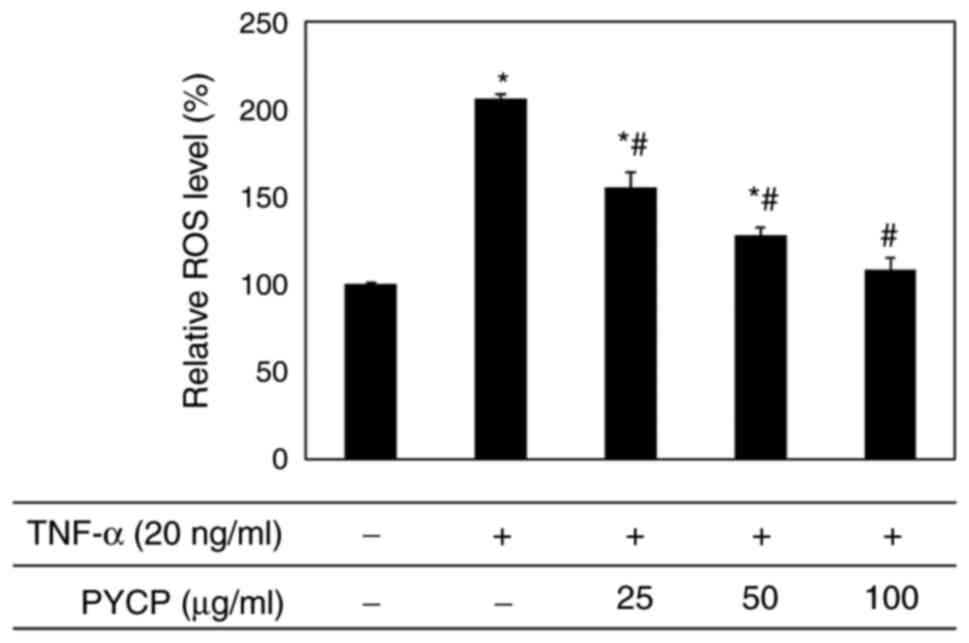
To examine whether PYCP functions as a scavenger of
TNF-α-induced ROS production, intracellular ROS levels were
measured. As shown in Fig. 3, the
production of ROS in TNF-α-treated myotubes was significantly
increased compared with non-treated myotubes. However,
TNF-α-treated myotubes exhibited a significant decrease in ROS
production in the presence of PYCP in a dose-dependent manner
(Fig. 3). These results indicated
that PYCP inhibited the intracellular accumulation of ROS in mouse
skeletal muscle cells damaged by TNF-α-mediated oxidative
stress.
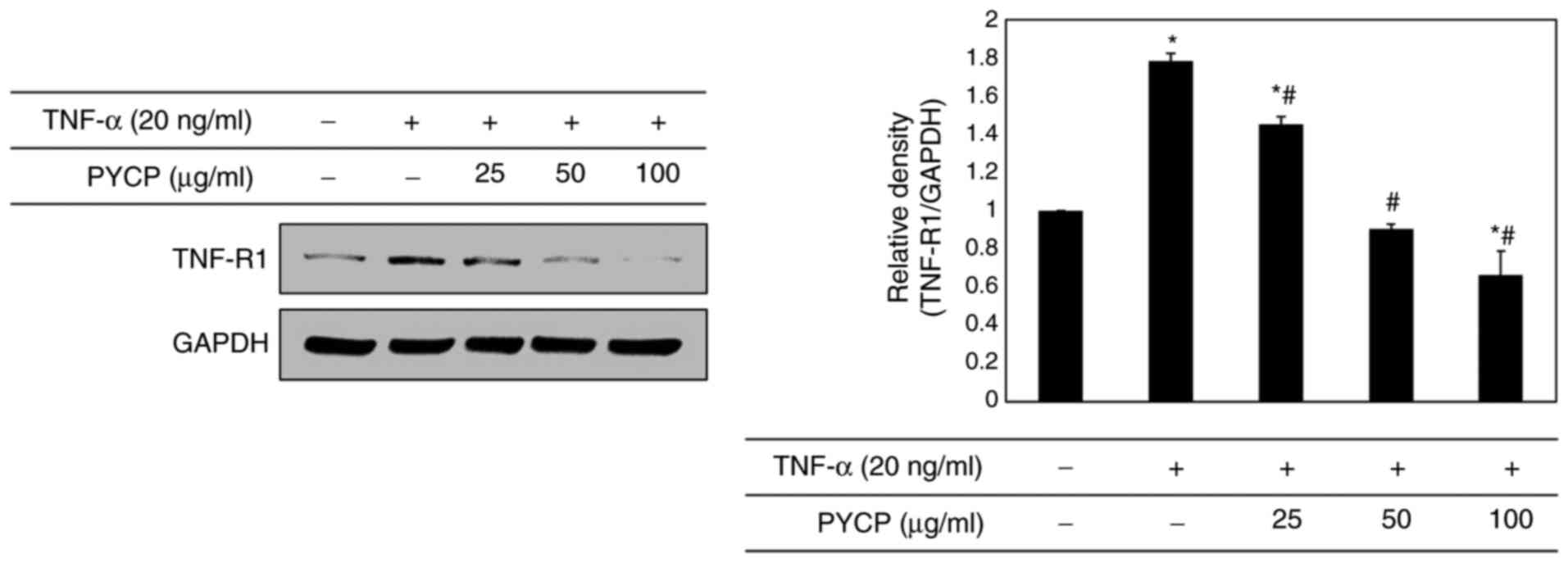
Effects of PYCP on TNF-R1
expression
To determine whether PYCP treatment altered the
expression levels of TNF-R1 in TNF-α-treated C2C12 myotubes, the
TNF-R1 protein expression levels were examined by western blot
analysis. As shown in Fig. 4, the
TNF-R1 protein expression levels were significantly increased in
TNF-α-treated C2C12 myotubes. However, the TNF-α-induced
upregulation of TNF-R1 was significantly attenuated by PYCP
treatment (Fig. 4).
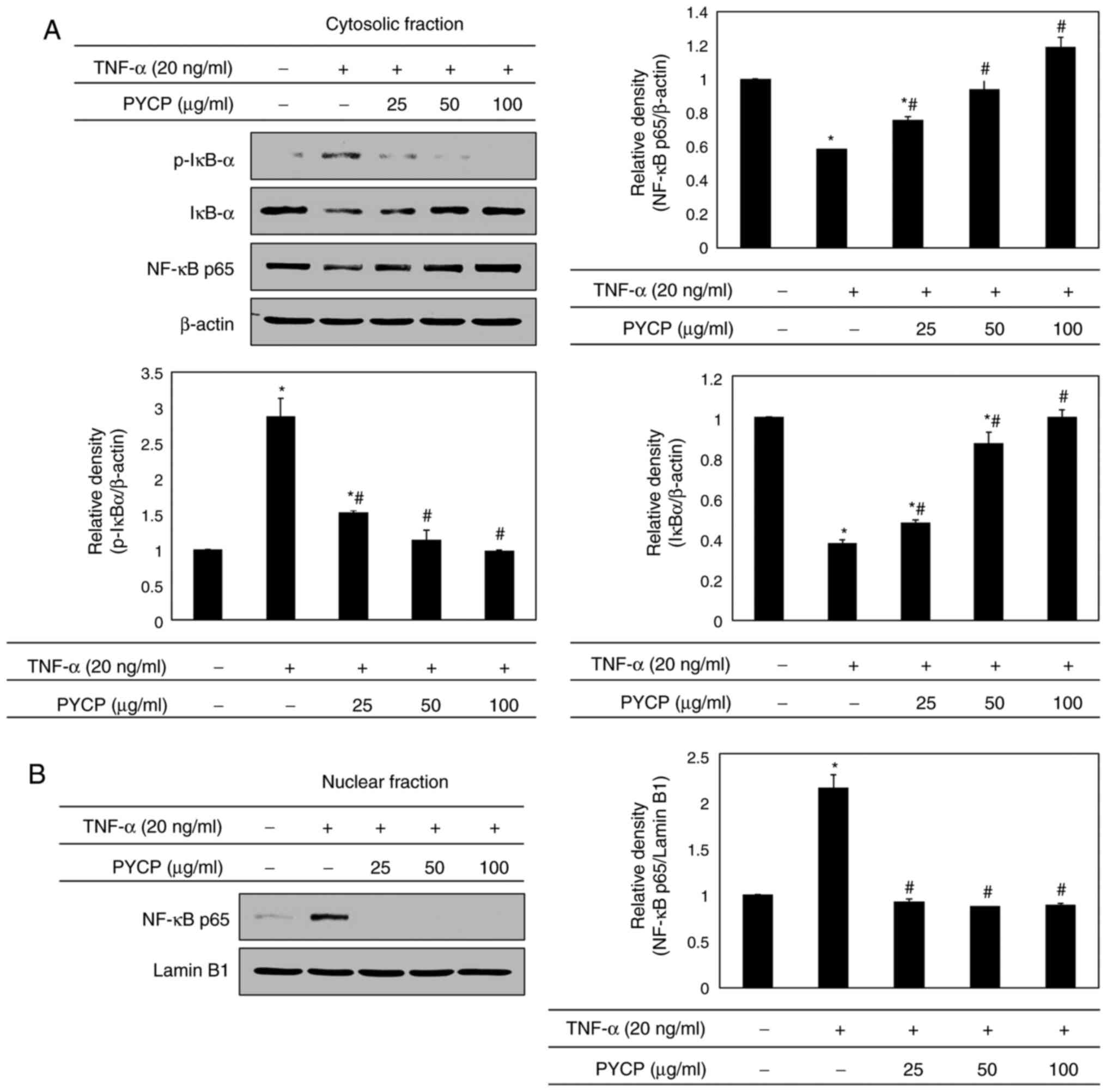
Effects of PYCP on the NF-κB signaling
pathway
To further explore the mechanism underlying
transcriptional control by PYCP, C2C12 myotubes were treated with
TNF-α (20 ng/ml) and PYCP (25, 50, or 100 µg/ml) for 48 h, and
proteins associated with NF-κB/p65 subunit nuclear translocation
were examined by western blot analysis. As shown in Fig. 5A, the phosphorylation of IκBα, which
induces NF-κB/p65 activation, was inhibited by PYCP treatment in
TNF-α-induced C2C12 myotubes. Consequently, the nuclear NF-κB/p65
levels were significantly decreased by PYCP treatment compared with
TNF-α-only treated C2C12 myotubes (Fig.
5B). The results indicated that PYCP treatment effectively
blocked the nuclear translocation and activation of NF-κB/p65 by
inhibiting TNF-α-induced IκBα phosphorylation.
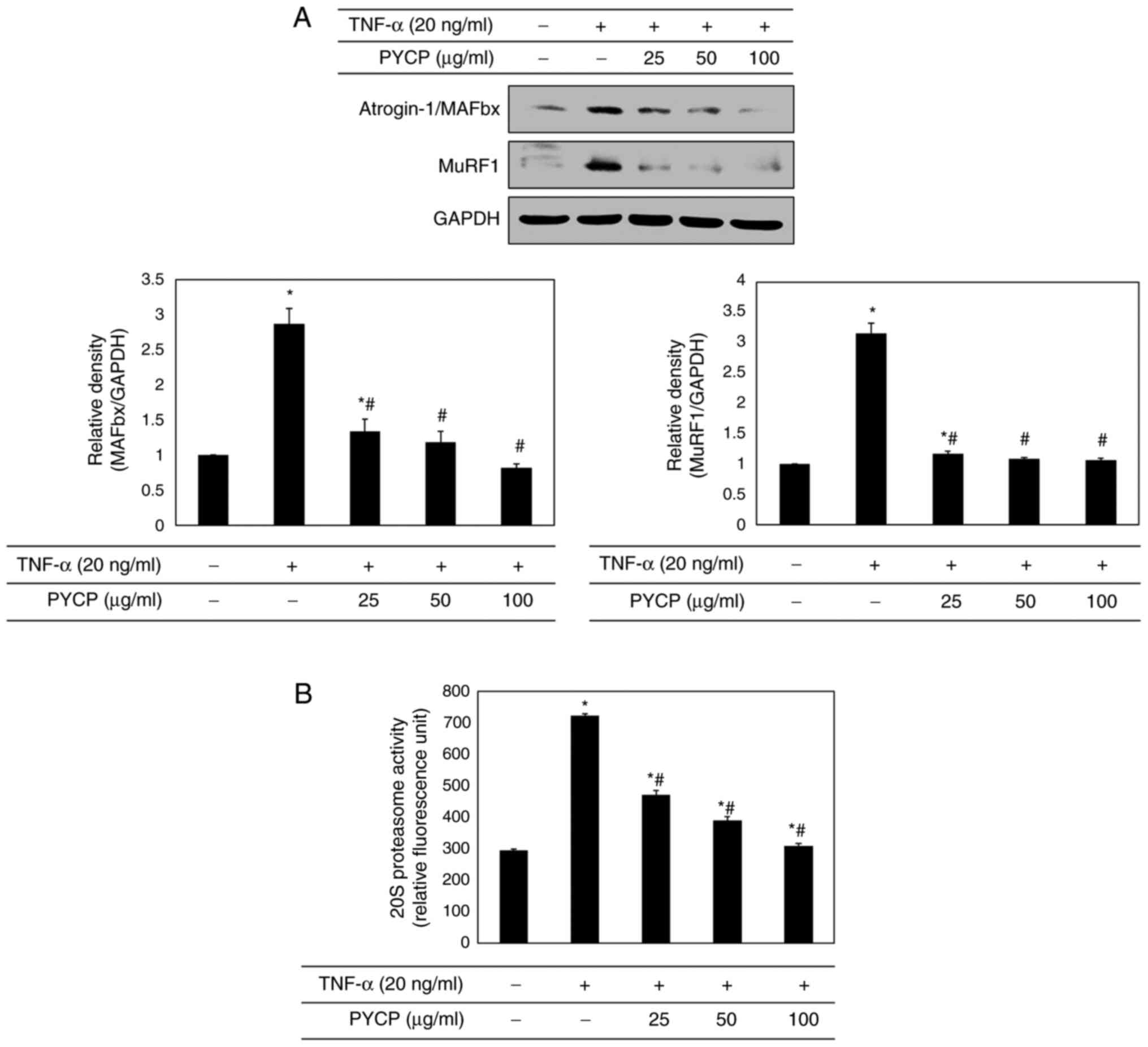
Effects of PYCP on the
ubiquitin-proteasome system
To confirm that the ubiquitin-proteasome system is
regulated by NF-κB, the protein expression levels of E3 ubiquitin
ligases in C2C12 myotubes were investigated by western blot
analysis. As shown in Fig. 6A,
TNF-α treatment significantly increased atrogin-1/MAFbx and MuRF1
expression levels. However, treatment with PYCP attenuated the
TNF-α-induced increase in the expression of atrogin-1/MAFbx and
MuRF1 (Fig. 6A). In addition, to
assess the protective effect of PYCP on 20S proteasome activity,
the proteolytic activity of 20S proteasome was monitored by
measuring the AMC fluorescence. As shown in Fig. 6B, TNF-α treatment significantly
increased 20S proteasome activity, but this was significantly
attenuated by PYCP treatment.
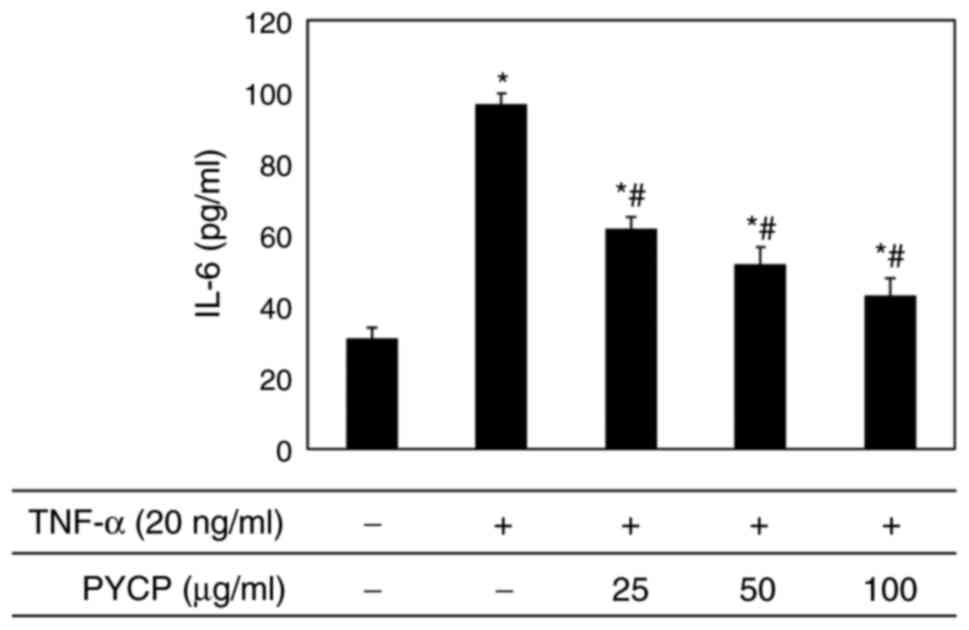
Effects of PYCP on IL-6
production
To evaluate the effect of PYCP on proinflammatory
cytokines regulated by the transcription factor NF-κB, IL-6
production in C2C12 myotubes was measured using an ELISA kit.
Increased IL-6 levels in TNF-α-treated C2C12 myotubes were
significantly reduced by PYCP treatment in a dose-dependent manner
(Fig. 7).
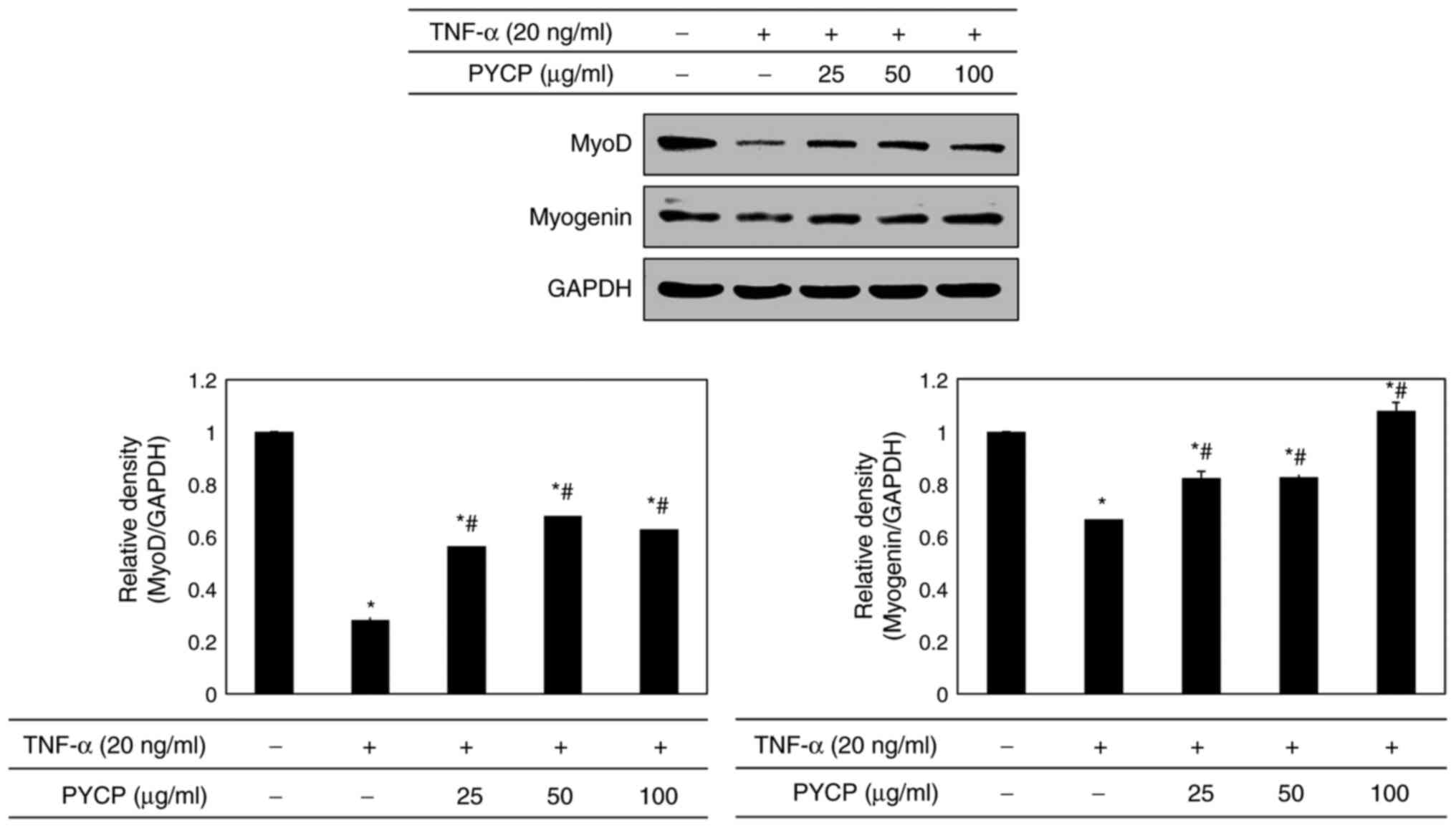
Effects of PYCP on myogenesis
To determine whether PYCP treatment altered myogenic
differentiation in TNF-α-treated C2C12 myotubes, MyoD and myogenin
protein expression levels were examined by western blot analysis.
As shown in Fig. 8, both MyoD and
myogenin protein expression levels were significantly decreased in
TNF-α-stimulated C2C12 myotubes compared with control untreated
myotubes. However, the TNF-α-mediated downregulation of MyoD and
myogenin was significantly reversed by treatment with PYCP
(Fig. 8).
Discussion
In the present study, the antiatrophic activities
and mechanisms of PYCP were investigated using TNF-α-treated C2C12
myotubes. The main findings demonstrated that PYCP efficiently
inhibited the expression of E3 ubiquitin ligases, 20S proteasome
activity, and IL-6 secretion via suppression of the NF-κB pathway
by inhibiting TNF-R1 expression and ROS production in TNF-α-induced
C2C12 myotubes. In addition, PYCP effectively increased MyoD and
myogenin expression in TNF-α-induced C2C12 myotubes. The present
results indicated that TNF-α-induced inflammation-related muscle
atrophy could be attenuated by PYCP treatment.
‘Inflammaging’, the chronic low-grade inflammation
that characterizes aging, is associated with muscle catabolism
(26). Additionally, an increase in
body fat due to aging causes fat to be deposited in muscle tissue,
and the accumulation of intramuscular fat can increase the
secretion of inflammatory cytokines, such as TNF-α and IL-6
(27,28). Muscle atrophy caused by the
inflammatory cytokine TNF-α is characterized by decreases in cell
survival, fiber diameter, and protein content (29,30).
In the present study, TNF-α reduced the viability and myotube
diameter of C2C12 myotubes; these effects were recovered by PYCP
treatment. These results indicated that PYCP may protect C2C12
myotubes against myotube atrophy caused by TNF-α exposure.
The mechanisms underlying muscle atrophy are
complex. The majority of the biological functions of TNF-α occur by
TNF-α binding to TNF-R1 (31).
Muscle atrophy caused by TNF-α results in loss of total muscle
protein, reportedly regulated by NF-κB (32,33).
Li et al showed that overexpression of IκBα sequesters NF-κB
in an inactive state in skeletal muscle, resulting in resistance to
TNF-α-stimulated protein degradation (34). Furthermore, the suppression of NF-κB
activation in vivo improves skeletal muscle regeneration
following trauma (35). Therefore,
a clear association exists between TNF-α-induced muscle atrophy and
NF-κB activation. In addition, the loss of muscle protein caused by
NF-κB has been correlated with the stimulation of the
ubiquitin-proteasome system and augmented by the production of
intracellular ROS (36). In the
present study, PYCP treatment significantly decreased the
TNF-α-stimulated intracellular ROS production in C2C12 myotubes,
indicating that PYCP exerts antioxidant activity. Intracellular ROS
contributes to the induction of E3 ubiquitin ligases by stimulating
translocation of NF-κB to the nucleus through phosphorylation of
IκBα in cultured C2C12 myotubes (36,37).
In the present study, PYCP treatment inhibited the TNF-α-stimulated
TNF-R1 expression and significantly inhibited IκBα phosphorylation
and translocation of NF-κB to the nucleus. In addition, PYCP
inhibited the expression of atrogin-1/MAFbx and MuRF1, and 20S
proteasome activity stimulated by TNF-α. Therefore, it can be
speculated that inhibition of the ubiquitin-proteasome system in
the TNF-α-induced C2C12 myotubes was predominantly associated with
the regulation of the NF-κB pathway through intracellular ROS
removal by the antioxidant activity of PYCP.
Skeletal muscle is considered an endocrine organ
capable of releasing certain cytokines and chemokines (38). The expression and release of
inflammatory cytokines are promoted in TNF-α-treated C2C12 myotubes
(39,40). Furthermore, cytokine and chemokine
release from C2C12 myotubes is regulated by NF-κB, a transcription
factor mediated by the production of free radicals, such as ROS
(40). Inflammation due to the
release of catabolic cytokines, such as IL-6, may contribute to
muscle atrophy progression by causing muscle protein degradation
(38–40). Therefore, the plasma concentration
of IL-6 may be a biochemical indicator of muscle atrophy. In the
present study, the TNF-α-stimulated production of IL-6 was reduced
by PYCP treatment. Based on these results, the inhibitory effects
of PYCP on the production of cytokines may be regulated by the
NF-κB pathway through blockade of ROS production, which has a
similar intrinsic antioxidant activity to PYCP. Thus, the reduction
of IL-6 production from C2C12 myotubes may be primarily associated
with the regulation of the NF-κB pathway through ROS removal due to
the antioxidant activity of PYCP.
Regarding the mechanisms by which NF-κB influences
myogenic differentiation, activation of NF-κB by TNF-α inhibits
muscle differentiation through the destabilization of the MyoD and
myogenin protein via a post-transcriptional mechanism (16,41).
TNF-α and IL-1β have been shown to inhibit myogenic differentiation
of cultured C2C12 myoblasts via activation and translocation of
NF-κB (42). Furthermore, TWEAK, a
structural homologue of TNF-α, can block myoblast terminal
differentiation by activating NF-κB (43). In the present study, PYCP treatment
restored the TNF-α reduced MyoD and myogenin expression in C2C12
myotubes. These findings are consistent with previous studies that
activation of NF-κB by proinflammatory cytokines leads to the
destabilization of MyoD and myogenin expression in skeletal muscle
cells (44–46). The present data indicated that
muscle atrophy progression caused by TNF-α may be alleviated by the
PYCP-induced recovery of MyoD and myogenin expression suppressed by
activation of NF-κB.
Overall, the present study demonstrated that the
antiatrophic effects of PYCP in TNF-α-stimulated C2C12 myotubes
were associated with the inactivation of the NF-κB pathway through
intracellular ROS removal by antioxidant activity. Preliminary mass
spectrometry (Q-TOF MS/MS) analysis of PYCP revealed a photosystem
II 12-kDa extrinsic protein (PSBU_PYRYE, PsbU, P. yezoensis)
(data not shown). Thus, the active ingredient of PYCP may be PsbU,
an exogenous protein localized in the lumen side of photosystem II
and found mostly in red algae and cyanobacteria (47). PsbU has been demonstrated to protect
cells from ROS produced as an inevitable by-product of
photosynthesis, by improving the thermal stability of photosystem
II (48,49). Therefore, PYCP may perform the same
functions as those of PsbU. Further investigations are warranted to
fully elucidate the active components in PYCP and their functions
and mechanisms. The results of the present study indicated that the
antiatrophic effect of PYCP in C2C12 myotubes stimulated by TNF-α
was associated to the inactivation of NF-κB through intracellular
ROS removal.
Acknowledgements
Not applicable.
Funding
This research was supported by the Basic Science
Research Program through the National Research Foundation of Korea
funded by the Ministry of Education (grant nos. 2019R1A6A3A01095099
and 2012R1A6A1028677).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MKL and TJN conceived and designed the study. MKL
and YHC acquired and analyzed the data. MKL and TJN confirm the
authenticity of all the raw data. MKL prepared the draft of the
manuscript, including the figures and table. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tolosa L, Morla M, Iglesias A, Busquets X,
Llado J and Olmos G: IFN-gamma prevents TNF-alpha-induced apoptosis
in C2C12 myotubes through down-regulation of TNF-R2 and increased
NF-kappaB activity. Cell Signal. 17:1333–1342. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lecker SH, Solomon V, Mitch WE and
Goldberg AL: Muscle protein breakdown and the critical role of the
ubiquitin-proteasome pathway in normal and disease states. J Nutr.
129 (1S Suppl):227S–237S. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schiaffino S, Dyar KA, Ciciliot S, Blaauw
B and Sandri M: Mechanisms regulating skeletal muscle growth and
atrophy. FEBS J. 208:4294–4314. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kandarian SC and Jackman RW: Intracellular
signaling during skeletal muscle atrophy. Muscle Nerve. 33:155–165.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Egerman MA and Glass DJ: Signaling
pathways controlling skeletal muscle mass. Crit Rev Biochem Mol
Biol. 49:59–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kirillova I, Chaisson M and Fausto N:
Tumor necrosis factor induces DNA replication in hepatic cells
through nuclear factor kappaB activation. Cell Growth Differ.
10:819–828. 1999.PubMed/NCBI
|
|
7
|
Schutze S, Machleidt T and Kronke M:
Mechanisms of tumor necrosis factor action. Semin Oncol. 19 (2
Suppl 4):S16–S24. 1992.PubMed/NCBI
|
|
8
|
Basu A, Johnson DE and Woolard MD:
Potentiation of tumor necrosis factor-alpha-induced cell death by
rottlerin through a cytochrome-C-dependent pathway. Exp Cell Res.
278:209–214. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li YP: TNF-alpha is a mitogen in skeletal
muscle. Am J Physiol Cell Physiol. 285:C370–C376. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li YP, Schwarts RJ, Waddell ID, Holloway
BR and Reid MB: Skeletal muscle myocytes undergo protein loss and
reactive oxygen-mediated NF-kappaB activation in response to tumor
necrosis factor alpha. FASEB J. 12:871–880. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cai D, Frantz JD, Tawa NE Jr, Melendez PA,
Oh BC, Lidov HG, Hasselgren PO, Frontera WR, Lee J, Glass DJ and
Shoelson SE: IKKbeta/NF-kappaB activation causes severe muscle
wasting in mice. Cell. 119:285–298. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mourkioti F, Kratsios P, Luedde T, Song
YH, Delafontaine P, Adami R, Parente V, Bottinelli R, Pasparakis M
and Rosenthal N: Targeted ablation of IKK2 improves skeletal muscle
strength, maintains mass, and promotes regeneration. J Clin Invest.
116:2945–2954. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kumar A, Takada Y, Boriek AM and Aggarwasl
BB: Nuclear factor-kappaB: Its role in health and disease. J Mol
Med (Berl). 82:434–448. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guttridge DC, Albanese C, Reuther JY,
Pestell RG and Baldwin AS Jr: NF-kappaB controls cell growth and
differentiation through transcriptional regulation of cyclin D1.
Mol Cell Biol. 19:5785–5799. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mitin N, Kudla AJ, Konieczny SF and
Taparowsky EJ: Differential effects of Ras signaling through
NFkappaB on skeletal myogenesis. Oncogene. 20:1276–1286. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Berkes CA and Tapscott SJ: MyoD and the
transcriptional control of myogenesis. Semin Cell Dev Biol.
16:585–595. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Hwang JY, Park HB, Yadav D, Oda T
and Jin JO: Porphyran isolated from Pyropia yezoensis
inhibits lipopolysaccharide-induced activation of dendritic cells
in mice. Carbohydr Polym. 229:1154572020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee MK, Choi JW, Choi YH and Nam TJ:
Pyropia yezoensis protein prevents dexamethasone-induced
myotube atrophy in C2C12 myotubes. Mar Drugs. 16:4972018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim IH, Kwon MJ, Jung JH and Nam TJ:
Protein extracted porphyra yezoensis prevents cisplatin-induced
nephrotoxicity by downregulating the MAPK and NF-κB pathways. Int J
Mol Med. 41:511–520. 2018.PubMed/NCBI
|
|
20
|
Park SJ, Ryu J, Kim IH, Choi YH and Nam
TJ: Activation of the mTOR signaling pathway in breast cancer MCF-7
cells by a peptide derived from porphyra yezoensis. Oncol Rep.
33:19–24. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Choi JW, Kwon MJ, Kim IH, Kim YM, Lee MK
and Nam TJ: Pyropia yezoensis glycoprotein promotes the M1
to M2 macrophage phenotypic switch via the STAT3 and STAT6
transcription factors. Int J Mol Med. 38:666–674. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim CR, Kim YM, Lee MK, Kim IH, Choi YH
and Nam TJ: Pyropia yezoensis peptide promotes collagen
synthesis by activating the TGF-β/Smad signaling pathway in the
human dermal fibroblast cell line HS27. Int J Mol Med. 39:31–38.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Choi JW, Kim IH, Kim YM, Lee MK, Choi YH
and Nam TJ: Protective effect of Pyropia yezoensis
glycoprotein on chronic ethanol consumption-induced hepatotoxicity
in rats. Mol Med Rep. 14:4881–4886. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Castillero E, Alamdari N, Lecker SH and
Hasselgren PO: Suppression of atrogin-1 and MuRF1 prevents
dexamethasone-induced atrophy of cultured myotubes. Metabolism.
62:1495–1502. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Menconi M, Gonnella P, Petkova V, Lecker S
and Hasselgren PO: Dexamethasone and corticosterone induce similar,
but not identical, muscle wasting responses in cultured L6 and
C2C12 myotubes. J Cell Biochem. 105:353–364. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Malafarina V, Uriz-Otano F, Iniesta R and
Gil-Guerrero L: Sarcopenia in the elderly: Diagnosis,
physiopathology and treatment. Maturitas. 71:109–114. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kob R, Bollheimer LC, Bertsch T, Fellner
C, Djukic M, Sieber CC and Fischer BE: Sarcopenic obesity:
Molecular clues to a better understanding of its pathogenesis?
Biogerontology. 16:15–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jensen GL: Inflammation: Roles in aging
and sarcopenia. JPEN J Parenter Enteral Nutr. 32:656–659. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wing SS, Lecker SH and Jagoe RT:
Proteolysis in illness-associated skeletal muscle atrophy: From
pathways to networks. Crit Rev Clin Lab Sci. 48:49–70. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ventadour S and Attaix D: Mechanisms of
skeletal muscle atrophy. Curr Opin Rheumatol. 18:631–635. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen G and David V: TNF-R1 signaling: A
beautiful pathway. Science. 296:1634–1635. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li YP, Atkins CM, Sweatt JD and Reid MB:
Mitochondria mediate tumor necrosis factor-alpha/NF-kappaB
signaling in skeletal muscle myotubes. Antioxid Redox Signal.
1:97–104. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Thoma A and Lightfoot AP: NF-κB and
inflammatory cytokine signaling: Role in skeletal muscle atrophy.
Adv Exp Med Biol. 1088:267–279. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li YP and Reid MB: NF-kappaB mediates the
protein loss induced by TNF-alpha in differentiated skeletal muscle
myotubes. Am J Physiol Regul Integr Comp Physiol. 279:R1165–R1170.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thaloor D, Miller KJ, Gephart J, Mitchell
PO and Pavlath GK: Systemic administration of the NF-kappaB
inhibitor curcumin stimulates muscle regeneration after traumatic
injury. Am J Physiol. 277:C320–C329. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nisr RB, Shah DS, Ganley IG and Hundal HS:
Proinflammatory NFκB signaling promotes mitochondrial dysfunction
in skeletal muscle in response to cellular fuel overloading. Cell
Mol Life Sci. 76:4887–4904. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
McKinnell IW and Rudnicki MA: Molecular
mechanisms of muscle atrophy. Cell. 119:907–910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Legard GE and Pedersen BK: Muscle as an
endocrine organ. Muscle Exerc Physiol. 25:258–307. 2019.
|
|
39
|
Bhatnagar S, Panguluri SK, Gupta SK,
Dahiya S, Lundy RF and Kumar A: Tumor necrosis factor-α regulates
distinct molecular pathway and gene networks in cultured skeletal
muscle cells. PLoS One. 5:e132622010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lightfoot AP, Sakellariou GK, Nye GA,
McArdle F, Jackson MJ, Griffiths RD and McArdle A: SS-31 attenuates
TNF-α induced cytokine release from C2C12 myotubes. Redox Biol.
6:253–259. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kaliman P, Canicio J, Testar X, Palacin M
and Zorzano A: Insulin-like growth factor-II, phosphatidylinositol
3-kinase, nuclear factor-kappaB and inducible nitric-oxide synthase
define a common myogenic signaling pathway. J Biol Chem.
274:17437–17444. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Langen RC, Schols AM, Kelders MC, Wouters
EF and Janssen-Heininger YM: Inflammatory cytokines inhibit
myogenic differentiation through activation of nuclear
factor-kappaB. FASEB J. 15:1169–1180. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dogra C, Changotra H, Mohan S and Kumar A:
Tumor necrosis factor-like weak inducer of apoptosis inhibits
skeletal myogenesis through sustained activation of nuclear
factor-kappaB and degradation of MyoD protein. J Biol Chem.
281:10327–10336. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Guttridge DC, Mayo MW, Madrid LV, Wang CY
and Baldwin AS Jr: NF-kappaB-induced loss of MyoD messenger RNA:
Possible role in muscle decay and cachexia. Science. 289:2363–2366.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sitcheran R, Cogswell PC and Baldwin AS
Jr: NF-kappaB mediates inhibition of mesenchymal cell
differentiation through a posttranscriptional gene silencing
mechanism. Genes Dev. 17:2368–2373. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Di Marco S, Mazroui R, Dallaire P, Chittur
S, Tenenbaum SA, Radzioch D, Marette A and Gallouzi IE:
NF-kappB-mediated MyoD decay during muscle wasting requires nitric
oxide synthase mRNA stabilization, HuR protein, and nitric oxide
release. Mol Cell Biol. 25:6533–6545. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Roose JL, Wegener KM and Pakrasi HB: The
extrinsic proteins of photosystem II. Photosynth Res. 92:369–387.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Balint I, Bhattacharya J, Perelman A,
Schatz D, Moskovitz Y, Keren N and Schwarz R: Inactivation of the
extrinsic subunit of photosystem II, PsbU, in Synechococcus PCC
7942 results in elevated resistance to oxidative stress. FEBS Lett.
580:2117–2122. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Abasova L, Deak Z, Schwarz R and Vass I:
The role of the PsbU subunit in the light sensitivity of PSII in
the cyanobacterium Synechococcus 7942. J Photochem Photobiol B.
105:149–156. 2011. View Article : Google Scholar : PubMed/NCBI
|