Introduction
The entry of the severe acute respiratory syndrome
(SARS) coronavirus-2 (SARS-CoV-2) into cells is facilitated by its
spike (S) proteins, mainly via binding to key cell entry mediators,
such as the angiotensin-converting enzyme 2 (ACE2) (1,2).
In addition, the S proteins of SARS-CoV-2 are also primed/activated
by the transmembrane protease serine 2 and 4 (TMPRSS2 and TMPRSS4),
which appear to play a crucial role in the tropism of this virus
and the multi-organ infection in the context of coronavirus disease
2019 (COVID-19) (1,3,4).
Indeed, TMPRSS4, along with TMPRSS2, have been found to be capable
of activating the viral S proteins, consequently increasing the
SARS-CoV-2 infection of human enterocytes in the small intestine
(5). Accordingly, recent data
demonstrate the abundant expression of ACE2, TMPRSS2 and TMPRSS4 in
enterocytes of the lower gastrointestinal (GI) tract (6). Of note, COVID-19 can also present
with a wide spectrum GI symptoms, including diarrhoea, nausea and
vomiting (7–9).
Moreover, based on data derived from a systematic
review and meta-analysis for the risk and prognosis of patients
with COVID-19 (10,11), cancer has been identified as an
independent risk factor which holds a positive association with
severe COVID-19 infection and related adverse clinical outcomes
(12). In accordance with this
finding, previous studies, including data from the authors'
research group, have documented the differential expression of
SARS-CoV-2 infection host cell entry mediators in malignancies
(4,13). Indeed, the authors have previously
demonstrated that TMPRSS4 is overexpressed in 11 types of cancer,
including lung adenocarcinoma (LUAD), lung squamous cell carcinoma
(LUSC), cervical squamous cell carcinoma, thyroid carcinoma,
ovarian cancer, colorectal cancer, pancreatic cancer,
adenocarcinoma of the stomach, uterine carcinosarcoma and uterine
endometrial carcinoma (14).
Furthermore, a recent study on patients with chronic obstructive
pulmonary disease (COPD) also demonstrated that TMPRSS4 gene
expression was elevated in epithelial brushes and bronchial
biopsies of these patients compared to the controls (15).
The present study aimed to expand on previous
observations regarding the role of TMPRSS4 in SARS-CoV-2 infection
and to provide novel evidence of the abundant protein expression of
TMPRSS4 in both the GI tract and in COVID-19-affected lungs.
Patients and methods
Patient selection, autopsy and sample
acquisition
The lung tissue of 1 patient (77-year-old male with
advanced poorly differentiated bronchial carcinoma under radiation
therapy) with non-small cell lung carcinoma (NSCLC) with concurrent
COVID-19 infection was retrieved from a clinical autopsy (the
specimen was obtained at the Institute of Pathology ‘Pathologie
Grünstrasse’, Düsseldorf, Germany). Appropriate written consent of
the patient/next of kin and ethical approval were granted by the
Ethics Committee of Hannover Medical School (ethics reference no.
9621_BO_K_2021). The autopsy [77-year-old male with the following
comorbidities: COPD, heart-hypertrophy, bronchial carcinoma (NSCLC)
after radiation treatment] was performed in a room with adequate
airflow (>6 air changes per hour of total room volume) at
conditions similar to recommendations for autopsies of suspected
Creutzfeldt-Jakob disease (i.e., hazmat suits, boots, goggles and
FFP2/3 masks) with an in corpore technique analogous to that
used in forensic institutions. The thoracic organs were
eviscerated, and the heart was separately dissected in the
direction of blood flow. The lungs, trachea and larynx were wholly
exenterated and perfused via the trachea with phosphate-buffered
formalin. The trachea was then closed with a clamp and the
specimens were left in formalin at room temperature for 72 h prior
to further dissection. The lungs were subsequently cut into
0.5-1-cm-thick parasagittal slices and examined macroscopically.
The areas of interest (tumour tissue, lung tissue adjacent to the
tumour and peripheral lung tissue) for histology were identified
and fitting tissue samples embedded in paraffin and cut into
5-µm-thick slices on coated glass slides (SuperfrostPlus, Gerhard
Menzel B.V. & Co. KG) for immunohistochemical staining. The
included COVID-19 specimen was positively PCR-tested on a nasal
swab, as well as lung tissue, and was found to be positive from the
reverse transcription-PCR (RT-PCR) of formalin-fixed paraffin
embedded tissue.
Paraffin-embedded tissue microarray slides, each
containing 48 cores, were also purchased from US Biomax (US Biomax,
Inc.; cat. nos. BN114c32 and LC241L; Tables SI and SII). All tissue samples were collected
under the highest ethical standards with the donors giving informed
consent [under Health Insurance Portability and Accountability Act
(HIPAA)-approved protocols]. Additional control samples of lung
tissue were retrieved from archived routine samples from Hannover
Medical School (ethics reference no. 1741–2013).
Immunohistochemistry
The slides were deparaffinised and rehydrated,
followed by antigen retrieval. Briefly, 100 ml sodium citrate
(Thermo Fisher Scientific, Inc.) was heated in the microwave for 2
min, followed by the addition of slides and further heating at
1-min intervals for 10 min (keeping the slides just below boiling
temperature, at 90°C). Blocking was performed using 5% BSA (Thermo
Fisher Scientific, Inc.) in PBS for 40 min at room temperature.
This was followed by overnight incubation at 4°C with primary
rabbit monoclonal antibodies for TMPRSS4 (cat. no. ab188816, Abcam;
1:500 dilution 5% BSA in PBS). Following three washes at room
temperature with PBS (10 min each), the slides were incubated with
secondary antibody (1:200 in 1% rabbit serum; ZytoChem Plus HRP-DAB
kit, rabbit, cat. no. HRP008DAB-RB, Zytomed Systems GmbH) for 60
min at room temperature. After washing with 0.025% Triton X-100, to
remove any unbound secondary antibody, streptavidin-HRP conjugate
from the same kit was added to the slides and left to incubate at
room temperature for 30 min in a humidity chamber. At room
temperature, the slides were then washed and subjected to
3,3′-diaminobenzidine (DAB) (Vector Laboratories, Inc.) staining
for 5 min, counterstained with haematoxylin (Merck KGaA) for ~10
sec and washed with 0.1% sodium bicarbonate at room temperature.
The slides were then analysed for the immunoreactivity of TMPRSS4
protein using a light microscope (Carl Zeiss AG). A
pheochromocytoma (adrenal gland) core was used as a positive
control (Fig. S1A; part of the
array #BN114c32), and an ovarian cancer core (part of the array
#BC11115d045) where the primary antibody was omitted was used as a
negative control (Fig. S1B).
UALCAN database and statistical
analysis
UALCAN (http://ualcan.path.uab.edu/), an online transcriptomic
database, was used to investigate the mRNA expression levels of
TMPRSS4 for comparisons between LUAD and LUSC and normal tissues,
as well as stratifying for sex as a clinicopathological parameter.
The method used for differential analysis in the present study was
one-way ANOVA, using disease state (tumour or normal) as variables
for calculating differential expression: Gene expression-disease
state. A P-value <0.05 was considered to indicate a
statistically significant difference.
Results
TMPRSS4 expression in the GI tract
tissue microarray
In a recent study, SARS-CoV-2 virions were
identified in the intestines and colon tissue of 2 patients using
transmission electron microscopy, suggesting a causal role of this
virus in bowel damage (16).
Using immunohistochemistry, the present study demonstrated that
TMPRSS4 was abundantly expressed at the protein level in the
oesophagus, stomach, small intestine, jejunum, ileum, colon, liver
and pancreas (Fig. 1). In the
oesophagus, staining was evident in epithelial cells, submucosal
glands and the lower muscularis mucosae. This is of increasing
importance given oesophageal mucosal lesions caused by SARS-CoV-2
have been shown to result in upper GI bleeding in a 77-year-old man
(17). Another study also
presented the case of a patient with COVID-19 with acute
oesophageal necrosis (18). In
the present study, in the small intestine, there was weak staining
of the cytoplasm in goblet cells and enterocytes. In the jejunum
and ileum, cytoplasmic expression was more pronounced compared to
the large intestine. The colonic mucosa exhibited cytoplasmic and
weak nuclear membrane staining of goblet cells and the adjacent
enterocytes (Fig. S1C for higher
magnification). In the stomach, the gastric fundic mucosa exhibited
positivity in the surface epithelial cells and the mucinous
parietal cells. Finally, the observations of the expression of
TMPRSS4 were expanded to the liver and pancreas. The liver
exhibited the staining of hepatocytes together with bile ducts and
ductules, whereas in the pancreas, pronounced cytoplasmic staining
of the acinar cells was evident.
TMPRSS4 expression in lungs of
patients with NSCLC and in a patient with COVID-19 with COPD
It is currently known that COVID-19 severity
increases in patients with lung cancer (19). Moreover, previous meta-analyses
have revealed a higher mortality rate in patients with COVID-19 and
cancer (20–22). In a previous study, the authors
demonstrated that the TMPRSS4 gene was significantly upregulated in
LUAD and in LUSC (14). The
present study determined the protein expression of TMPRSS4 in 4
patients with NSCLC; 2 patients diagnosed with LUAD and 2 patients
with LUSC (Fig. 2). The
expression of TMPRSS4 was evident in the tumour cells of patients
with LUSC (Fig. 2A and C) and
LUAD (Fig. 2E and G) and to a
varying degree in the peritumoral stroma. Of note, TMPRSS4 was also
highly expressed in tumour adjacent morphological normal lung
tissue, particularly on bronchial and alveolar epithelial cells
(Fig. 2B,D,F and H). This finding
corroborates mRNA data demonstrating a similar gene expression of
TMPRSS4 in males and females in LUAD and LUSC (Fig. S1D and E).
 | Figure 2.Immunohistochemical staining for
TMPRSS4 in human non-small cell lung cancer at ×20 magnification.
High expression of TMPRSS4 in LUSC cells (black arrowhead) with a
lesser expression in (A) the adjacent peritumoral tissue, and a
high expression in alveolar epithelial cells (red arrowhead) in (B)
the NAT of a male patient (53 years of age, T2N0M0; grade 2, stage
IB). (C) Lesser, but detectable staining of TMPRSS4 in lung
squamous cell carcinoma cells (black arrowhead) with minor staining
in the adjacent peritumoral tissue, and (D) a high expression on
alveolar epithelial cells (red arrowhead) in the NAT of a female
patient (56 years of age, T2N0M0; grade 3, stage IIIA). Protein
expression in lung adenocarcinoma cells (black arrowhead) with (E)
a lesser expression in the adjacent peritumoral tissue (LUAD) and
(F) a high expression on alveolar epithelial cells (red arrowhead)
in cancer adjacent lung tissue (AT) of a male patient (65 years of
age, T2N2M0, grade 3, stage IIIA). (G) Expression in LUAD (black
arrowhead) and a (H) high expression on both bronchial (blue
arrowhead) and alveolar epithelial cells (red arrowhead) in cancer
adjacent lung tissue of a female patient (35 years of age, T4N1M0,
grade 3, stage IIIA). Scale bar, 500 nm. TMPRSS4, transmembrane
protease serine 4; LUSC, lung squamous cell carcinoma; LUAD, lung
adenocarcinoma; NAT, normal adjacent tissue; AT, adjacent
tissue. |
A recent systematic review and meta-analysis
demonstrated that patients with COVID-19 with COPD have a
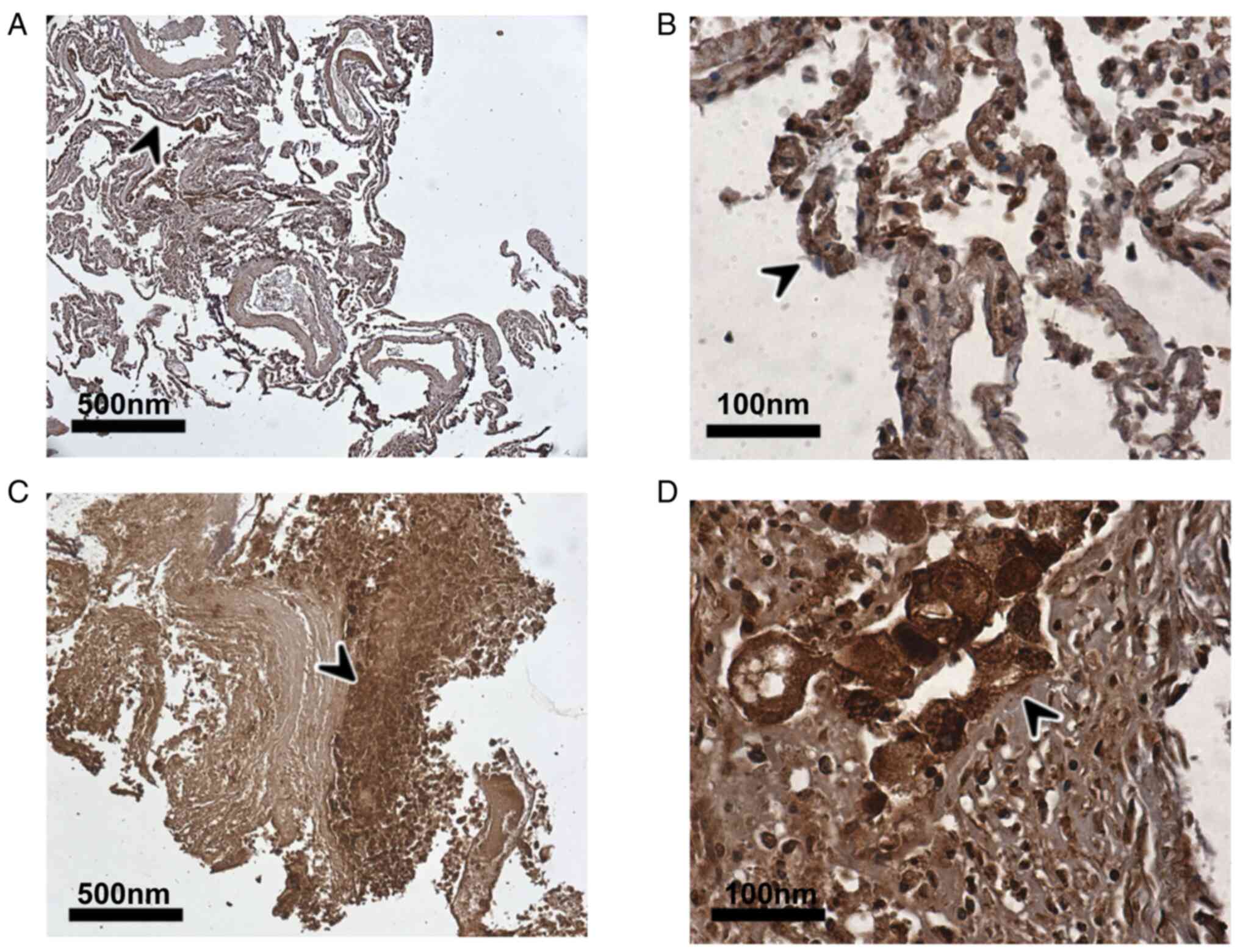
significantly increased risk of poor clinical outcomes (23). The present study demonstrates for
the first time, to the best of our knowledge, the extensive protein
expression of TMPRSS4 in tumour cells in the lungs of a deceased
patient with COVID-19 with COPD and bronchial carcinoma (Fig. 3C and D), as well as in alveolar
epithelial cells of the adjacent non-tumour tissue (Fig. 3A and B), comparable to the results
obtained for the LUAD and LUSC samples without COVID-19
infection.
Discussion
The present study provides novel (to the best of our
knowledge), comprehensive evidence regarding the protein
distribution of TMPRSS4 in the GI tract and lungs. Recently, two
different groups described the involvement of TMPRSS4 as a
SARS-CoV-2 entry mediator in the GI tract (5,6),
suggesting that a leaky gut may allow SARS-CoV-2 to spread to other
organs (e.g., the liver) (5).
Based on the findings of the present study using a tissue
microarray, evidence of TMPRSS4 protein expression across the GI
tract (oesophagus, stomach, small intestine and colon), as well as
in the pancreas and liver is provided. In relation to the latter,
it is noteworthy that up to 70% of patients with COVID-19 exhibit
abnormal liver function tests upon admission to hospital, a finding
which is transient in the majority of cases (24). As such, this high hepatic TMPRSS4
protein expression suggests that this mediator may be involved in
the SARS-CoV-2 infectivity of the liver, explaining, at least in
part, why immunocompromised patients with liver cirrhosis or
hepatic cancer are more susceptible to COVID-19 infection (25,26). In addition to the liver, recent
research interests have also focused on the involvement of the
pancreas in the context of COVID-19, particularly since it has been
suggested that pancreatic ACE2 expression can cause damage of this
vital exocrine and endocrine organ following SARS-CoV-2 infection
(27). Indeed, an association
between pancreatitis and COVID-19 has been reported (28). In accordance with such findings,
the present study provides novel data (at least to the best of our
knowledge) on widespread TMPRSS4 protein expression in the
pancreas. Given that the main cell entry mediator (ACE2) is present
in the pancreas, it is plausible that the co-expression of these
two SARS-CoV-2 cell entry mediators may facilitate increased local
viral infectivity, which can lead to pancreatic damage. Of note,
previous research on SARS-CoV has demonstrated that this
coronavirus can bind to ACE2 and damage the pancreatic islets
(endocrine portion of the pancreas), thus causing acute diabetes
(29).
Furthermore, the authors, as well as other
researchers have demonstrated that TMPRSS4 is overexpressed in LUAD
and LUSC, which are both conditions that predispose to cases of
severe COVID-19 infection (30–33). Recently, the cellular distribution
of ACE2, TMPRSS2 and TMPRSS4 was examined in normal and LUAD
samples using single-cell RNA-sequencing (33). Han et al (33) demonstrated that TMPRSS4 expression
was highest and most frequently detected (75%) in malignant
pulmonary cells, primarily of epithelial origin. Notably, ACE2
appears to be co-expressed in the same cells in LUAD, alluding to
potential higher infectivity, but also to implications for the
management of patients with lung cancer and severe COVID-19.
Moreover, as aforementioned, patients with COPD are also at higher
risk of poor COVID-19-related outcomes. The exact mechanisms
contributing to this higher risk are still under investigation,
with data demonstrating that both ACE2 and TMPRSS4 expression
levels are upregulated in the epithelial brushing and bronchial
biopsy samples of patients with COPD (15). The present study demonstrated the
abundant expression of TMPRSS4 in the lung tissue (both
adenocarcinoma and normal adjacent lung tissue) of a patient with
COVID-19 with COPD and cardiac hypertrophy, who succumbed to the
disease.
However, it should be acknowledged that there are a
number of limitations to the present study. The present study did
not use an in vitro model to further study the interactions
between TMPRSS4 and other SARS-CoV-2 cell entry mediators. In
addition, a quantitative analysis of the protein expression on
tissue microarrays was not performed. Finally, there was a limited
number of patients used in the present study.
In conclusion, the findings of the present study
demonstrate the widespread protein expression of TMPRSS4 in the GI
tract, liver and pancreas, and provide further evidence regarding
the expression of TMPRSS4 in the lungs. Indeed, collectively, the
data of the present data study suggest that TMPRSS4 may be
implicated in both the pulmonary and extra-pulmonary COVID-19
symptomatology/manifestations, functioning as another host cell
SARS-CoV-2 entry mediator, contributing to the tropism of this new
coronavirus. The pharmacological inhibition of viral entry via
TMPRSS4 may thus present an interesting additional target for the
development of antiviral agents; however, additional studies using
larger cohorts of patients with COVID-19 and lung carcinoma
patients are required.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was funded by the Cancer Treatment and
Research Trust and University Hospitals Coventry and Warwickshire
NHS Trust (grant no. 12899).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HSR, IK, EK, DJ, DAS and CW were involved in the
conceptualization of the study. RK, PK, CW, PJ and JLR were
involved in the study methodology (immunohistochemistry and
statistical analyses). RK, PK, DJ, PJ, CW and EK were involved in
formal analysis. RK, IK, JLR and EK were involved in the writing
and preparation of the original draft. RK, HSR, DJ, CW, JLR, PK,
PJ, IK, DAS and EK were involved in the writing, reviewing and
editing of the manuscript. DJ and EK supervised the study. EK was
involved in project administration. HSR and DJ was involved in
funding acquisition. EK and IK are the guarantors of this work and,
as such, had full access to all the data in the study and take
responsibility for the integrity of the data and the accuracy of
the data analysis. EK and IK confirm the authenticity of all the
raw data. All authors have read and agreed to the published version
of the manuscript.
Ethics approval and consent to
participate
The present study was conducted according to the
guidelines of the Declaration of Helsinki. Appropriate patient/next
of kin written consent and ethical approval were ensued by the
Ethics Committee of Hannover Medical School (OE 9515; ethics
reference no. 9621_BO_K_2021); Institute for Pathology, Hannover
Medical School, Hannover, Germany. All tissue samples were
collected under the highest ethical standards with the donors
giving informed consent (under Health Insurance Portability and
Accountability Act (HIPAA) approved protocols). Additional control
samples of lung tissue were retrieved from archived routine samples
from Hannover Medical School (ethics reference no. 1741–2013).
Patient consent for publication
Not applicable.
Competing interests
DAS is the Editor-in-Chief for the journal, but had
no personal involvement in the reviewing process, or any influence
in terms of adjudicating on the final decision, for this article.
The other authors declare that they have no competing
interests.
References
|
1
|
Hoffmann M, Kleine-Weber H, Schroeder S,
Kruger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH,
Nitsche A, et al: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2
and is blocked by a clinically proven protease inhibitor. Cell.
181:271–280.e8. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara
H, Geng Q, Auerbach A and Li F: Structural basis of receptor
recognition by SARS-CoV-2. Nature. 581:221–224. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Iwata-Yoshikawa N, Okamura T, Shimizu Y,
Hasegawa H, Takeda M and Nagata N: TMPRSS2 contributes to virus
spread and immunopathology in the airways of murine models after
coronavirus infection. J Virol. 93:e01815–18. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katopodis P, Anikin V, Randeva HS,
Spandidos DA, Chatha K, Kyrou I and Karteris E: Pan-cancer analysis
of transmembrane protease serine 2 and cathepsin L that mediate
cellular SARS-CoV-2 infection leading to COVID-19. Int J Oncol.
57:533–539. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zang R, Gomez Castro MF, McCune BT, Zeng
Q, Rothlauf PW, Sonnek NM, Liu Z, Brulois KF, Wang X, Greenberg HB,
et al: TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human
small intestinal enterocytes. Sci Immunol. 5:eabc35822020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee JJ, Kopetz S, Vilar E, Shen JP, Chen K
and Maitra A: Relative Abundance of SARS-CoV-2 entry genes in the
enterocytes of the lower gastrointestinal tract. Genes (Basel).
11:6452020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ng SC and Tilg H: COVID-19 and the
gastrointestinal tract: More than meets the eye. Gut. 69:973–974.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Almeida JFM and Chehter EZ: COVID-19 and
the gastrointestinal tract: What do we already know? Einstein (Sao
Paulo). 18:eRW59092020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Syed A, Khan A, Gosai F, Asif A and
Dhillon S: Gastrointestinal pathophysiology of SARS-CoV2-a
literature review. J Community Hosp Intern Med Perspect.
10:523–528. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
ElGohary GM, Hashmi S, Styczynski J,
Kharfan-Dabaja MA, Alblooshi RM, de la Cámara R, Mohmed S,
Alshaibani A, Cesaro S, Abd El-Aziz N, et al: The risk and
prognosis of COVID-19 infection in cancer patients: A systematic
review and meta-analysis. Hematol Oncol Stem Cell Ther. Jul
30–2020.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tian Y, Qiu X, Wang C, Zhao J, Jiang X,
Niu W, Huang J and Zhang F: Cancer associates with risk and severe
events of COVID-19: A systematic review and meta-analysis. Int J
Cancer. 148:363–374. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang K, Sheng Y, Huang C, Jin Y, Xiong N,
Jiang K, Lu H, Liu J, Yang J, Dong Y, et al: Clinical
characteristics, outcomes, and risk factors for mortality in
patients with cancer and COVID-19 in Hubei, China: A multicentre,
retrospective, cohort study. Lancet Oncol. 21:904–913. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chai P, Yu J, Ge S, Jia R and Fan X:
Genetic alteration, RNA expression, and DNA methylation profiling
of coronavirus disease 2019 (COVID-19) receptor ACE2 in
malignancies: A pan-cancer analysis. J Hematol Oncol. 13:432020.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Katopodis P, Kerslake R, Davies J, Randeva
HS, Chatha K, Hall M, Spandidos DA, Anikin V, Polychronis A,
Robertus JL, et al: COVID-19 and SARS-CoV-2 host cell entry
mediators: Expression profiling of TMRSS4 in health and disease.
Int J Mol Med. 47:642021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Watson A, Öberg L, Angermann B, Spalluto
CM, Hühn M, Burke H, Cellura D, Freeman A, Muthas D, Etal D, et al:
Dysregulation of COVID-19 related gene expression in the COPD lung.
Respir Res. 22:1642021. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Martin-Cardona A, Lloreta Trull J,
Albero-González R, Paraira Beser M, Andújar X, Ruiz-Ramirez P,
Tur-Martínez J, Ferrer C, De Marcos Izquierdo JA, Pérez-Madrigal A,
et al: SARS-CoV-2 identified by transmission electron microscopy in
lymphoproliferative and ischaemic intestinal lesions of COVID-19
patients with acute abdominal pain: Two case reports. BMC
Gastroenterol. 21:3342020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li X, Huang S, Lu J, Lai R, Zhang Z, Lin
X, Zheng X and Shan H: Upper gastrointestinal bleeding caused by
SARS-CoV-2 infection. Am J Gastroenterol. 115:1541–1542. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mustafa NF, Jafri NS, Holtorf HL and Shah
SK: Acute oesophageal necrosis in a patient with recent SARS-CoV-2.
BMJ Case Rep. 14:e2441642021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Luo J, Rizvi H, Preeshagul IR, Egger JV,
Hoyos D, Bandlamudi C, McCarthy CG, Falcon CJ, Schoenfeld AJ,
Arbour KC, et al: COVID-19 in patients with lung cancer. Ann Oncol.
31:1386–1396. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kulkarni AA, Wilson G, Fujioka N and Patel
MR: Mortality from COVID-19 in patients with lung cancer. J Cancer
Metastasis Treat. 7:312021.
|
|
21
|
Desai A, Gupta R, Advani S, Ouellette L,
Kuderer NM, Lyman GH and Li A: Mortality in hospitalized patients
with cancer and coronavirus disease 2019: A systematic review and
meta-analysis of cohort studies. Cancer. 127:1459–1468. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang H, Han H, He T, Labbe KE, Hernandez
AV, Chen H, Velcheti V, Stebbing J and Wong KK: Clinical
characteristics and outcomes of COVID-19-Infected cancer patients:
A systematic review and meta-analysis. J Natl Cancer Inst.
113:371–380. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gerayeli FV, Milne S, Cheung C, Li X, Yang
CWT, Tam A, Choi LH, Bae A and Sin DD: COPD and the risk of poor
outcomes in COVID-19: A systematic review and meta-analysis.
EClinicalMedicine. 33:1007892021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang C, Shi L and Wang FS: Liver injury
in COVID-19: Management and challenges. Lancet Gastroenterol
Hepatol. 5:428–430. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chan SL and Kudo M: Impacts of COVID-19 on
liver cancers: During and after the Pandemic. Liver Cancer.
9:491–502. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chai X, Hu L, Zhang Y, Han W, Lu Z, Ke A,
Zhou J, Shi G, Fang N, Fan J, et al: Specific ACE2 expression in
cholangiocytes may cause liver damage after 2019-nCoV Infection.
bioRxiv. https://doi.org/10.1101/2020.02.03.931766
|
|
27
|
Liu F, Long X, Zhang B, Zhang W, Chen X
and Zhang Z: ACE2 expression in pancreas may cause pancreatic
damage after SARS-CoV-2 infection. Clin Gastroenterol Hepatol.
18:2128–2130.e2. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
de-Madaria E and Capurso G: COVID-19 and
acute pancreatitis: Examining the causality. Nat Rev Gastroenterol
Hepatol. 18:3–4. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang JK, Lin SS, Ji XJ and Guo LM: Binding
of SARS coronavirus to its receptor damages islets and causes acute
diabetes. Acta Diabetol. 47:193–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
de Aberasturi AL, Redrado M, Villalba M,
Larzabal L, Pajares MJ, Garcia J, Evans SR, Garcia-Ros D, Bodegas
ME, Lopez L, et al: TMPRSS4 induces cancer stem cell-like
properties in lung cancer cells and correlates with ALDH expression
in NSCLC patients. Cancer Lett. 370:165–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Larzabal L, Nguewa PA, Pio R, Blanco D,
Sanchez B, Rodríguez MJ, Pajares MJ, Catena R, Montuenga LM and
Calvo A: Overexpression of TMPRSS4 in non-small cell lung cancer is
associated with poor prognosis in patients with squamous histology.
Br J Cancer. 105:1608–1614. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Voinsky I and Gurwitz D: Smoking and
COVID-19: Similar bronchial ACE2 and TMPRSS2 expression and higher
TMPRSS4 expression in current versus never smokers. Drug Dev Res.
Aug 5–2020.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Han G, Sinjab A, Hara K,
Treekitkarnmongkol W, Brennan P, Chang K, Bogatenkova E,
Sanchez-Espiridion B, Behrens C, Solis LM, et al: Single-Cell
expression landscape of SARS-CoV-2 Receptor ACE2 and host proteases
in normal and malignant lung tissues from pulmonary adenocarcinoma
patients. Cancers (Basel). 13:12502021. View Article : Google Scholar : PubMed/NCBI
|