Introduction
T-cell lymphoblastic lymphoma (T-LBL) is an
aggressive cancer of precursor T cells, which accounts for ~90% of
lymphoblastic lymphoma cases worldwide (1). The majority of T-LBL cases occur in
children and young adults, particularly in male patients (1). T-LBL is predicted to originate from
malignant thymocytes at a specific stage during intrathymic T-cell
differentiation (1). The
development of T-LBL can lead to metastases of lymphoblasts to the
spleen, bone marrow and central nervous system, which can be
life-threatening (2). Although
the 5-year event-free survival rate has been greatly improved by
contemporary chemotherapy (~85%), salvage remains poor with <25%
event-free and overall survival rates for relapsed disease in
Australia (3). Therefore,
exploration of the mechanisms underlying T-LBL malignancy, and the
identification of novel and effective therapeutic targets is
crucial.
Transglutaminase 2 (TGM2) is a ubiquitously
expressed member of the transglutaminase family of
Ca2+-dependent enzymes. TGM2 is responsible for
catalysing the formation of covalent bonds between peptide-bound
glutamine and a variety of primary amines, including the γ amino
group of peptide-bound lysine, or mono- and polyamines, thus
producing cross-linked or aminated proteins, respectively (4–6).
Previous studies have suggested a close association between TGM2
and cancer progression, including apoptosis, angiogenesis, cell
stemness and drug resistance (7–9).
The effects of TGM2 on various types of cancer have previously been
reported, including osteosarcoma (8), colorectal cancer (CRC) (7), glioma (9) and endometrial cancer (10). As for lymphoma, TGM2 has been
shown to facilitate the survival of malignant B cells and promote
lymphoma progression (11).
Additionally, TGM2 may modulate autophagy in mantle cell lymphoma
cells by activating NF-κB, which can confer cell survival (11).
IL-6 is a cytokine that mediates the homeostatic
process, and has a great impact on various immune and non-immune
cell types (12). IL-6 has been
reported to modulate the survival, proliferation, maturation and
cytokine secretion of B and T cells (12) and, has an important role in
activating oncogenic signalling pathways in cancer (13,14). Furthermore, IL-6 can activate
multiple intracellular signalling pathways, such as GTPase RAS and
mitogen-activated protein kinase signalling, which modulate cell
proliferation and differentiation, as well as the well-known JAK
family and STAT family of transcription factors (15). Mounting evidence has suggested
that IL-6 may mediate resistance against lymphoma treatment.
Notably, levels of IL-6 and phosphorylated (p)-STAT3 were revealed
to be increased in duvelisib [a phosphatidylinositol-3-kinase
(PI3K) PI3K inhibitor]-resistant T-cell lymphoma cells, and the
upregulated expression of IL-6/STAT3 possibly mediated the
resistance in lymphoma (16).
ERK-dependent IL-6 positive feedback regulatory signalling has also
been shown to cause resistance of lymphoma to the PI3K inhibitor
BKM120, whereas blockade of ERK activity could reduce IL-6
secretion and improve antitumor outcomes (17).
The present study aimed to explore the novel
mechanisms underlying T-LBL progression. Bioinformatics analysis of
microarray data from the GSE132550 and GSE143382 datasets revealed
a significantly higher expression of TGM2 in T-LBL samples compared
with that in normal samples. The effects of TGM2 on T-LBL cell
proliferation and apoptosis were determined. Furthermore, whether
TGM2 exerted its effects on T-LBL cells through regulating
IL-6/JAK2/STAT3 signalling was revealed. The findings of present
study may present a novel therapeutic target for T-LBL
treatment.
Materials and methods
Bioinformatics analysis
The GSE132550 (18) and GSE143382 datasets (19) were selected for targeted chip
research from the Gene Expression Omnibus (GEO) database
(www.ncbi.nlm.nih.gov/geo). The GSE132550
dataset comprised 80 formalin-fixed paraffin-embedded (FFPE) tumour
cases, including 68 peripheral T-cell lymphoma, not otherwise
specified cases and 12 Lennert lymphoma cases. Peripheral T-cell
lymphoma cases and 10 CD4+ T-cell samples from healthy
donors were subjected to gene expression profiling in the GSE132550
dataset. The mRNA profiling was carried out on all tumour samples
and 10 normal CD4+ T-cell samples from healthy donors
and analysed using GeneSpring version GX 12 (Agilent Technologies,
Inc.). The GSE143382 dataset comprised mRNA information from
early-stage mycosis fungoides biopsies (43 cases), dermatitis
biopsies (29 cases) and 12 healthy skin tissue controls, which were
analysed by an 800 gene NanoString panel (NanoString Technologies,
Inc.). Differentially expressed genes were screened using the
‘limma’ package in R (Linear Models for Microarray Data, http://www.bioconductor.org/packages/release/bioc/html/limma.html).
The mechanism underlying TGM2-mediated T-cell lymphoma was analysed
based on microarray data using the gene set enrichment analysis
(GSEA) software (version 2.0.14, http://www.broadinstitute.org/gsea/index.jsp). Gene
Ontology (GO) term function annotation and Kyoto Encyclopaedia of
Genes and Genomes (KEGG) pathway enrichment analysis were performed
using the ClusterProfiler package in R language (20) to annotate differentially expressed
genes under the ‘biological process’, ‘cellular component’ and
‘molecular function’ categories, and the enrichment pathways were
analysed. Biological processes and pathways with significant
differences were selected, and their enrichment scores and P-values
were ranked. Visualisation of these pathways with P<0.05 was
completed using the R software package.
Tissue sample collection
This study was approved by the ethics committee of
Shengli Oilfield Central Hospital (Dongying, China; approval no.
DYSLYT20191201). Written informed consent was obtained from all the
enrolled individuals. The skin biopsies were collected from 36
patients with stage III~IV cutaneous T-cell lymphoma who underwent
tumor resection or biopsy procedure at the Shengli Oilfield Central
Hospital between January 2020 and January 2021. In total, 21
patients were male and 15 were female with a median age of 36 years
(ranged from 19 to 64 years). The skin biopsies were fixed by using
10% formalin at room temperature (22±2°C) for 24 h, embedded in
paraffin, and sectioned at 4-µm thickness. No samples were
collected following the treatment of first-line therapy, such as
hyper-CVAD. T-cell lymphoma was diagnosed by two physicians
depending on the skin biopsy. A total of 36 normal skin tissues
collected from sex- and age-matched healthy individuals were used
as controls. Peripheral blood was collected from four healthy human
adults (two male and two female subjects) with a medium age of 45
years for the following CD4+ T cell isolation.
CD4+ cell collection
For isolation of peripheral blood mononuclear cells
(PBMCs), peripheral blood diluted in phosphate buffer saline (PBS;
1:1) was added into a 15 ml centrifuge tube together with 5 ml
Ficoll-Paque media (Cytiva). After centrifugation at 400 × g for 20
min, the supernatant was collected and washed in PBS twice. A
sample of 1×1010/l PBMCs was incubated with anti-CD4
primary antibody (cat. no. 612761; BD Biosciences) at 25°C for 30
min and loaded on a FACSCalibur (BD Biosciences) to obtain
CD4+ T cells. The obtained CD4+ T cells were
used in the following reverse transcription-quantitative polymerase
chain reaction (RT-qPCR) and western blotting.
Cell culture
T-cell lymphoma cell lines, including Jurkat, MJ, HH
and SUP-T1, were purchased from American Type Culture Collection
(ATCC). MOTN-1 cells were purchased from Leibniz Institute
DSMZ-German Collection of Microorganisms and Cell Cultures GmbH.
ATN-1 cells were purchased from RIKEN BioResource Center. Jurkat,
MOTN-1, ATN-1, HH and SUP-T1 cells were maintained in RPMI-1640
medium (HyClone; Cytiva) containing 10% foetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (MilliporeSigma). MJ cells were maintained
in Iscove's modified Dulbecco's medium (ATCC) supplemented with 20%
FBS and 1% penicillin/streptomycin. All cells were cultured in a
humidified incubator containing 5% CO2 at 37°C.
siRNA transfection
siRNAs targeting TGM2 (si-TGM2-1 and si-TGM2-2) and
the scrambled control were synthesised by Shanghai GenePharma Co.,
Ltd. Jurkat and SUP-T1 cells in 6-well plates at a density of
1×105 cells/well were transfected with 30 nM siRNAs
using Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 48 h at 37°C. Following transfection, the
cells were cultured for 48 h at 37°C and used in subsequent
experimentation. The siRNA sequences were as follows: si-TGM2-1,
5′-GAGCTGGTCTTAGAGAGGTGT −3′; si-TGM2-2, 5′-GCAACCTTCTCATCGAGTACT
−3′. A scrambled sequence (5′-GATGGCATGGCGGAGTGATTT −3′) was used
as the negative control. Following transfection, Jurkat and SUP-T1
cells were treated with 60 ng/ml IL-6 (MedChemExpress) for 6 h or
100 µM AG490 (MedChemExpress) for 24 h at 37°C.
Cell Counting Kit-8 (CCK-8)
CCK-8 (Dojindo Molecular Technologies, Inc.) was
used to evaluate cell viability. Briefly, 3×103 Jurkat
and SUP-T1 cells were inoculated in a 96-well plate. After
incubation for the 24, 48 and 72 h at 37°C, 10 µl CCK-8 reagent was
added to each well and incubated for a further 2 h at 37°C. The
absorbance values at 450 nm were measured using a microplate
detector (PerkinElmer, Inc.).
5-ethynyl-2′-deoxyuridine (EdU)
assay
An EdU-labelling kit (Thermo Fisher Scientific,
Inc.) was used to determine cells in the S phase. Briefly,
3×103 Jurkat and SUP-T1 cells were seeded in 96-well
plates overnight, followed by incubation with 50 µM EdU reagent for
1 h at 37°C. Nuclei were stained with 0.1 µg/ml DAPI
(MilliporeSigma) for 20 min at room temperature (22±2°C) and the
samples were observed under a fluorescence microscope (Leica
Microsystems GmbH).
Colony formation
Jurkat or SUP-T1 cells (1×103) were
seeded in 6-well plates as single cell suspensions and were
cultured for 2 weeks. Subsequently, visible colonies with >50
cells were stained with 0.1% crystal violet (Beyotime Institute of
Biotechnology) for 20 min at room temperature (22±2°C), and were
observed and captured under a light microscope (Leica Microsystems
GmbH).
Cell apoptosis
Apoptosis was detected using an Annexin V-FITC/PI
kit (Beyotime Institute of Biotechnology). A sample of
1×105 Jurkat or SUP-T1 cells were collected after the
indicated treatment, suspended in binding buffer, and incubated
with FITC-conjugated Annexin V and PI reagent in the dark for 10
min at room temperature (22±2°C). The samples were then analysed
using flow cytometry (BD LSRFortessa™ X-20; BD Biosciences) and the
cytometry data were analysed using FlowJo software version 7.6.1
(FlowJo LLC).
RT-qPCR
RNA was extracted from FFPE biopsy samples,
CD4+ T cells and T-cell lymphoma cells lines using
RecoverAll™ Total Nucleic Acid Isolation Kit (Thermo Fisher
Scientific, Inc.) and TRIzol® Reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), respectively. Subsequently, cDNA
was obtained using a High-Capacity cDNA RT kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) in accordance with the
manufacturer's protocol. mRNA expression was evaluated using SYBR
Green/ROX qPCR Master Mix (Thermo Fisher Scientific, Inc.) and
normalised to GAPDH using the 2−ΔΔCq method (21). The following thermocycling
conditions were used: Enzyme activation at 95°C for 10 min,
followed by 40 cycles of denaturation at 95°C for 15 sec, annealing
at 60°C for 1 min and extension at 72°C for 20 sec. The primers
used were as follows: TGM2, forward 5′-GAGGAGCTGGTCTTAGAGAGG −3′
and reverse 5′-CGGTCACGACACTGAAGGTG-3′; and GAPDH, forward
5′-GTTGCAACCGGGAAGGAAAT-3′ and reverse 5′-GCCCAATACGACCAAATCAGA
−3′.
Western blotting
Proteins were extracted from Jurkat or SUP-T1 cells
using ice-cold radioimmunoprecipitation assay buffer (Thermo Fisher
Scientific, Inc.) containing protease and phosphatase inhibitors
(Thermo Fisher Scientific, Inc.). Protein concentration of the
extracts was determined by using a Bicinchoninic Acid (BCA) Protein
Assay Kit (Beyotime Institute of Biotechnology). Equal amounts (20
µg) of protein were separated via 10~12% SDS-PAGE and blotted onto
PVDF membranes, which were blocked with 5% non-fat milk at room
temperature (22±2°C) for 1 h. Subsequently, the membranes were
incubated with specific primary antibodies against TGM2 (1:1,000;
cat. no. ab2386; Abcam), cleaved caspase-3 (1:500; cat. no. ab2302;
Abcam), cleaved poly ADP-ribose polymerase (PARP) (1:1,000; cat.
no. ab32561; Abcam), JAK2 (1:5,000; cat. no. ab108596; Abcam),
p-JAK2 (1:2,000; cat. no. ab195055; Abcam), STAT3 (1:2,000; cat.
no. ab68153; Abcam), p-STAT3 (1:2,000; cat. no. ab32143; Abcam) and
GAPDH (1:2,000; cat. no. ab9485; Abcam) overnight at 4°C. The next
day, the blots were probed with secondary goat anti-mouse (1:2,000;
cat. no. ab6789; Abcam) or goat anti-rabbit antibodies (1:2,000;
cat. no. ab205718; Abcam) at room temperature (22±2°C) for 1 h. The
blots were visualised using an ECL system (Thermo Fisher
Scientific, Inc.) and analysed via Image Lab software (version 5.0,
Bio-Rad Laboratories, Inc.).
Statistical analysis
Data are presented as the mean ± SD from three
independent experiments. Statistical analysis was performed using
SPSS 22.0 (IBM Corp.). An unpaired t-test was used for two-group
comparisons. One-way ANOVA followed by Dunnett's multiple
comparisons test was used for multiple-group comparisons. P<0.05
was considered to indicate a statistically significant
difference.
Results
TGM2 is highly expressed in T-cell
lymphoma samples and cells
To explore the potential oncogenes in the
development of T-cell lymphoma, an overlap analysis was performed
based on the upregulated genes from the GSE132550 and GSE143382
datasets. Eight overlapping genes were identified, namely MMP12,
CCL19, MMP9, CD180, CCL18, FPR3, TNC and TGM2. TGM2 was selected
for further analysis, because its role in lymphoma is not well
understood (Fig. 1A).
Bioinformatics analysis based on the GEO database from the
GSE132550 dataset revealed that TGM2 was significantly upregulated
in T-cell lymphoma samples compared with that in the normal group
(P<0.05; Fig. 1B).
Furthermore, the present study validated that the expression of
TGM2 was elevated in the FFPE skin biopsies from patients with
T-cell lymphoma (n=36) relative to the skin tissue from healthy
patients (n=36) (P<0.05; Fig.
1C). Further analysis revealed that the mRNA and protein
expression levels of TGM2 were increased in T-cell lymphoma cell
lines, including Jurkat, MOTN-1, ATN-1, MJ, HH and SUP-T1 cells,
compared with CD4+ T cells collected from four healthy
controls (P<0.05; Fig. 1D and
E). Collectively, these results suggested that TGM2 may be
highly expressed in T-cell lymphoma samples and cells.
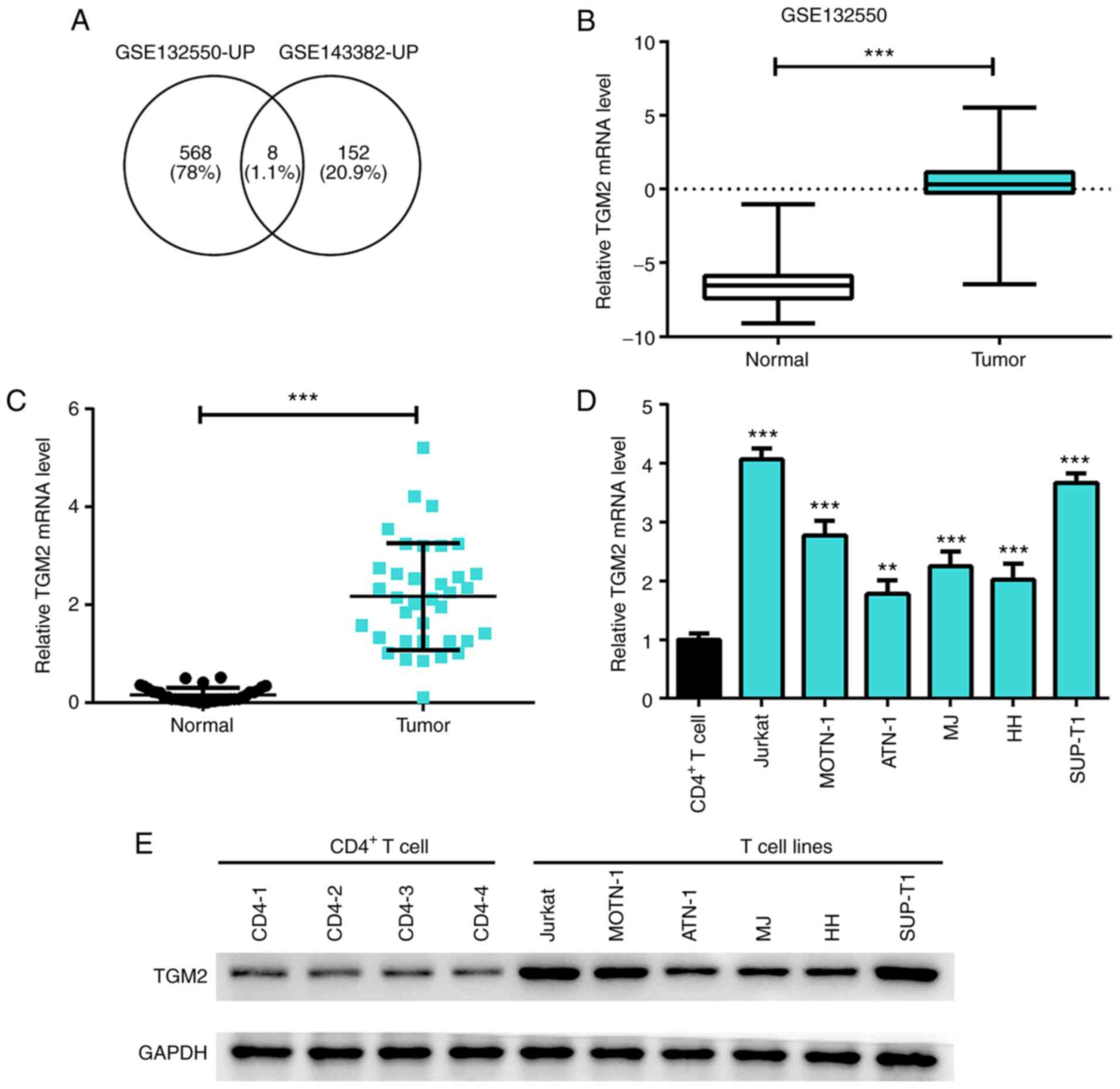 | Figure 1.TGM2 is highly expressed in T-cell
lymphoma samples and cell lines. (A) Overlap analysis was conducted
by comparing the upregulated genes in microarray data from the
GSE132550 and GSE143382 datasets; eight genes were identified,
including MMP12, CCL19, MMP9, CD180, CCL18, FPR3, TNC and TGM2. (B)
Expression of TGM2 was analysed in T-cell lymphoma samples and
normal samples from the Gene Expression Omnibus database. (C)
Formalin-fixed paraffin-embedded skin biopsies from patients with
T-cell lymphoma (n=36) were collected and skin tissues from healthy
individuals were used as controls (n=36). The expression of TGM2 in
the samples was detected by RT-qPCR. (D) mRNA expression levels of
TGM2 in CD4+ T cells and T-cell lymphoma cell lines
(Jurkat, MOTN-1, ATN-1, MJ, HH and SUP-T1) were measured by
RT-qPCR. (E) Protein expression levels of TGM2 in CD4+ T
cells and T-cell lymphoma cell lines (Jurkat, MOTN-1, ATN-1, MJ, HH
and SUP-T1) was detected by western blotting. Data are presented as
the mean ± SD. **P<0.01, ***P<0.01 as indicated or vs.
CD4+ T cells. RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; TGM2,
transglutaminase 2. |
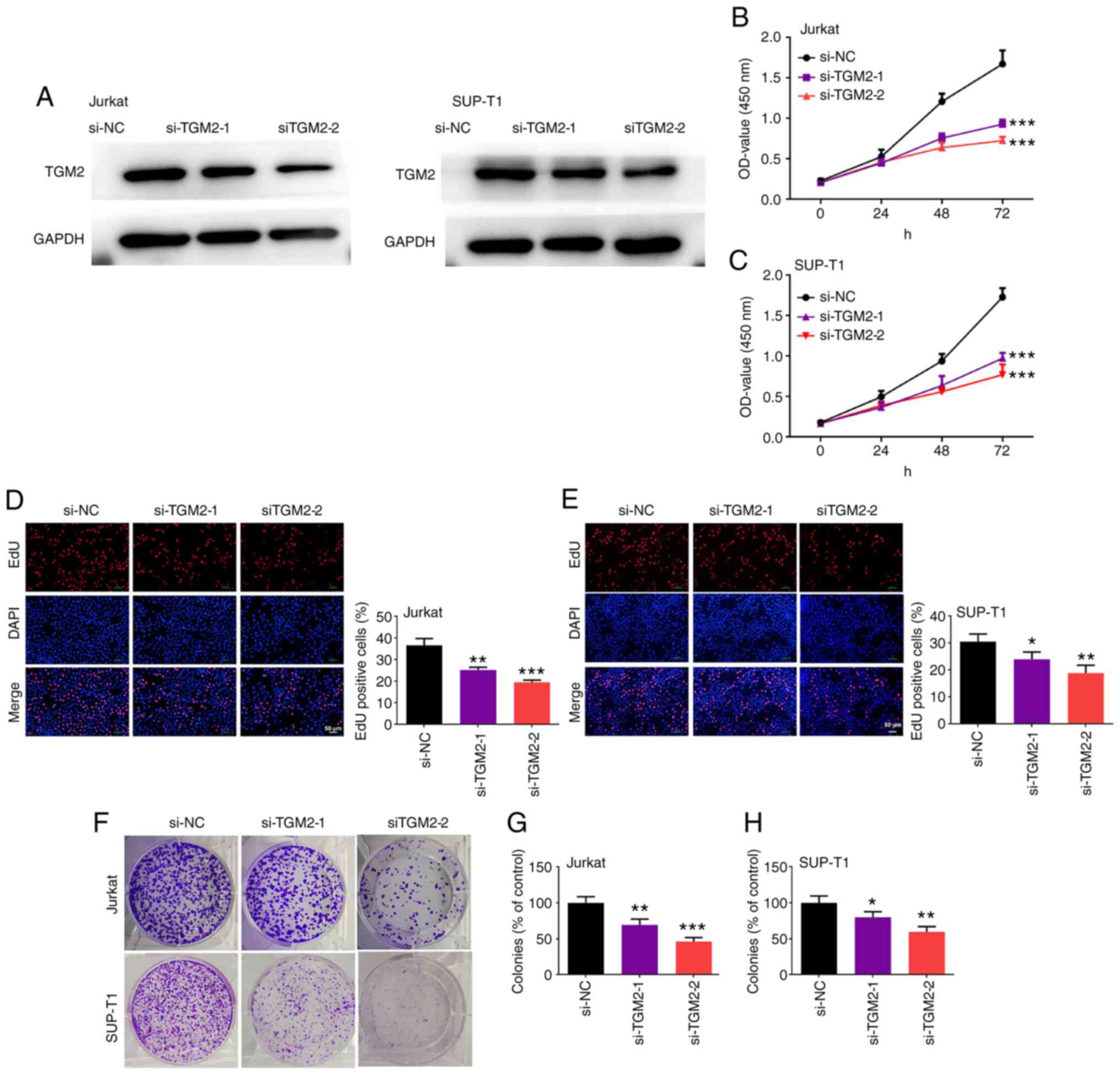
TGM2 siRNAs suppress the viability and
proliferation of T-cell lymphoma cells
The function of TGM2 in T-cell lymphoma cell
proliferation was studied. For this purpose, T-cell lymphoma Jurkat
and SUP-T1 cells, which had higher expression of TGM2, were
transfected with TGM2 siRNAs. As shown in Fig. 2A, TGM2 siRNAs markedly reduced the
protein expression levels of TGM2 in both Jurkat and SUP-T1 cells,
indicating that TGM2 was silenced successfully. In addition, TGM2
siRNAs decreased the viability of Jurkat and SUP-T1 cells
(P<0.05; Fig. 2B and C).
Furthermore, the number of EdU-positive Jurkat and SUP-T1 cells was
reduced by TGM2 siRNAs (P<0.05; Fig. 2D and E). Consistently, TGM2 siRNAs
suppressed the colony-forming ability of Jurkat and SUP-T1 cells
(P<0.05; Fig. 2F-H). Taken
together, these results suggested that TGM2 siRNAs were effective
at suppressing the viability and proliferation of T-cell lymphoma
cells in vitro.
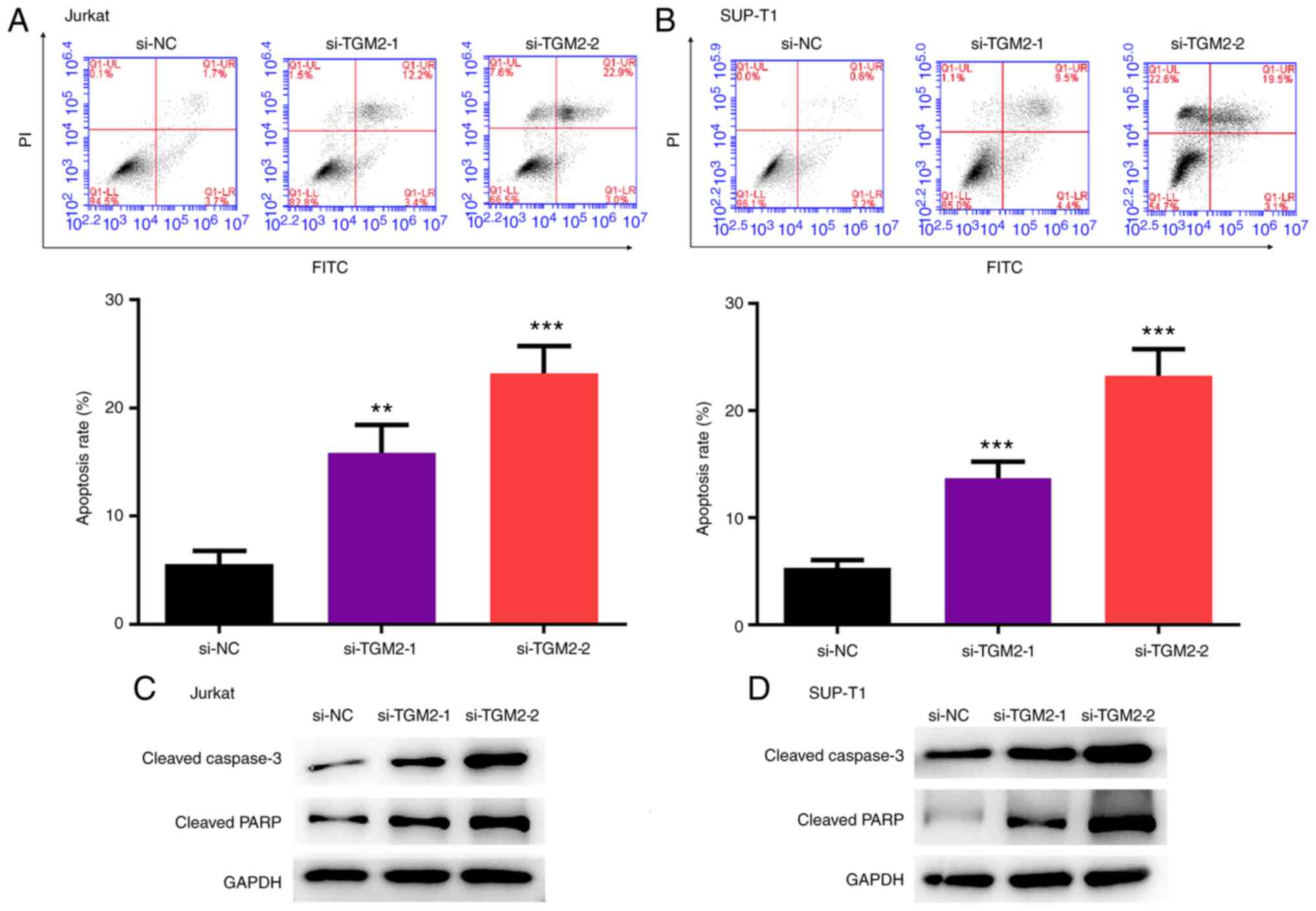
TGM2 siRNAs stimulate the apoptosis of
T-cell lymphoma cells
The present study analysed the effect of TGM2 on the
apoptosis of T-cell lymphoma cells. TGM2 siRNAs enhanced the
apoptosis of Jurkat and SUP-T1 cells (P<0.05; Fig. 3A and B). Furthermore, the
expression levels of cleaved caspase-3 and cleaved PARP were
increased by TGM2 siRNAs in Jurkat and SUP-T1 cells (Fig. 3C and D).
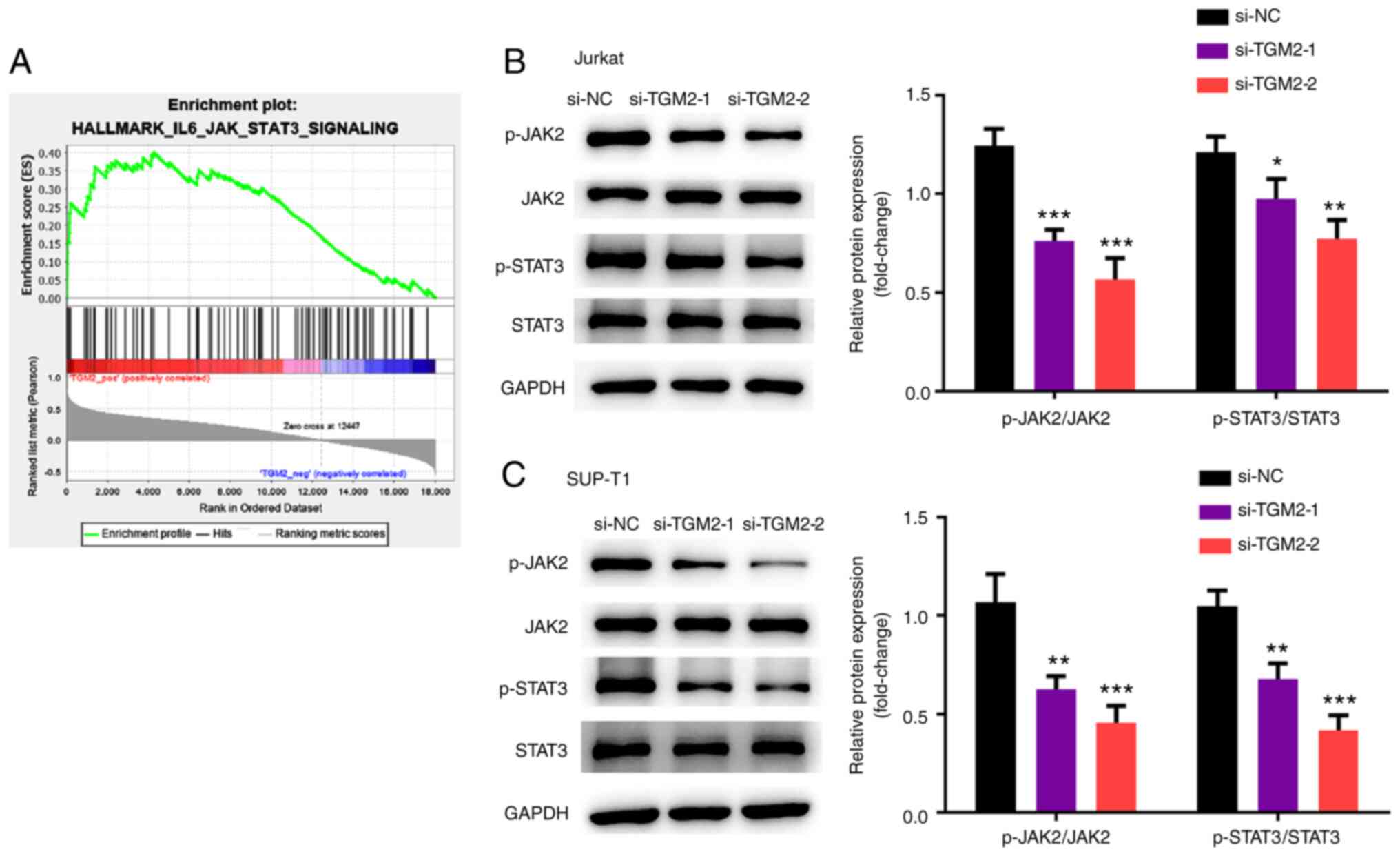
IL-6/JAK/STAT3 signalling is a
downstream pathway of TGM2
The mechanism underlying TGM2-mediated T-cell
lymphoma was studied. To this end, bioinformatics analysis based on
microarray data using the GSEA software was performed. GO and KEGG
pathway enrichment analyses were conducted using the
ClusterProfiler package, and the activation and suppression
pathways of TGM2 were analysed. The IL-6/JAK/STAT3 pathway was
predicted to be activated by TGM2 and was selected for subsequent
analysis (Fig. 4A). Subsequently,
it was confirmed that TGM2 siRNAs suppressed the phosphorylation of
JAK2 and STAT3 in Jurkat and SUP-T1 cells (P<0.05; Fig. 4B and C). Collectively, these data
suggested that IL-6/JAK/STAT3 signalling may be a downstream
pathway of TGM2.
TGM2 siRNAs suppress T-cell lymphoma
by regulating the IL-6/JAK/STAT3 pathway
The present study validated whether TGM2 affected
T-cell lymphoma by regulating the IL-6/JAK/STAT3 pathway. TGM2
siRNAs suppressed the number of EdU-positive cells and the
colony-forming ability of Jurkat and SUP-T1 cells, whereas the
IL-6/JAK/STAT3 activator IL-6 reversed these effects and the
IL-6/JAK/STAT3 inhibitor AG490 enhanced these effects (P<0.05;
Fig. 5A and B). Furthermore,
apoptosis of Jurkat and SUP-T1 cells was induced by TGM2 siRNAs,
whereas IL-6 reversed it; however, AG490 enhanced the effect of
TGM2 siRNAs on apoptosis (P<0.05; Fig. 5C). Taken together, these results
suggested that TGM2 siRNAs may suppress T-cell lymphoma by
regulating the IL-6/JAK/STAT3 pathway.
Discussion
T-LBL is a malignant tumour of precursor T cells and
accounts for ~90% of lymphoblastic lymphoma cancer cases worldwide
(1). In the present study, an
overlap analysis based on the upregulated genes from the GSE132550
and GSE143382 datasets was performed. Eight overlapping genes were
identified, namely MMP12, CCL19, MMP9, CD180, CCL18, FPR3, TNC and
TGM2. Among these genes, the roles of MMP12, CCL19, MMP9, CD180 and
CCL18 have been well established in various types of cancer,
including lymphoma (22–24). FPR3 is a member of the formyl
peptide receptor family, which is mainly expressed in bone marrow
cells and mature neutrophils (25). Its main function is relative to
immune responses (25) and less
is currently known regarding its role in cancer. TNC is a member of
the extracellular matrix (ECM) protein family, the main function of
which is tissue regeneration via its participation in wound healing
and inflammation (26). Since TNC
is a glycoprotein of the ECM, its roles in several solid tumours
have been demonstrated (27).
TGM2 is a member of the transglutaminase family, and its effects on
various types of cancer have been previously reported, including
osteosarcoma (8), CRC (7), glioma (9) and endometrial cancer (10). However, less is currently known
about its role in lymphoma; therefore, TGM2 was selected for
further studies. The present study identified the innovative role
of TGM2 in T-cell lymphoma proliferation and apoptosis.
TGM2 has been shown to participate in the modulation
of cancer development. It has been reported that TGM2 may serve as
a biomarker for cisplatin resistance in non-small cell lung cancer
(28). In a previous study,
inhibition of TGM2 suppressed docetaxel resistance and
epithelial-to-mesenchymal transition in breast cancer (29). Furthermore, kaempferol has been
reported to promote reactive oxygen species-related apoptosis via
the TGM2-regulated Akt/mTOR pathway in pancreatic cancer cells
(30). TGM2 was also revealed to
be correlated with the prognosis of CRC and may serve as a
potential treatment target (31).
In addition, TGM2 depletion can inhibit cisplatin chemoresistance
in osteosarcoma cells (8), and
TGM2 may modulate apoptosis and angiogenesis via Wnt/β-catenin
signalling in CRC (7).
Furthermore, microRNA (miR)-532-3p has been shown to attenuate the
progression of CRC by targeting the ETS1/TGM2-regulated
Wnt/β-catenin pathway (32).
These previous studies suggested the key roles of TGM2 in human
cancer. In the present study, TGM2 was revealed to be highly
expressed in FFPE skin biopsies from patients with T-cell lymphoma
(n=36) relative to the skin tissue from healthy individuals (n=36).
The mRNA and protein expression levels of TGM2 were also higher in
T-cell lymphoma cell lines compared with those in CD4+ T
cells. Silencing of TGM2 via siRNA transfection decreased cell
viability, EdU-positive numbers and colony formation of T-cell
lymphoma cells. In addition, silencing of TGM2 was able to enhance
the apoptosis of T-cell lymphoma cells potentially by regulating
the cleavage of caspase-3 and PARP. These data indicated a novel
function of TGM2 in regulating proliferation and apoptosis in
T-cell lymphoma, suggesting that TGM2 may be considered a new
therapeutic target of T-cell lymphoma. The clinical significance of
TGM2 needs to be confirmed by further studies.
The function of the IL-6/JAK/STAT3 pathway in
lymphoma has been widely studied. It has been reported that
JAK/STAT3 signalling is activated in mantle cell lymphoma (33). Furthermore, benzoxathiole was
shown to inhibit the proliferation and survival of lymphoma cells
by inactivation of the JAK3/STAT3 pathway (34). In a previous study, miR-155
modulated the apoptosis and proliferation of lymphoma cells by
regulating JAK/STAT3 signalling (35). Generation sequencing has also
revealed a novel STAT3-JAK2 fusion in anaplastic large cell
lymphomas (36). Furthermore,
inhibition of JAK3/STAT3 signalling may serve as a treatment option
for NK/T-cell lymphoma (37).
Kinase fusion and convergent mutations may result in activation of
STAT3 in the anaplastic large cell lymphoma (38). The results of the present study
revealed that the IL-6/JAK/STAT3 pathway may be a potential
downstream signalling pathway of TGM2. Silencing of TGM2 suppressed
the phosphorylation of JAK2 and STAT3 in T-cell lymphoma cells.
Further study confirmed that the silencing of TGM2 reduced the
number of EdU-positive cells and the colony-forming ability of
T-cell lymphoma cells, whereas the IL-6/JAK/STAT3 activator IL-6
blocked this effect and the IL-6/JAK/STAT3 inhibitor AG490 enhanced
it. Consistently, the apoptosis of T-cell lymphoma cells was
induced by TGM2 silencing, whereas IL-6 reversed it and AG490
enhanced the effect of TGM2 silencing on the cells. These findings
indicated that TGM2 may regulate T-cell lymphoma proliferation and
apoptosis, possibly through the activation of IL-6/JAK/STAT3
signalling. A previous study demonstrated that activation of
IL-6/IL-6 receptor/STAT3 can promote TGM2 expression to induce
epithelial-mesenchymal transition in hepatocellular carcinoma
(39). Although the results
cannot be compared, these findings indicate an association between
TGM2 and IL-6/JAK/STAT3 signalling in human cancer. IL-6/JAK/STAT3
signalling may be one of the downstream pathways of TGM2-mediated
cancer progression and other mechanisms should be explored in
future studies.
In conclusion, silencing of TGM2 inhibited the
growth of T-cell lymphoma by regulating IL-6/JAK/STAT3 signalling.
These findings indicated that TGM2 may function as a potential
therapeutic target for T-cell lymphoma.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YW and NZ designed the study. TS, YC and CL
performed the experiments. YW and HZ analysed the data. NZ prepared
the manuscript. YW and NZ confirm the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The protocol of this research has been approved by
the Ethics Committee of Shengli Oilfield Central Hospital (approval
no. DYSLYT20191201; Dongying, China). All patients have signed
written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cortelazzo S, Ferreri A, Hoelzer D and
Ponzoni M: Lymphoblastic lymphoma. Crit Rev Oncol Hematol.
113:304–317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Burkhardt B and Hermiston ML:
Lymphoblastic lymphoma in children and adolescents: Review of
current challenges and future opportunities. Br J Haematol.
185:1158–1170. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Raetz EA and Teachey DT: T-cell acute
lymphoblastic leukemia. Hematology Am Soc Hematol Educ Program.
2016:580–588. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leicht DT, Kausar T, Wang Z, Ferrer-Torres
D, Wang TD, Thomas DG, Lin J, Chang AC, Lin L and Beer DG: TGM2: A
cell surface marker in esophageal adenocarcinomas. J Thorac Oncol.
9:872–881. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tovar-Vidales T, Clark AF and Wordinger
RJ: Transforming growth factor-beta2 utilizes the canonical
Smad-signaling pathway to regulate tissue transglutaminase
expression in human trabecular meshwork cells. Exp Eye Res.
93:442–451. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lai TS and Greenberg CS: TGM2 and
implications for human disease: Role of alternative splicing. Front
Biosci (Landmark Ed). 18:504–519. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang P, Yu D, Zhou J, Zhuang S and Jiang
T: TGM2 interference regulates the angiogenesis and apoptosis of
colorectal cancer via Wnt/β-catenin pathway. Cell Cycle.
18:1122–1134. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li C, Cai J, Ge F and Wang G: TGM2
knockdown reverses cisplatin chemoresistance in osteosarcoma. Int J
Mol Med. 42:1799–1808. 2018.PubMed/NCBI
|
|
9
|
Fu J, Yang QY, Sai K, Chen FR, Pang JC, Ng
HK, Kwan AL and Chen ZP: TGM2 inhibition attenuates ID1 expression
in CD44-high glioma-initiating cells. Neuro Oncol. 15:1353–1365.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Torres A, Pac-Sosińska M, Wiktor K,
Paszkowski T, Maciejewski R and Torres K: CD44, TGM2 and EpCAM as
novel plasma markers in endometrial cancer diagnosis. BMC Cancer.
19:4012019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang H and McCarty N: Tampering with
cancer chemoresistance by targeting the TGM2-IL6-autophagy
regulatory network. Autophagy. 13:627–628. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hunter CA and Jones SA: IL-6 as a keystone
cytokine in health and disease. Nat Immunol. 16:448–457. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
West AJ, Tsui V, Stylli SS, Nguyen HPT,
Morokoff AP, Kaye AH and Luwor RB: The role of interleukin-6-STAT3
signalling in glioblastoma. Oncol Lett. 16:4095–4104.
2018.PubMed/NCBI
|
|
14
|
Burger R: Impact of interleukin-6 in
hematological malignancies. Transfus Med Hemother. 40:336–343.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Heinrich PC, Behrmann I, Haan S, Hermanns
HM, Muller-Newen G and Schaper F: Principles of interleukin
(IL)-6-type cytokine signalling and its regulation. Biochem J.
374:1–20. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kim JH, Kim WS and Park C: Interleukin-6
mediates resistance to PI3K-pathway-targeted therapy in lymphoma.
BMC Cancer. 19:9362019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu J, Hong J, Ahn KS, Go J, Han H, Park
J, Kim D, Park H, Koh Y, Shin DY and Yoon SS: ERK-dependent IL-6
positive feedback loop mediates resistance against a combined
treatment using danusertib and BKM120 in Burkitt lymphoma cell
lines. Leuk Lymphoma. 60:2532–2540. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Etebari M, Navari M, Agostinelli C, Visani
A, Peron C, Iqbal J, Inghirami G and Piccaluga PP: Transcriptional
analysis of lennert lymphoma reveals a unique profile and
identifies novel therapeutic targets. Front Genet. 10:7802019.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nielsen PR, Eriksen JO, Lindahl LM,
Wehkamp U, Bzorek M, Andersen G, Woetmann A, Iversen L, Ødum N,
Litman T and Gjerdrum LMR: Diagnostic two-gene classifier in
early-stage mycosis fungoides: A retrospective multicenter study. J
Invest Dermatol. 141:213–217.e5. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu G, Wang LG, Han Y and He QY:
ClusterProfiler: An R package for comparing biological themes among
gene clusters. Omics. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gobin E, Bagwell K, Wagner J, Mysona D,
Sandirasegarane S, Smith N, Bai S, Sharma A, Schleifer R and She
JX: A pan-cancer perspective of matrix metalloproteases (MMP) gene
expression profile and their diagnostic/prognostic potential. BMC
Cancer. 19:5812019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Korbecki J, Kojder K, Barczak K and
Simińska D: Hypoxia alters the expression of CC chemokines and CC
chemokine receptors in a tumor-A literature review. Int J Mol Sci.
21:56472020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mayeur-Rousse C, Guy J, Miguet L, Bouyer
S, Geneviève F, Robillard N, Solly F, Maar A, Bené MC and Mauvieux
L; GEIL (Groupe d'Etude Immunologique des Leucémies), . CD180
expression in B-cell lymphomas: A multicenter GEIL study. Cytometry
B Clin Cytom. 90:462–466. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stempel H, Jung M, Pérez-Gómez A,
Leinders-Zufall T, Zufall F and Bufe B: Strain-specific loss of
formyl peptide receptor 3 in the murine vomeronasal and immune
systems. J Biol Chem. 291:9762–9775. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tucker RP and Chiquet-Ehrismann R: The
regulation of tenascin expression by tissue microenvironments.
Biochim Biophys Acta. 1793:888–892. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lowy CM and Oskarsson T: Tenascin C in
metastasis: A view from the invasive front. Cell Adh Migr.
9:112–124. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Park KS, Kim HK, Lee JH, Choi YB, Park SY,
Yang SH, Kim SY and Hong KM: Transglutaminase 2 as a cisplatin
resistance marker in non-small cell lung cancer. J Cancer Res Clin
Oncol. 136:493–502. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
He W, Sun Z and Liu Z: Silencing of TGM2
reverses epithelial to mesenchymal transition and modulates the
chemosensitivity of breast cancer to docetaxel. Exp Ther Med.
10:1413–1418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang F, Wang L, Qu C, Chen L, Geng Y,
Cheng C, Yu S, Wang D, Yang L, Meng Z and Chen Z: Kaempferol
induces ROS-dependent apoptosis in pancreatic cancer cells via
TGM2-mediated Akt/mTOR signaling. BMC Cancer. 21:3962021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Miyoshi N, Ishii H, Mimori K, Tanaka F,
Hitora T, Tei M, Sekimoto M, Doki Y and Mori M: TGM2 is a novel
marker for prognosis and therapeutic target in colorectal cancer.
Ann Surg Oncol. 17:967–972. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gu C, Cai J, Xu Z, Zhou S, Ye L, Yan Q,
Zhang Y, Fang Y, Liu Y, Tu C, et al: MiR-532-3p suppresses
colorectal cancer progression by disrupting the ETS1/TGM2
axis-mediated Wnt/β-catenin signaling. Cell Death Dis. 10:7392019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yared MA, Khoury JD, Medeiros LJ,
Rassidakis GZ and Lai R: Activation status of the JAK/STAT3 pathway
in mantle cell lymphoma. Arch Pathol Lab Med. 129:990–996. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kim BH, Min YS, Choi JS, Baeg GH, Kim YS,
Shin JW, Kim TY and Ye SK: Benzoxathiol derivative BOT-4-one
suppresses L540 lymphoma cell survival and proliferation via
inhibition of JAK3/STAT3 signaling. Exp Mol Med. 43:313–321. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li XD, Li XM, Gu JW and Sun XC: MiR-155
regulates lymphoma cell proliferation and apoptosis through
targeting SOCS3/JAK-STAT3 signaling pathway. Eur Rev Med Pharmacol
Sci. 24:75772020.PubMed/NCBI
|
|
36
|
Quesada AE, Zhang Y, Ptashkin R, Ho C,
Horwitz S, Benayed R, Dogan A and Arcila ME: Next generation
sequencing of breast implant-associated anaplastic large cell
lymphomas reveals a novel STAT3-JAK2 fusion among other activating
genetic alterations within the JAK-STAT pathway. Breast J.
27:314–321. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Liu J, Liang L, Li D, Nong L, Zheng Y,
Huang S, Zhang B and Li T: JAK3/STAT3 oncogenic pathway and PRDM1
expression stratify clinicopathologic features of extranodal
NK/T-cell lymphoma, nasal type. Oncol Rep. 41:3219–3232.
2019.PubMed/NCBI
|
|
38
|
Crescenzo R, Abate F, Lasorsa E, Tabbo' F,
Gaudiano M, Chiesa N, Di Giacomo F, Spaccarotella E, Barbarossa L,
Ercole E, et al: Convergent mutations and kinase fusions lead to
oncogenic STAT3 activation in anaplastic large cell lymphoma.
Cancer Cell. 27:516–532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jia C, Wang G, Wang T, Fu B, Zhang Y,
Huang L, Deng Y, Chen G, Wu X, Chen J, et al: Cancer-associated
Fibroblasts induce epithelial-mesenchymal transition via the
Transglutaminase 2-dependent IL-6/IL6R/STAT3 axis in hepatocellular
carcinoma. Int J Biol Sci. 16:2542–2558. 2020. View Article : Google Scholar : PubMed/NCBI
|