Introduction
Pancreatic adenocarcinoma (PAAD) is a common
gastrointestinal cancer associated with a high mortality rate,
resulting in >227,000 deaths annually worldwide (1–3). The
most common type of PAAD is pancreatic ductal adenocarcinoma,
accounting for 80–90% of all pancreatic tumors (4). PAAD is among the most invasive types
of carcinoma, and clinical diagnosis is often delayed due to a lack
of known PAAD-specific symptoms (5). PAAD is therefore often diagnosed
following metastasis and/or increases in cancer aggression, both of
which greatly reduce the survival rate (6). The prognosis of PAAD is poor with a
1-year survival rate of <10% (2). However, a definitive diagnosis of PAAD
is not currently possible due to a lack of reliable tumor markers,
which are urgently needed for initial detection and intervention
(5). Currently, chemotherapy is the
only available option for the treatment of advanced end-stage or
metastatic PAAD (5).
MicroRNAs (miRNAs/miRs) are small, endogenous,
non-coding RNAs that range in length from 19–25 nucleotides
(7). miRNA can degrade or inhibit
translation of mRNA by partial or total binding of the
3′-untranslated region (UTR) of the target molecule (7). miRNAs are key regulatory factors in
various types of tumors, functioning as tumor oncogenes or tumor
suppressors to regulate cell proliferation, differentiation,
metastasis and apoptosis (7,8).
miR-32-5p, a member of the miR-32 family and located on chromosome
band Xq26.2 (9), is involved in the
regulation and development of numerous types of carcinoma. For
example, Ye et al (10)
reported that miR-32-5p inhibits the migration, invasion and
proliferation of colorectal carcinoma cells; Wang et al
(11) demonstrated that miR-32-5p
represses the migration and invasion of clear cell renal cell
carcinoma cells, and is positively correlated with a good
prognosis; and Liu et al (12) indicated that miR-32-5p is
downregulated in cervical carcinoma tissues and inhibits the
proliferation, migration and invasion of cervical carcinoma
cells.
TBC/LysM-associated domain-containing 1 (TLDC1) is
an evolutionarily conserved protein 1 (13). Bioinformatics databases were
analyzed to gain insight into the molecular functions of TLDC1,
also known through genomic and proteomic studies as KIAA1609
(14), LOC57707 (15), or mEAK-7 (mammalian EAK-7 or MTOR
associated protein, eak-7 homolog) (16). Independent reports reveal that TLDC1
mRNA is overexpressed in diseases, such as hepatocellular carcinoma
(17) and lymph node-positive
breast cancer (18). TLDC1
activates an alternative mTOR signaling pathway through S6K2 and
4E-BP1 to regulate cell proliferation and migration (13). miR-1911-3p targets mEAK-7 to
suppress mTOR signaling in human lung cancer cells (19). it was hypothesized that miR-32-5p
and TLDC1 may serve an important role in the development of
PAAD.
Relatively few previous studies have investigated
the role and underlying mechanisms of miR-32-5p in the pathogenesis
of PAAD. For example, to the best of our knowledge, only one
previous study has reported that growth arrest specific 5 could
positively regulate PTEN-induced tumor suppressor pathway via
miR-32-5p, thus inhibiting the metastasis of pancreatic cancer
(20). Therefore, the aim of the
present study was to investigate the relationship between miR-32-5p
and TLDC1, and the underlying mechanisms in the onset and
progression of PAAD.
Materials and methods
Cell culture
Human PAAD cell lines (BxPC-3, AsPC-1 and PANC-1)
were obtained from American Type Culture Collection. PANC-1 and
AsPC-1 cells were incubated in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS, whereas BxPC-3 cells
were incubated in RPMI medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS. The human pancreatic HPDE6 cell
line was purchased from Shanghai Gefan Biotechnology Co., Ltd. and
incubated in keratinocyte serum-free medium (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with human recombinant
epidermal growth factor (Gibco; Thermo Fisher Scientific, Inc.) and
bovine pituitary extract (Sigma-Aldrich; Merck KGaA). All cells
were maintained under 5% CO2 at 37°C.
Cell transfection
Human AsPC-1 cells were transfected with a negative
control (NC) mimic, miR-32-5p mimic, NC inhibitor, miR-32-5p
inhibitor (100 nM; all synthesized by Suzhou GenePharma Co., Ltd.),
pcDNA control (empty vector) or pcDNA/TLDC1 (0.5 µg; both obtained
from GenScript) using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's protocol. After incubation at 37°C for 48 h, the
cells were collected for subsequent assays. The sequences were as
follows: NC mimic, 5′-UAUUGCACAUUACUAAGUUGCA-3′,
5′-UAUUGCACAUUACUAAGUUGCA-3′; miR-32-5p mimic (double-stranded RNA
oligonucleotides without chemically modification),
5′-UAUUGCACAUUACUAAGUUGCA-3′, 5′-CAACUUAGUAAUGUGCAAUAUU-3′; NC
inhibitor, 5′-GUAUUCGAGUAAUCAACGAUAU-3′; and miR-32-5p inhibitor
miR-96-5p inhibitor (single-stranded oligonucleotides chemically
modified by 2′-Ome), 5′-UGCAACUUAGUAAUGUGCAAUA-3′. To verify
successful transfection, reverse transcription-quantitative PCR
(RT-qPCR) was performed to detect miR-32-5p expression levels at 12
h post-transfection, and western blotting was performed to detect
protein expression levels 48 h post-transfection.
Bioinformatic analysis
miRNAs that regulate TLDC1 were predicted using
starBase (http://starbase.sysu.edu.cn/) with TLDC1 as the key
word.
Dual-luciferase reporter assay
The binding site between TLDC1 3′-UTR fragment and
miR-32-5p were predicted using starBase (http://starbase.sysu.edu.cn/). To validate the
microRNA-binding sequence of miR-32-5p, fragments containing
wild-type or mutated TLDC1 3′-untranslated region (3′UTRs) of the
predicted binding site were cloned into the pGL3 vector (Promega
Corporation), which were constructed by Shanghai GeneChem Co., Ltd.
AsPC-1 cells were seeded (5×104 cells/well) into 24-well
plates. Subsequently, cells were co-transfected with the reporter
plasmid (TLDC1 3′UTR construct; 500 ng) and miR-32-5p mimic or NC
mimic (900 ng) using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h. Luciferase activity was detected using
the Dual-Luciferase® Reporter Assay System (Promega
Corporation) according to the manufacturer's protocol. Firefly
luciferase activity was normalized to Renilla luciferase
activity. The co-transfection systems were as follows: miR-32-5p
mimic + TLDC1-WT; NC-mimic + TLDC1-WT; miR-32-5p mimic + TLDC1-MUT;
and NC-mimic + TLDC1-MUT.
Cell Counting Kit-8 (CCK-8) assay
AsPC-1 cells (4×103cells/well) were
seeded into a 96-well culture plate following incubation for 24,
48, 72 and 96 h at 37°C with the CCK-8 reagent (ImmunoWay
Biotechnology Company) according to the manufacturer's protocol,
cell proliferation was measured at a wavelength of 450 nm using a
microplate reader.
Wound-healing assay
Transfected human AsPC-1 cells were seeded
(2×104 cells/well) into the wells of 6-well plates and
incubated in serum-free medium for ~24 h. At 100% confluence, a
linear wound to the cell monolayer was made using the sterilized
tip of a liquid pipette gun and the cells were cultured for 48 h at
37°C. Subsequently, the media was discarded and the plates were
washed three times in PBS. Plates were imaged and the serum-free
medium was added prior to incubation for an additional 24 h at
37°C. The plates were then imaged again after the 24-h incubation.
Cells in randomly selected fields of view were counted using a
light microscope (magnification, ×100). The migratory ability of
cells was indicated by gap closure. The assay was performed in
triplicate.
Transwell assay
At 48 h post-transfection, AsPC-1 cells
(5×104) were transferred to a Transwell chamber
containing Matrigel (BD Biosciences). Matrigel pre-coating was
conducted for 30 min at 37°C. Prior to conducting the assay, the
lower and upper chambers were filled with pre-warmed media for
hydration. Cells were then digested and resuspended in serum-free
media. The upper chamber was loaded with cells suspended in
serum-free media and the lower chamber with media supplemented with
10% FBS. The assay was conducted for 48 h at 37°C. Following
completion of the assay, the cells in the lower chamber were fixed
for 30 min with 40% methanol at 25°C and then stained using 0.1%
crystal violet for 20 min at 25°C. Stained cells were visualized
using an inverted light microscope (Olympus Corporation;
magnification, ×100).
Western blotting
Total protein was extracted from AsPC-1 cells using
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.) containing a protease inhibitor cocktail (Roche Diagnostics)
and quantified using a BCA kit (Beyotime Institute of
Biotechnology,). Total protein (30 µg; 25 µl/well) was separated
via 12% SDS-PAGE acrylamide gel. The separated proteins were then
transferred onto a PVDF membrane, which was blocked with 5% skimmed
milk in TBS containing 0.1% Tween-20 for 1 h at 25°C. The membranes
were incubated overnight at 4°C with primary antibodies against
TLDC1 (1:1,000; cat. no. AA426-456; 4A Biotech Co., Ltd.) and
β-actin (1:2,000; Cell Signaling Technology Inc.). Following the
primary incubation, membranes were washed three times with TBST
(0.05% Tween-20) and incubated at 25°C for 2 h with secondary
antibodies (horseradish peroxidase-conjugated goat anti-rabbit
immunoglobulin G; 1:2,000; cat. no. ab6721; Abcam). The membranes
were washed a further three times before visualization using ECL
chemiluminescent detection system (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. β-actin (1:2,000; cat.
no. ab6276; Abcam) was used as the loading control. ImageJ version
1.46 software (National Institutes of Health) was used to
semi-quantify protein expression.
RT-qPCR
Total RNA was extracted from AsPC-1 cells using
TRIzol® reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. Total RNA was reverse
transcribed into cDNA using the PrimeScript™ RT Reagent kit (Takara
Biotechnology Co., Ltd.) according to the manufacturer's
instructions. The expression of miR-32-5p was determined using a
SYBR® Premix Ex Taq TM II (Tli RNaseH Plus) kit (Takara
Bio, Inc.) according to the manufacturer's protocol. The following
thermocycling conditions were used for qPCR: Pre-denaturation at
95°C for 30 sec; denaturation at 95°C for 10 sec; annealing and
extension at 60°C for 30 sec for 40 cycles. miR-32-5p expression
levels were quantified using the 2−ΔΔCq method (21) and normalized to the internal
reference gene, U6. The primers used for RT-qPCR are shown in
Table 1.
 | Table I.Sequences of primers used for reverse
transcription-quantitative PCR. |
Table I.
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene | Sequence
(5′→3′) |
|---|
| miR-32-5p | F: TAT TGC ACA TTA
CTA AGC CTT |
|
| R: GAA TAC CTC GGA
CCC TGC |
| U6 | F: GCT TCG GCA GCA
CAT ATA CTA AAA |
|
| R: CGC TTC ACG AAT
TTG CGT GTCAT |
Statistical analysis
All statistical analyses were performed using SPSS
version 24.0 (IBM Corp.). Data are presented as the mean ± SD. The
unpaired Student's t-test was used to compare the means between two
groups, whereas one-way ANOVA followed by Tukey's post-hoc test was
used for multiple comparisons. Kaplan-Meier survival curves were
analyzed using the log-rank test to determine statistical
significance. Pearson's correlation coefficient test was used to
determine whether there was a linear correlation between miR-32-5p
and TLDC1 expression in PAAD. All experiments were repeated three
times independently. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-32-5p is lowly expressed in PAAD
tissues and cells
The starBase database was used to assess the
expression pattern of miR-32-5p in PAAD and to identify the
relationship between miR-32-5p expression levels and PAAD
prognosis. The results revealed that miR-32-5p expression was
downregulated in the PAAD patient group compared with the healthy
group (Fig. 1A; P<0.05). Low
miR-32-5p expression was associated with a poor prognosis of
patients with PAAD compared with high miR-32-5p expression
(Fig. 1B; P=0.12). Furthermore,
miR-32-5p expression was measured in HPDE6 and PAAD cell lines
(AsPC-1, PANC-1 and BxPC-3). The RT-qPCR results demonstrated that
miR-32-5p expression levels were significantly reduced in the PAAD
cells compared with those in HDPE6 cells (Fig. 1C; P<0.001). These findings
indicated that miR-32-5p might be involved in the development and
progression of PAAD.
miR-32-5p inhibits the proliferation,
migration and invasion of PAAD cells
To assess the effect of miR-32-5p overexpression and
knockdown, the AsPC-1 cell line was selected due to moderate
miR-32-5p expression levels. To determine whether miR-32-5p was
associated with the proliferation, migration and invasion of PAAD
cells, functional studies of AsPC-1 cells were performed. The
results demonstrated that miR-32-5p mimic caused a significant
increase in miR-32-5p mRNA expression compared with NC mimic,
whereas miR-32-5p inhibitor significantly reduced miR-31-5p
expression compared with NC inhibitor. This indicated the
successful establishment of miR-32-5p overexpression and knockdown
in AsPC-1 cells (Fig. 2A; both
P<0.001). The CCK-8 assay results revealed that miR-32-5p mimic
significantly impaired the proliferation of AsPC-1 compared with NC
mimic (Fig. 2B; P<0.001 at 48 h;
P<0.01 at 72 h; P<0.05 at 96 h). The metastatic potential of
AsPC-1 cells was measured using wound-healing and Transwell assays.
Cell migration was significantly impaired in miR-32-5p
mimic-transfected cells compared with NC mimic-transfected cells
(Fig. 2C; P<0.01). miR-32-5p
knockdown significantly increased cell proliferation (Fig. 2B; P<0.01 at 72 h; P<0.001 at
96 h), and significantly increased cell migration (Fig. 2C; P<0.001) and invasion compared
with NC inhibitor (Fig. 2D;
P<0.001). Collectively, these findings demonstrated that
miR-32-5p inhibited the proliferation and metastatic potential of
PAAD cells.
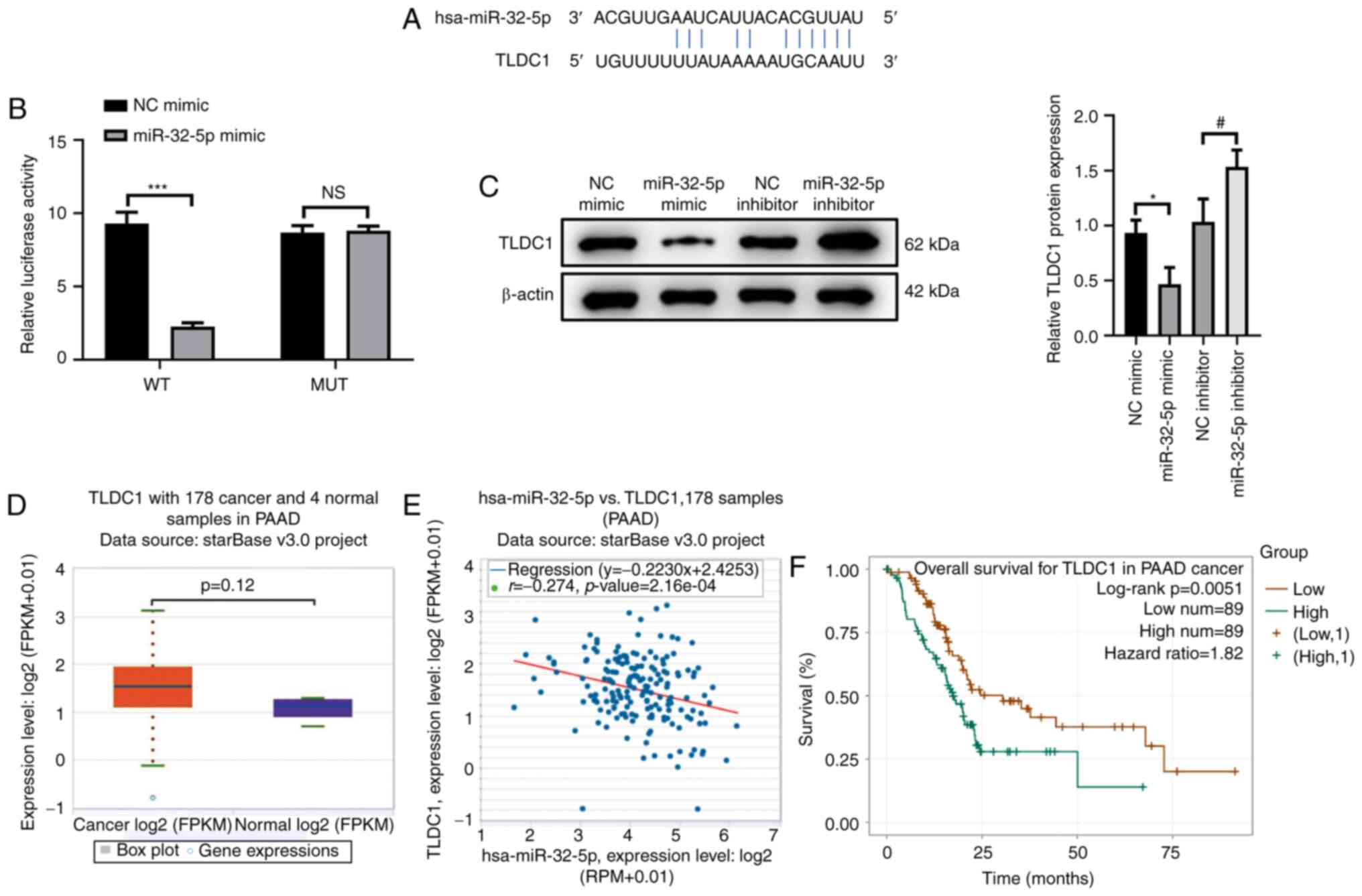
TLDC1 is a target of miR-32-5p
To elucidate the downstream molecular mechanisms of
miR-32-5p in the proliferation, migration and invasion of PAAD
cells, the starBase database was searched to identify the target
sequence of miR-32-5p. The results demonstrated that miR-32-5p
targeted the 3′-UTR of TLDC1 (Fig.
3A). The dual-luciferase reporter assay was performed to
confirm the interaction between miR-32-5p and TLDC1. The results
demonstrated that luciferase activity was significantly reduced in
cells co-transfected with TLDC1-WT and miR-32-5p mimic compared
with those co-transfected with NC mimic (P<0.001). No
significant difference in luciferase activity was observed in the
MUT TLDC1 co-transfection system, confirming that miR-32-5p
targeted TLDC1 (Fig. 3B;
P>0.05). Moreover, TLDC1 protein expression levels were
significantly reduced in cells transfected with miR-32-5p mimic
compared with the NC mimic group, whereas miR-32-5p inhibitor
significantly increased the protein expression levels of TLDC1
compared with the NC inhibitor group (Fig. 3C; both P<0.05). These results
demonstrated that miR-32-5p could both target and regulate TLDC1 in
PAAD cells, which was further confirmed by bioinformatics analyses.
The starBase database demonstrated that TLDC1 expression was
increased in PAAD cells compared with the NC group (Fig. 3D; P=0.12). Furthermore, TLDC1
expression levels were negatively correlated with miR-32-5p in PAAD
tissues (Fig. 3E; P<0.05). High
TLDC1 expression levels were significantly associated with a better
prognosis of patients with PAAD compared with lower TLDC expression
levels (Fig. 3F; P<0.05).
Overall, these results indicated that TLDC1 may serve a regulatory
role in the proliferation, migration and invasion of PAAD cells via
miR-32-5p.
TLDC1 reverses the suppressive effect
of miR-32-5p on the proliferation, migration and invasion of PAAD
cells
To explore the effect of TLDC1 on the proliferation,
migration and invasion of PAAD cells mediated by miR-32-5p,
simultaneous overexpression of TLDC1 and miR-32-5p was induced. The
results demonstrated that TLDC1 protein expression levels were
significantly increased in cells transfected with NC + pcDNA/TLDC1
compared with those transfected with pcDNA/control, indicating that
TLDC1 was successfully overexpressed (Fig. 4A; all P<0.001). Transfection with
miR-32-5p mimic significantly reduced TLDC1 protein expression
levels compared with the NC + pcDNA/control group. However,
co-transfection with miR-32-5p and TLDC1 significantly reversed the
decrease in TLDC1 protein expression levels induced by miR-32-5p
compared with miR-32-5p + pcDNA/control (Fig. 4B; P<0.01). Compared with the NC +
pcDNA/control group, miR-32-5p overexpression was also shown to
significantly impair the proliferation of AsPC-1 cells, whereas
TLDC1 overexpression significantly reversed miR-32-5p
mimic-mediated effects (Fig. 4C;
both P<0.001). The wound-healing assay results showed that cell
migration was significantly decreased in the miR-32-5p mimic +
pCDNA/control group compared with that in the NC mimic +
pCDNA/control group (P<0.05), and TLDC1 overexpression
significantly reversed miR-32-5p mimic-mediated inhibition of
migration (Fig. 4D; P<0.01).
Furthermore, the Transwell assay results indicated miR-32-5p
overexpression significantly decreased the number of invasive cells
compared with the NC mimic + pCDNA/control group (P<0.001), and
TLDC1 overexpression partially restored this decrease (Fig. 4E; P<0.001). In summary, these
data suggested that TLDC1 was a target of miR-32-5p and reversed
miR-32-5p-mediated inhibition of the proliferation, migration and
invasion of PAAD cells.
Discussion
PAAD is characterized by poor prognosis and
resistance to therapy, which explains the relatively high mortality
rate compared with other types of cancer (22–24).
At present, the treatment options for PAAD are very limited, with
surgical resection of the tumor remaining the most effective
(25–27). However, for the majority of patients
with PAAD, surgery is not a viable option due to a high chance of a
relapse occurring (23,28,29).
Therapeutic prospects for PAAD remain poor even with improved
treatment strategies because of the high risk of metastasis
(23,30,31).
Due to the complicated configuration of the adjoining anatomy of
the pancreas, whereby the pancreas is in contact with numerous
neighboring structures, pancreatic cancer cells tend to also
aggressively attack the adjacent tissues (24,25,30,32).
Therefore, it is essential to identify the mechanisms underlying
the proliferation and metastasis of PAAD.
Previous studies have determined that multiple
miRNAs can influence the occurrence and progression of PAAD by
modulating the expression of various target genes. Zhang et
al (33) reported that
miR-135b-5p contributed to the epithelial-mesenchymal transition,
migration and invasion of PAAD cells by targeting the nuclear
receptor protein, nuclear receptor subfamily 3 group C member 2.
Furthermore, Daniel et al (34) revealed that miR-32-5p is
downregulated in prostate cancer tissues, whereas Wang et al
(35) reported that miR-32-5p is
lowly expressed in triple-negative breast cancer. Collectively,
these previous reports suggest that miR-32-5p regulates the
development of various types of cancer to a certain degree. Hence,
the aim of the present study was to investigate the role, and
identify the target genes and physiological functions of miR-32-5p
in PAAD. The results demonstrated that miR-32-5p expression levels
were markedly reduced in PAAD tissues and cell lines compared with
healthy tissues and healthy human pancreatic cells. Moreover,
elevated miR-32-5p expression levels were associated with a better
prognosis of patients with PAAD compared with those with lower
miR-32-5p expression levels, which indicated that miR-32-5p may
display a tumor-suppressive effect in the onset and progression of
PAAD.
Based on the differential expression of miR-32-5p in
PAAD, it was speculated that miR-32-5p may be associated with the
proliferation, migration and invasion of PAAD. The present study
demonstrated that overexpression of miR-32-5p significantly
suppressed the proliferation and markedly decreased the migration
and invasion of PAAD cells. Collectively, these results suggested
that miR-32-5p had an antitumor effect in PAAD cells, serving an
oncogenic role in the progression of PAAD. Therefore, miR-32-5p may
have the potential to function as a diagnostic biomarker and
therapeutic target for PAAD.
Various miRNAs have been shown to serve roles in
tumor development and progression by mediating cell proliferation
and metastasis. miR-337-3p has been reported to regulate the
proliferation, invasion, migration and apoptosis of cervical cancer
cells by targeting Ras-related protein Rap1A expression (36), whereas miR-124 was found to regulate
metastasis and the epithelial-mesenchymal transition in
triple-negative breast cancer by targeting zinc finger
E-box-binding homeobox 2 protein expression (37). Furthermore, miR-335 was demonstrated
to suppress the proliferation of lung cancer cells by targeting
transformer-2 protein homolog β expression (38). Further identification of miR-32-5p
target genes will help to further elucidate the role of miR-32-5p
in the pathogenesis of PAAD. miRNAs generally bind to the 3′-UTR of
target mRNAs (39). In the present
study, the starBase database was searched to predict the direct
target gene of miR-32-5p in PAAD, which identified TLDC1.
Furthermore, these results demonstrated that TLDC1 expression was
relatively higher in PAAD tissue compared with that in healthy
tissue and negatively correlated with miR-32-5p expression in
patients with PAAD. Patients with lower TLDC1 protein expression
levels exhibited a better prognosis compared with those displaying
higher TLDC protein expression levels. The present study therefore
indicated that TLDC1 may serve a negative regulatory function in
the development of PAAD induced by miR-32-5p.
miR-32-5p serves a key biological role in the
tumorigenesis of various types of cancer by regulating multiple
target genes. In cervical cancer, homeobox B8 is reported to
reverse the proliferation, invasion and migration of cells
inhibited by miR-32-5p (12). In
colorectal cancer, the transducer of ERRB2-1 protein was found to
reverse the inhibitory effect of miR-32-5p on cell sensitization
and the promoting effect of miR-32-5p on cell migration and
invasion (7). In ovarian cancer,
SMG1 nonsense mediated mRNA decay-associated PI3K-related kinase
was shown to reduce the promoting effect of miR-32-5p on cell
proliferation and motility (9). The
present study demonstrated that miR-32-5p suppressed the
proliferation, migration and invasion of PAAD cells, which was
rescued by TLDC1 overexpression.
The present study revealed that miR-32-5p was lowly
expressed in PAAD. miR-32-5p was shown to negatively target TLDC1,
inhibiting the proliferation, migration, and invasion of PAAD
cells. As a tumor suppressor gene, miR-32-5p may serve as a
promising biomarker for the diagnosis and prognosis of PAAD.
However the present study had a number of limitations. For example,
the results were not verified using animal models or other PAAD
cells. Moreover, the upstream regulation of miR-32-5p and the
pathways involved in TLDC1 requires further investigation.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
PY and CT designed the study and wrote the
manuscript with contributions from all authors. All authors
participated in performing the experiments. BC, PL, JS, GX, ZW, and
GZ were responsible for data acquisition and the interpretation of
data. YH, WY and GW were responsible for statistical analysis, the
literature search and revising the article critically for important
intellectual content. All authors read and approved the final
manuscript. PY, CT and GZ confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Xu B, Liu J, Xiang X, Liu S, Zhong P, Xie
F, Mou T and Lai L: Expression of miRNA-143 in pancreatic cancer
and its clinical significance. Cancer Biother Radiopharm.
33:373–379. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu Z, Zhao S, Wang L, Wang J and Zhou J:
MiRNA-339-5p plays an important role in invasion and migration of
pancreatic cancer cells. Med Sci Monit. 25:7509–7517. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Du W, Lei C, Wang Y, Ding Y and Tian P:
LINC01232 sponges multiple miRNAs and its clinical significance in
pancreatic adenocarcinoma diagnosis and prognosis. Technol Cancer
Res Treat. 20:15330338209885252021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang C, Huang Y, Zhang J and Fang Y:
MiRNA-339-5p suppresses the malignant development of gastric cancer
via targeting ALKBH1. Exp Mol Pathol. 115:1044492020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hessmann E, Buchholz SM, Demir IE, Singh
SK, Gress TM, Ellenrieder V and Neesse A: Microenvironmental
determinants of pancreatic cancer. Physiol Rev. 100:1707–1751.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Leinwand J and Miller G: Regulation and
modulation of antitumor immunity in pancreatic cancer. Nat Immunol.
21:1152–1159. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liang H, Tang Y, Zhang H and Zhang C:
MiR-32-5p regulates radiosensitization, migration and invasion of
colorectal cancer cells by targeting TOB1 gene. Onco Targets Ther.
12:9651–9661. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Du B, Zhang P, Tan Z and Xu J: MiR-1202
suppresses hepatocellular carcinoma cells migration and invasion by
targeting cyclin dependent kinase 14. Biomed Pharmacother.
96:1246–1252. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zeng S, Liu S, Feng J, Gao J and Xue F:
MicroRNA-32 promotes ovarian cancer cell proliferation and motility
by targeting SMG1. Oncol Lett. 20:733–741. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ye T, Zhang N, Wu W, Yang B, Wang J, Huang
W and Tang D: SNHG14 promotes the tumorigenesis and metastasis of
colorectal cancer through miR-32-5p/SKIL axis. In Vitro Cell Dev
Biol Anim. 55:812–820. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang M, Sun Y, Xu J, Lu J, Wang K, Yang
DR, Yang G, Li G and Chang C: Preclinical studies using miR-32-5p
to suppress clear cell renal cell carcinoma metastasis via altering
the miR-32-5p/TR4/HGF/Met signaling. Int J Cancer. 143:100–112.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu YJ, Zhou HG, Chen LH, Qu DC, Wang CJ,
Xia ZY and Zheng JH: MiR-32-5p regulates the proliferation and
metastasis of cervical cancer cells by targeting HOXB8. Eur Rev Med
Pharmacol Sci. 23:87–95. 2019.PubMed/NCBI
|
|
13
|
Nguyen JT, Ray C, Fox AL, Mendonça DB, Kim
JK and Krebsbach PH: Mammalian EAK-7 activates alternative mTOR
signaling to regulate cell proliferation and migration. Sci Adv.
4:eaao58382018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nagase T, Kikuno R, Nakayama M, Hirosawa M
and Ohara O: Prediction of the coding sequences of unidentified
human genes. XVIII. The complete sequences of 100 new cDNA clones
from brain which code for large proteins in vitro. DNA Res.
7:273–281. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schröder B, Wrocklage C, Pan C, Jäger R,
Kösters B, Schäfer H, Elsässer HP, Mann M and Hasilik A: Integral
and associated lysosomal membrane proteins. Traffic. 8:1676–1686.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nguyen JT, Haidar FS, Fox AL, Ray C,
Mendonça DB, Kim JK and Krebsbach PH: mEAK-7 forms an alternative
mTOR complex with DNA-PKcs in human cancer. iScience. 17:190–207.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Riou P, Saffroy R, Comoy J, Gross-Goupil
M, Thiéry JP, Emile JF, Azoulay D, Piatier-Tonneau D, Lemoine A and
Debuire B: Investigation in liver tissues and cell lines of the
transcription of 13 genes mapping to the 16q24 region that are
frequently deleted in hepatocellular carcinoma. Clin Cancer Res.
8:3178–3186. 2002.PubMed/NCBI
|
|
18
|
Ellsworth RE, Field LA, Love B, Kane JL,
Hooke JA and Shriver CD: Differential gene expression in primary
breast tumors associated with lymph node metastasis. Int J Breast
Cancer. 2011:1427632011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mendonça DB, Nguyen JT, Haidar F, Fox AL,
Ray C, Amatullah H, Liu F, Kim JK and Krebsbach PH:
MicroRNA-1911-3p targets mEAK-7 to suppress mTOR signaling in human
lung cancer cells. Heliyon. 6:e057342020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao ZQ, Wang JF, Chen DH, Ma XS, Wu Y,
Tang Z and Dang XW: Long non-coding RNA GAS5 suppresses pancreatic
cancer metastasis through modulating miR-32-5p/PTEN axis. Cell
Biosci. 7:662017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cao J, Yang J, Ramachandran V, Arumugam T,
Deng DF, Li ZS, Xu LM and Logsdon CD: TM4SF1 regulates pancreatic
cancer migration and invasion in vitro and in vivo. Cell Physiol
Biochem. 39:740–750. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu Q, Zong L, Chen X, Jiang Z, Nan L, Li
J, Duan W, Lei J, Zhang L, Ma J, et al: Resveratrol in the
treatment of pancreatic cancer. Ann N Y Acad Sci. 1348:10–19. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cao L, Chen X, Xiao X, Ma Q and Li W:
Resveratrol inhibits hyperglycemia-driven ROS-induced invasion and
migration of pancreatic cancer cells via suppression of the ERK and
p38 MAPK signaling pathways. Int J Oncol. 49:735–743. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koay EJ, Amer AM, Baio FE, Ondari AO and
Fleming JB: Toward stratification of patients with pancreatic
cancer: Past lessons from traditional approaches and future
applications with physical biomarkers. Cancer Lett. 381:237–243.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sabater L, Muñoz E, Roselló S, Dorcaratto
D, Garcés-Albir M, Huerta M, Roda D, Gómez-Mateo MC,
Ferrández-Izquierdo A, Darder A and Cervantes A: Borderline
resectable pancreatic cancer. Challenges and controversies. Cancer
Treat Rev. 68:124–135. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hartwig W, Werner J, Jäger D, Debus J and
Büchler MW: Improvement of surgical results for pancreatic cancer.
Lancet Oncol. 14:e476–e485. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kamisawa T, Wood LD, Itoi T and Takaori K:
Pancreatic cancer. Lancet. 388:73–85. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qian B, Wei L, Yang Z, He Q, Chen H, Wang
A, Yang D, Li Q, Li J, Zheng S and Fu W: Hic-5 in pancreatic
stellate cells affects proliferation, apoptosis, migration,
invasion of pancreatic cancer cells and postoperative survival time
of pancreatic cancer. Biomed Pharmacother. 121:1093552020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Middleton G, Palmer DH, Greenhalf W,
Ghaneh P, Jackson R, Cox T, Evans A, Shaw VE, Wadsley J, Valle JW,
et al: Vandetanib plus gemcitabine versus placebo plus gemcitabine
in locally advanced or metastatic pancreatic carcinoma (ViP): A
prospective, randomised, double-blind, multicentre phase 2 trial.
Lancet Oncol. 18:486–499. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hartwig W, Vollmer CM, Fingerhut A, Yeo
CJ, Neoptolemos JP, Adham M, Andrén-Sandberg A, Asbun HJ, Bassi C,
Bockhorn M, et al: Extended pancreatectomy in pancreatic ductal
adenocarcinoma: Definition and consensus of the International Study
Group for Pancreatic Surgery (ISGPS). Surgery. 156:1–14. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang Z, Che X, Yang N, Bai Z, Wu Y, Zhao
L and Pei H: miR-135b-5p Promotes migration, invasion and EMT of
pancreatic cancer cells by targeting NR3C2. Biomed Pharmacother.
96:1341–1348. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Daniel R, Wu Q, Williams V, Clark G,
Guruli G and Zehner Z: A Panel of MicroRNAs as diagnostic
biomarkers for the identification of prostate cancer. Int J Mol
Sci. 18:12812017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang R, Huang Z, Qian C, Wang M, Zheng Y,
Jiang R and Yu C: LncRNA WEE2-AS1 promotes proliferation and
inhibits apoptosis in triple negative breast cancer cells via
regulating miR-32-5p/TOB1 axis. Biochem Biophys Res Commun.
526:1005–1012. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cao XM: Role of miR-337-3p and its target
Rap1A in modulating proliferation, invasion, migration and
apoptosis of cervical cancer cells. Cancer Biomark. 24:257–267.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ji H, Sang M, Liu F, Ai N and Geng C:
MiR-124 regulates EMT based on ZEB2 target to inhibit invasion and
metastasis in triple-negative breast cancer. Pathol Res Pract.
215:697–704. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu J, Bian T, Feng J, Qian L, Zhang J,
Jiang D, Zhang Q, Li X, Liu Y and Shi J: MiR-335 inhibited cell
proliferation of lung cancer cells by target Tra2β. Cancer Sci.
109:289–296. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cannell IG, Kong YW and Bushell M: How do
microRNAs regulate gene expression? Biochem Soc Trans. 36((Pt 6)):
1224–1231. 2008. View Article : Google Scholar : PubMed/NCBI
|