Introduction
The brain utilizes high levels of energy to maintain
its physiological function. The mammalian brain depends on glucose
as its main source of energy but lacks oxygen and glucose reserves
and therefore requires a continuous supply of glucose from blood
(1). Two independent groups of
transporter proteins, glucose transporters (GLUTs) and solute
carrier family members, mediate glucose transport into the brain
(2). GLUT protein 1 (GLUT-1),
also called solute carrier family 2 member 1, was first identified
in a fetal skeletal muscle cell line (3) and is ubiquitously expressed in all
types of tissue (4). It is also
reported to be highly expressed in endothelial cells (ECs) of the
central nervous system and is considered to be the principal
glucose transporter of the blood-brain barrier (5). Functional deficiency of GLUT-1
decreases the amount of glucose available to brain cells, which
affects brain function (6).
Danshen (Salvia miltiorrhiza Bunge), a
medicinal herb, has been used in traditional Chinese medicine to
treat conditions associated with the cardiovascular and
cerebrovascular systems, such as stroke (7). Tanshinones are the primary active
component of Danshen. Tanshinone IIA (Tan 2A), the most abundant
diterpenoid quinone in S. miltiorrhiza, a natural inhibitor
of monoacylglycerol lipase, is a fat-soluble component and is
reported to exert anti-inflammation, anti-cancer, neuroprotection
and hypolipidemic activity (8,9),
and have a neuroprotective and cardiovascular protective effect.
Tan 2A increases blood flow in the heart and improves myocardial
metabolic disorder by increasing the tolerance of cardiomyocytes to
hypoxia (10). Tan 2A protects
neuron and microvascular ECs against hypoxia/ischemia both in
vitro and in vivo (11). Tan 2A sodium sulfonate decreases
atherosclerotic lesion area by inhibiting expression of
intracellular chloride channel 1 in ECs (12) and activating Kruppel-like factor 4
in macrophages (13,14). The combination of Danshen and
Sanqi (a Chinese herbal medicine) prescription has been reported to
markedly increase expression of liver glycogen synthesis genes,
such as GLUT-1, and improve fat and glucose metabolism (15). To the best of our knowledge,
however, whether GLUT-1 expression of ECs is regulated by Tan 2A is
not yet known. It was hypothesized that Tan 2A may induce glucose
uptake via regulation of GLUT1 expression in vascular ECs.
Previous studies have reported that
hypoxia-inducible factor 1 α (HIF-1α) may mediate the therapeutic
actions of Tan 2A (16,17). For example, Tan 2A is reported to
inhibit breast cancer growth by inhibiting HIF-1α expression via
the mammalian target of rapamycin signaling pathway (18). Tan 2A also blocks
epithelial-mesenchymal transition in breast cancer cell lines by
regulating the HIF-1α signaling pathway (19). Tan 2A decreases the inflammatory
response in LPS-induced lung injury via the HIF-1α signaling
pathway (20). Furthermore,
overexpression of GLUT-1 enhances the effect of Tan 2A on the
treatment of middle cerebral artery occlusion (21). The present study assessed the
regulatory effect of Tan 2A on expression of GLUT-1 in ECs and
evaluated whether the effect of Tan 2A was associated with
HIF-1α.
Materials and methods
Cell culture and reagents
Human umbilical vein endothelial cells (HUVECs) were
purchased from ScienCell Research Laboratories, Inc. and cultured
in EC medium containing EC growth supplement (ScienCell Research
Laboratories, Inc.), 5% fetal bovine serum (FBS; ScienCell Research
Laboratories, Inc.), 100 U/ml penicillin and 100 U/ml streptomycin
in 37°C. HUVECs at passages 3–5 were used for all experiments. Tan
2A was purchased from Selleck Chemicals (Cat. no. S2365, Lot no.
S2767130005001). DMSO (sigma-Aldrich; Merck KGaA) was used as
vehicle control. Lipofectamine™ RNAiMAX Transfection Reagent and
Lipofectamine® 3000 Transfection Reagent were purchased
from Invitrogen (Thermo Fisher Scientific, Inc.). Antibody against
GLUT-1 (ab115730, 1:2,000) was purchased from Abcam. Antibodies
against HIF-1α (#36169, 1:1,000), RBPJκ (#5313, 1:1,000) and GAPDH
(#5174, 1:2,000) and horseradish peroxidase-conjugated goat
anti-rabbit (#7074, 1:2,000) or anti-mouse secondary antibodies
(#7076, 1:2,000) were purchased from Cell Signaling Technology,
Inc.
Transfection and gene silencing
Small interfering (si)RNAs targeting human GLUT-1,
HIF-1α and recombination signal-binding protein for immunoglobulin
κ J region (RBPJκ) were synthesized by Shanghai GenePharma Co.,
Ltd. siRNA (100 nM) were transfected into cells using Lipofectamine
RNAiMAX Transfection Reagent in room temperature for 10 min. About
48 h later, the cells were extracted for RNA or protein detection.
Scrambled siRNA was used as a negative control (Shanghai GenePharma
Co., Ltd.). siRNA sequences are presented in Table SI.
Reverse transcription-quantitative
(RT-q)PCR
Total cellular RNA was extracted using RNAiso Plus
(Code No.: 9109, Takara Biotechnology Co., Ltd.). Complementary
(c)DNA was synthesized from total RNA using PrimeScript RT reagent
Kit with gDNA Eraser (Takara Biotechnology Co., Ltd. TKR-RR047) as
follows: 42°C for 2 min, 37°C for 15 min and then 85°C for 5 sec.
Primer sequences are presented in Table SI. qPCR was performed using
SYBR-Green Master Mix (Takara Biotechnology Co., Ltd.). The
thermocycling conditions were as follows: Pre-denaturation at 94°C
for 5 sec; followed by 40 cycles of denaturation, 95°C for 5 sec,
annealing at 60°C and extension for 34 sec. The 18s rRNA was used
as an internal control (Sangon Biotech Co., Ltd.). The mRNA
expression level of each target gene was determined using the
2−ΔΔCq method (22).
Plasmid and luciferase activity
assay
The DNA fragment containing the hypoxia-responsive
element (HRE) sequence (TGTCACGTCCTGCACGACTCTAGT) was subcloned
into the pGL3 basic reporter vector (Promega Corporation). The
plasmid luciferase reporters were electroporated into HUVECs using
an ECM 839 Electroporation System (Harvard Apparatus) as previously
reported (23). Briefly, HUVECs
were resuspended in EC medium (without serum; ScienCell Research
Laboratories, Inc.), 20 µg plasmid was added and mixed in
MicroPulse Cuvettes (0.4 cm, Bio-Rad). The cuvettes are one-piece
injection molded chambers with embedded aluminum electrodes and
square sealing caps. An electrical pulse (160 mA, 1 ms, 1 pulse)
was applied. Cells (2×104) were then seeded in 24 wells
plates immediately using EC medium at room temperature for 24 h.
The transactivation activity was analyzed using the Dual-Luciferase
Reporter Assay System (Promega Corporation) according to the
manufacturer's instructions. Briefly, 1X Passive Lysis Buffer was
dispensed into each culture vessel for 15 min at room temperature.
Then the cell lysis was mixed with Luciferase Assay Substrate and
measured firefly luciferase activity in GloMax 20/20 Luminometer
(Promega).
Western blotting
HUVECs were lysed with RIPA buffer (Thermo Fisher
Scientific, Inc.) with protease inhibitors (Cell Signaling
Technology, Inc.). The protein concentration was determined using
the bicinchoninic acid protein assay (Thermo Fisher Scientific,
Inc.). A total of 20 µg/lane protein sample was resolved on 10%
SDS-PAGE and electrotransferred to polyvinylidene fluoride
membranes. The immunoblots were blocked with 5% BSA (Sangon Biotech
Co., Ltd) for 1 h at room temperature, and probed with the
aforementioned antibodies overnight at 4°C, followed by incubation
with the corresponding secondary antibodies for 1 h at room
temperature. Subsequently, the membranes were washed with TBS-Tween
(0.1%) three times, 10 min each. The blots were visualized using
Clarity Western ECL Substrate (Bio-Rad Laboratories, Inc.). GAPDH
was used as an internal reference. Densitometry analysis was
performed using ImageJ (V1.8.0.112; National Institutes of
Health).
Glucose uptake assay
Glucose uptake assay was performed using the
Colorimetric Glucose Uptake Assay kit (Abcam) according to the
manufacturer's protocol. Briefly, 2-deoxyglucose (2-DG) was added
to HUVECs and incubated for 20 min at 37°C. After 2-DG was taken up
by glucose transporters and metabolized to 2-DG-6-phosphate, the
cells were washed with phosphate-buffered saline (PBS) to remove
exogenous 2-DG. Cells were lysed using repeated pipetting,
freeze/thaw and heating at 85°C for 40 min. The level of
2-DG-6-phosphate in each sample was determined by enzymatic
recycling amplification reaction and analyzed using a microplate
reader (optical density, 412 nm) in kinetic mode at 37°C.
Co-immunoprecipitation (Co-IP)
assay
HUVEC protein was collected using Pierce™ IP lysis
buffer (Thermo Fisher Scientific, Inc.) and centrifugation at
12,000 g, 4°C for 10 min, supernatant including protein were
collected. Protein lysate (400 µg) was immunoprecipitated with the
RBPJκ antibody (2 µg/well; CST, Inc.; cat. no. #5313) or control
Rabbit IgG control Polyclonal antibody (2 µg/well; ProteinTech
Group, Inc.; cat. no. 30,000-0-AP) bound to Dynabeads Protein G (50
µl/tube) by using Immunoprecipitation Kit-Dynabeads Protein G
(Thermo Fisher Scientific, Inc.) according to manufacturer's
instructions. Completes were isolated by addition of 20 µl IP lysis
buffer (Thermo Fisher Scientific, Inc.) and 5 µl SDS-PAGE SDS
Sample Loading Buffer (5X, Beyotime, cat. no. P0015L) and heat for
5 min at 100°C, then place the tube on the DynaMag-Spin (Thermo
Fisher, cat. 12320D) and load the supernatant/sample onto a gel and
assessed using western blotting as per the aforementioned
method.
Chromatin IP (ChIP) assay
ChIP assay was performed according to the
manufacturer's instructions using Magna ChIP G-Chromatin
Immunoprecipitation kit; cat. no. 17-611; Millipore). Briefly,
HUVECs were treated with Tan 2A (20 µM) for 16 h in 37°C. 275 µl
37% formaldehyde (Sangon Biotech Co., Ltd.) was added to 10 ml of
growth media to cross-link protein to chromatin for 10 min at room
temperature, then quenched with glycine at a final concentration of
125 mM immediately at room temperature for 5 min. Cells were
harvested in cold PBS. The cellular nuclear pellets were
resuspended with sonication buffer. The resulting cell suspension
was sheared by sonication (MiSonix Sonicator 3000) on ice using
30/30-sec on/off for 10 min (Output Power: 4). ChIP assay was
performed using 1.5 µg Rabbit IgG (1.15 mg/ml, ProteinTech Group,
30000-0-AP) or anti-HIF-1α antibodies (CST, #14179) bounded to the
Magnetic Protein G beads. Each IP requires the addition of 500 µl
Dilution Buffer. Wash the Protein G bead-antibody/chromatin complex
by resuspending beads in 0.5 ml each of the cold buffers in the
order listed below [Low Salt Immune Complex Wash Buffer (cat.# 20-1
54), High Salt Immune Complex Wash Buffer (cat.# 20-1 55), High
Salt Immune Complex Wash Buffer (cat.# 20-1 55), High Salt Immune
Complex Wash Buffer (cat.# 20-1 55)]. The DNA fragment containing
the HRE site was amplified by qPCR (SYBR-Green Master Mix, Takara
Biotechnology Co., Ltd.) using the following primers: forward:
5′-TGGGAAAAGGCATAGACTGG-3′, reverse: 5′-ATGCACGAATGAGTGAGCAG-3′)
specific for the HRE site were synthesized for this purpose.
Reference gene primer sequences are as follows: forward:
5′-GTAACCCGTTGAACCCCATT-3′, reverse: 5′-CCATCCAATCGGTAGTAGCG-3′.
PCR conditions were Pre-denaturation, 94°C for 5 sec; followed by
40 cycles of denaturation, 95°C for 5 sec, annealing at 60°C and
extension for 34 sec. The DNA expression level of each target gene
was determined using the 2-ΔΔCq method (22).
RNA sequencing (RNAseq)
Total RNA was extracted using mirVana miRNA
Isolation Kit (Ambion) following the manufacturer's protocol. RNA
purity and quantification were evaluated using a NanoDrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.). RNA integrity
was assessed using an Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc.). Libraries were constructed. The transcriptome
sequencing and analysis were performed by OE Biotech Co. Ltd. The
libraries were constructed using TruSeq Stranded mRNA LTSample Prep
Kit (RS-122-2101/RS-122-2102/RS-122-2103, Illumina, Inc.) according
to the manufacturer's instructions. A total of 10 pM loading
concentration was used. Sequencing kits were as follows: NovaSeq
6000 S4 Reagent Kit V1.5 (300 cycles; Illumina, Inc., Catalog no.
20028312) and NovaSeq Xp 4-Lane kit v1.5 (Illumina, Inc., cat. no.
20043131). The libraries were sequenced on the Illumina sequencing
platform (Illumina HiSeq X Ten) and 125 or 150 bp paired-end reads
were generated. Fragments Per Kilobase of transcript per million
fragments mapped value of each gene was calculated using cufflinks
and read counts of each gene were obtained by HTSeqcount.
Differential expression analysis was performed using the DESeq2
(1.20.0) R package. P<0.05 and fold-change >2 or <0.5 was
set as the threshold for significantly different expression.
Bioinformatics
Hierarchical cluster analysis of differentially
expressed genes was performed to explore genes expression pattern.
Gene Ontology (GO, http://geneontology.org/) enrichment and Kyoto
encyclopedia of genes and genome (KEGG, http://www.genome.jp/kegg/) pathway enrichment
analysis of differentially expressed genes were performed using R
(R-v3.4.2, r-project.org) based on the hypergeometric distribution.
GO datasets used in this research were as follows: response to
hypoxia (GO:0001666), regulation of GPCR signaling (GO:0008277),
protein homooligomerization (GO:0051260), brain development
(GO:0007420), angiogenesis (GO:0001525), negative regulation of
cell proliferation (GO:0008285), positive regulation of cell
proliferation (GO:0008284), GPCR signaling (GO:0007186),
transcription from RNA pol 2 promoter (GO:0045944), transcription,
DNA-templated (GO:0006351).
Statistical analysis
The data are presented as the mean ± SEM. Each
experiment was independently repeated three times. Statistical
significance was calculated using one-way ANOVA and Tukey's
multiple comparison test for ≥3 groups. Unpaired Student's t-test
was used for comparison between two groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
Tan 2A increases GLUT-1 expression and
glucose uptake in HUVECs
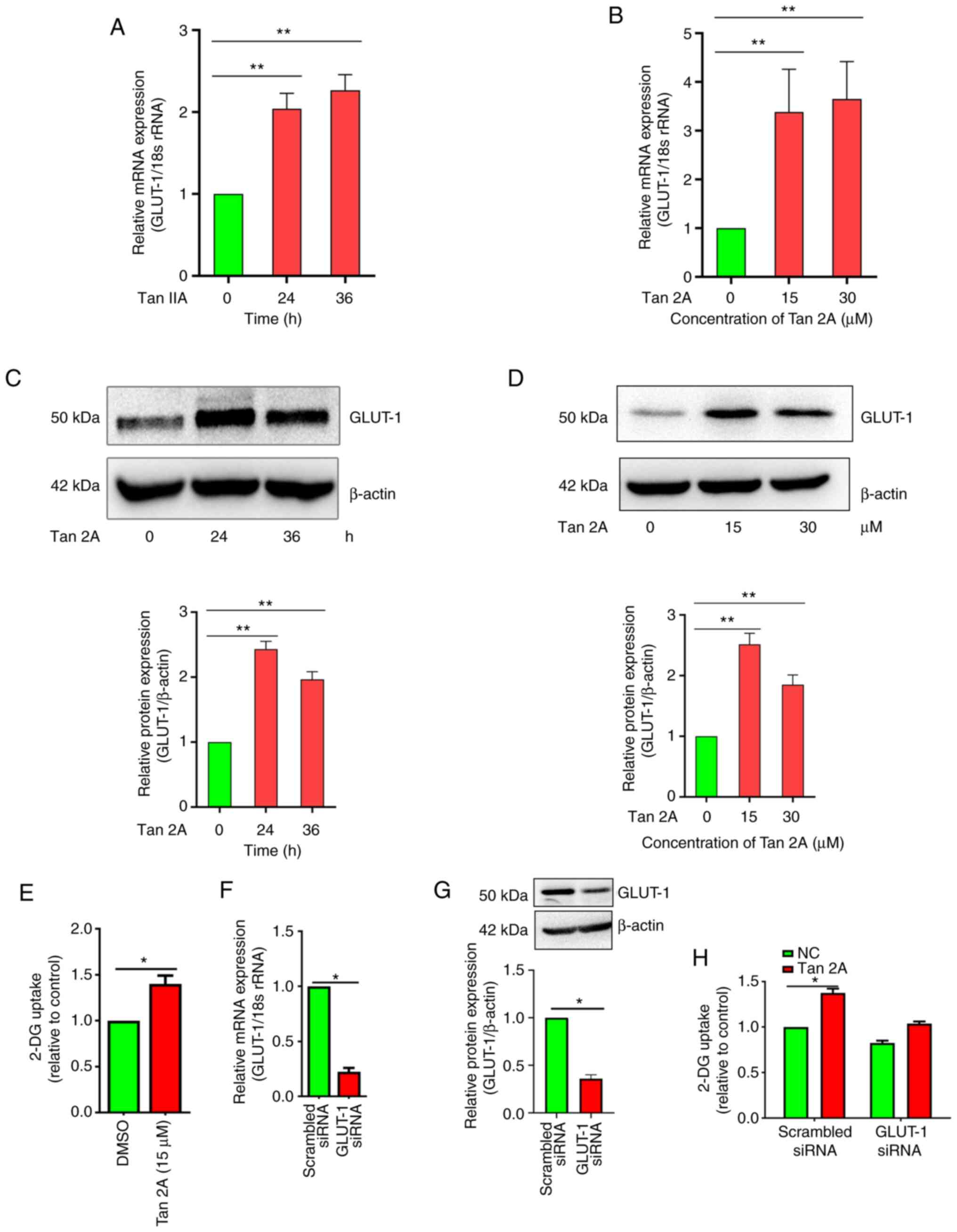
To assess the effect of Tan 2A on GLUT-1 expression,
HUVECs, a commonly used model for ECs (24), were treated with Tan 2A at 0, 15
and 30 µM for 24 h and 15 µM for 24 and 36 h. RT-qPCR results
demonstrated that mRNA expression levels of GLUT-1 in HUVECs
exposed to 15 µM Tan 2A increased significantly at 24 and 36 h
compared with 0 h control (Fig.
1A). The relative mRNA expression levels of GLUT-1 increased
significantly to 3.18±0.32 and 3.65±0.51 following 15 and 30 µM
treatment for 24 h, respectively, compared with the 0 h control.
And (Fig. 1B). Furthermore,
GLUT-1 protein expression levels were assessed using western
blotting. The protein expression levels were consistent with the
mRNA expression levels of GLUT-1, also exhibiting upregulation by
Tan 2A (Fig. 1C and D). Moreover,
endothelial glucose uptake was assessed using glucose analog 2-DG.
Glucose uptake in HUVECs was significantly enhanced by Tan 2A
compared with DMSO control (Fig.
1E). siRNA mediated knockdown of GLUT-1 was evaluated using
RT-qPCR and western blotting (Fig.
1F, G), there was a significant decrease compared with the
scrambled siRNA. GLUT-1 knockdown significantly decreased the
promotive effect of Tan 2A on glucose uptake compared to the
scrambled siRNA control (Fig.
1H). These result demonstrated that Tan 2A induced expression
of GLUT-1 and thus promoted glucose uptake in HUVECs.
Tan 2A induces expression of GLUT-1
via HIF-1α in HUVECs
RNAseq was used to evaluate the potential mechanisms
of the aforementioned effect of Tan 2A in HUVECs. Gene Ontology
analysis demonstrated that ‘response to hypoxia’ was markedly
induced by Tan 2A (Fig. 2A).
Kyoto Encyclopedia of Genes and Genomes enrichment analysis
demonstrated that genes were markedly enriched in ‘HIF-1α signaling
pathway’ (Fig. 2B). Furthermore,
RNA-seq analysis demonstrated that seven genes were differentially
expressed (>1.5 fold) in ‘HIF-1 signaling pathway’, including
GLUT-1, vascular endothelial growth factor A (VEGFA), BCL2
interacting protein 3 (BNIP3) and enolase 2 (ENO2) in HUVECs
(Fig. 2C). The results of the
RNAseq were evaluated using RT-qPCR. There were significant
increases in mRNA expression levels of VEGF, BNIP3 and ENO2
following treatment with Tan 2A compared with the control (Fig. 2D-F). Knockdown of HIF-1α
significantly inhibited Tan 2A-induced mRNA expression of GLUT-1,
VEGFA, BNIP3 and ENO2 in HUVECs (Fig.
2G-K). Western blotting demonstrated that Tan 2A-induced
protein expression of GLUT-1 was also significantly reversed by
HIF-1α knockdown (Fig. 2L). These
results suggested that Tan 2A regulated GLUT-1 expression in HUVECs
via the HIF-1α signaling pathway.
 | Figure 2.Effect of Tan 2A on transcriptional
activity of HIF-1α in HUVECs. (A) Gene Ontology analysis of genes
upregulated in Tan 2A-treated compared with control cells. (B) KEGG
pathway enrichment analysis of DEGs. Color, P-value of differential
gene enrichment; size of the circle, enrichment score. (C) RNA
sequencing heat map for HIF-1α target genes. Color, relative
expression of protein-coding genes from high (red) to low (gray).
mRNA expression levels of (D) VEGF, (E) BNIP3 and (F) ENO2, PGAM1,
PKM and ALDOA in control and Tan 2A-treated HUVECs analyzed using
RT-qPCR (n=3). The mRNA expression levels of (G) GLUT-1, (H) VEGF,
(I) BNIP3, (J) ENO2 and (K) HIF-1α in control or HIF-1α-silenced
HUVECs treated with Tan 2A for 24 h were analyzed using RT-qPCR
(n=3). (L) HIF-1α silenced HUVECs were treated with Tan 2A for 24 h
before western blotting of GLUT-1. Data from 3 independent
experiments are presented. *P<0.05 and **P<0.01. Tan 2A,
Tanshinone 2A; HUVEC, human umbilical vein endothelial cell; RT-q,
reverse transcription-quantitative; GLUT-1, glucose transporter 1;
KEGG, Kyoto Encyclopedia of Genes and Genomes; VEGF, vascular
endothelial cell growth factor; BNIP3, BCL2 interacting protein 3;
ENO2, enolase 2; HIF-1α, hypoxia-inducible factor-1α; PGAM1,
phosphoglycerate mutase 1; PKM, pyruvate kinase M1/2; ALDOA,
aldolase; CTRL, control; si, small interfering; G Protein-Coupled
Receptor, GPCR, RAGE, Receptor for AGE; stem cell (SC). |
Tan 2A increases HRE activity in
HUVECs
To assess regulation of HIF-1α by Tan 2A in HUVECs,
luciferase reporter assay was performed using constructs with the
regulatory region of the HRE. The HRE displayed significantly
increased luciferase activity in response to Tan 2A compared with
the control (Fig. 3A). ChIP assay
was performed to assess the regulatory effect of Tan 2A on the
binding ability of HIF-1α to the potential HRE within the promoter
region of the GLUT-1 gene and demonstrated that binding of HIF-1α
to the promoter region of GLUT-1 gene was significantly enhanced by
Tan 2A in HUVECs (Fig. 3B).
HIF-1α is hydroxylated by prolyl hydroxylases (25). RT-qPCR demonstrated that Tan 2A
significantly increased expression of EGL-9 family hypoxia
inducible factor 3 (EGLN3), which belongs to prolyl hydroxylase
family, compared with the control, these data suggested that EGLN3
mediated Tan 2A-induced activation of HIF-1α in HUVECs (Fig. 3C). These results showed Tan 2A
induced HIF-1/HRE pathway in HUVECs.
 | Figure 3.Tan 2A induces binding of HIF-1α to
the HRE in the GLUT-1 promoter. (A) HUVECs were transfected with
HRE-luc for 24 h, then treated with Tan 2A for 24 h and luciferase
reporter assay was used to assess HRE activity (n=3). (B) ChIP-PCR
analysis of the HIF-1α binding site in the GLUT-1 promoter. Fold
enrichment of the HRE site was calculated (n=3). (C) HUVECs were
treated with Tan 2A (0, 15 and 30 µM) for 24 h and the mRNA
expression of EGLN1, EGLN2 and EGLN3 was analyzed by RT-qPCR (n=3).
*P<0.05 and **P<0.01. Tan 2A, Tanshinone 2A; HIF-1α,
hypoxia-inducible factor-1α; HRE, hypoxia-responsive element;
GLUT-1, glucose transporter 1; HUVEC, human umbilical vein
endothelial cell; luc, luciferase; ChIP, chromatin
immunoprecipitation; EGLN, EGL-9 family hypoxia inducible factor;
RT-q, reverse transcription-quantitative. |
RBPJκ mediates regulation of GLUT-1 by
Tan 2A in HUVECs
RBPJκ binds with HIF-1α to regulate expression of
downstream target genes, such as VEGFA (26). Whether RBPJκ mediates Tan
2A-regulated expression of GLUT-1 was therefore examined. RT-qPCR
demonstrated that knockdown of RBPJκ significantly inhibited Tan
2A-induced mRNA expression of GLUT-1, BNIP3 and ENO2 in HUVECs
(Fig. 4A-D). Furthermore, Tan 2A
markedly decreased protein expression levels of GLUT-1, as
demonstrated by western blotting (Fig. 4E). These data suggested that RBPJκ
potentiated Tan 2A-induced GLUT-1 expression in HUVECs.
 | Figure 4.Knockdown of RBPJκ decreases
expression of HIF-1α target genes induced by Tan 2A. RBPJκ-silenced
human umbilical vein endothelial cells were stimulated with Tan 2A
(15 µM) for 24 h and mRNA expression levels of (A) GLUT-1, (B)
ENO2, (C) BNIP3 and (D) RBPJκ were analyzed using RT-qPCR (n=3).
*P<0.05. (E) Representative western blots for GLUT-1 from 3
independent experiments. Tan 2A, Tanshinone 2A; HIF-1α,
hypoxia-inducible factor-1α; RT-q, reverse
transcription-quantitative; GLUT-1, glucose transporter 1; BNIP3,
BCL2 interacting protein 3; ENO2, enolase 2; RBPJκ, recombination
signal-binding protein for immunoglobulin κJ region; si, small
interfering. |
Tan 2A enhances the interaction
between HIF-1α and RBPJκ protein
It has been reported that RBPJκ physically interacts
with HIF protein (27). Whether
Tan 2A regulates coupling between HIF-1 and RBPJκ was assessed.
Western blotting of Co-IP samples demonstrated that HIF-1α was
co-immunoprecipitated with RBPJκ, which suggested a physical
interaction between the proteins. Furthermore, Tan 2A treatment
markedly enhanced the physically interaction of RBPJκ with HIF-1α
(Fig. 5).
Discussion
Under both normal and pathological conditions, the
concentration of glucose in neurons is strictly controlled and
relies on sustained blood flow and glucose transport (28). Therefore, altered expression of
GLUT-1 at the blood-brain barrier affects the function of neurons.
In the present study, Tan 2A regulated GLUT-1 expression and
glucose uptake in HUVECs. Furthermore, Tan 2A enhanced the
interaction between HIF-1α and RBPJκ and HIF-1α and RBPJκ mediated
the effect of Tan 2A on mRNA and protein expression levels of
GLUT-1. These results suggested that Tan 2A may facilitate recovery
of brain function via upregulation of the GLUT-1 to increase
glucose absorption.
The energy demand of neurons in the brain is served
by glucose transported from the blood. Glucose is transported into
brain cells via members of the GLUT family. GLUT-3 is primarily
expressed in neurons, while GLUT-1 gene expression is limited to
ECs in the healthy brain (29)
and regulates glucose transport into the brain (30,31). Two subtypes of GLUT-1, a 55 kDa
isoform in brain ECs and a 45 kDa isoform in adjacent astrocytes,
have been reported previously (32). Previous studies have reported that
changes in expression of GLUT affect the function of neurons and
that GLUT-1 haploid deficiency induces decreased brain weight,
activated astrocytosis, impaired motor performance and diminished
brain glucose metabolism in mice (28,33). Our results showed that Tan 2A
upregulated GLUT-1 expression and 2-DG uptake in HUVECs. In
addition to its role in transporting glucose into the brain
(34), GLUT-1 is needed for
maintenance of proper brain capillary networks, blood flow and
blood-brain barrier integrity (35). Furthermore, inhibition of GLUT-1
decreases EC glucose uptake and glycolysis, which leads to energy
depletion, activation of the cellular energy sensor AMPK and
decreased EC proliferation (5),
as well as decreased nitric oxide-dependent endothelial relaxation
(36).
Tan 2A is used in the treatment of cardio- and
cerebrovascular disorders, such as coronary heart disease and
cerebral infarction (37). Tan 2A
exerts anti-atherosclerosis, anti-cardiac hypertrophy and
antioxidant effects via regulation of the expression of numerous
molecules, including transcription factors, scavenger receptors,
ion channels, pro- and anti-apoptotic proteins, growth factors,
inflammatory mediators and microRNAs (38). Tan 2A protects against
cardiovascular disease via regulation of Akt/glycogen synthase
kinase-3β, nitric oxide and JNK signaling (39–41). Tan 2A treatment combined with
overexpression of GLUT-1 increases glucose uptake and thus promotes
viability of neurons and recovery of brain function (42). It has also been reported that
treatment with Danshen and Sanqi (a kind of Chinese herbal
medicine) ameliorates hyperlipidemia and hyperglycemia phenotypes
in mice with diet-induced obesity via regulation of liver glycogen
synthesis and cholesterol anabolism genes, including GLUT-1
(15). The present study
demonstrated that Tan 2A significantly increased GLUT-1 mRNA and
markedly increased GLUT-1 protein expression levels in HUVECs.
Therefore, Tan 2A may serve as an adjuvant drug to treat
GLUT-1-associated neuron disorder.
HIF-1 is a heterodimeric transcription factor
comprising an α and β subunit. Its activity is dependent on the
stability of HIF-1α. It is relatively stable under hypoxia and
interacts with HIF-1β to form the HIF-1 active heterodimer
(25). HIF-1α is considered to
mediate cardio- and neuroprotection, partly by reprogramming
cellular metabolism (43). The
expression of key downstream genes, including VEGF (44), carbonic anhydrase (45) and GLUT-1 (46), is induced by HIF-1 via binding to
the enhancer sequence located HRE in these genes. Our results
showed that Tan 2A increased the activity of HRE of HIF-1α and the
expression of VEGFA and GLUT-1. Tan 2A decreases astrocyte
proliferation and significantly attenuates oxygen-glucose
deprivation-induced accumulation of HIF-1α and C-X-C motif
chemokine ligand 12, which blocks downstream signaling via Erk1/2
and Akt activation. By contrast with previous reports that Tan 2A
inhibits transcription of HIF-1 in tumor cells, the present study
demonstrated an increase in HIF-1α mRNA and protein expression
levels in Tan 2A-treated HUVECs and that the binding of HIF-1α to
the HRE in GLUT-1 promotor sequence was induced by Tan 2A. But how
the Tan 2A activate the HIF-1α is still underdetermined. And
whether Tan 2A interacts with HIF-1α or DNA sequences in target
genes is not known. Previous studies have reported Tan 2A is a
natural monoacylglycerol lipase (MAGL) inhibitor that interacts
with the MAGL binding pocket (47). Furthermore, it has been reported
that imidazole-based Tan 2A derivatives improve the selectivity and
binding affinity with G-quadruplex DNA and enhances inhibition of
breast cancer metastasis (48).
The present study further demonstrates that HIF-1α partially
mediated the regulatory effect of Tan 2A on GLUT-1 expression in
ECs.
The DNA binding transcription factor RBPJκ, a key
mediator of Notch signaling, recruits additional co-factors,
including mastermind proteins 1–3, to activate target genes. RBPJκ
is also a putative cofactor of HIF-1α (27). The association of RBPJκ with the
HIF-1α complex promotes expression of replication and
transcriptional activators (49).
HIF-1 and HIF-2 competitively bind to the intracellular domain of
the Notch receptor, an RBPJκ partner, and dynamically regulate
activation of Notch signaling in glioma stem cells (50). However, RBPJκ may control gene
expression of angiogenic factors by antagonizing the activity of
HIF-1α (27). Deficiency of RBPJκ
in mice leads to activation of HIF-1α-mediated regulation of Notch
targets in hematopoietic stem and progenitor cells (51). The present study demonstrated that
RBPJκ and HIF-1α knockdown both significantly reversed Tan
2A-induced mRNA expression levels of GLUT-1 and markedly decreased
protein expression levels of GLUT-1 in HUVECs. Furthermore, Tan 2A
markedly increased binding of HIF-1α and RBPJκ. It was therefore
hypothesized that RBPJκ affects binding of HIF-1α to the HRE in the
GLUT-1 promotor and that both RBPJκ and HIF-1α may be involved in
regulation of GLUT-1 gene expression induced by Tan 2A. Whether Tan
2A plays the same role under anoxic conditions is still to be
studied.
In summary, the present study suggested that Tan 2A
induced expression of GLUT-1 via the HIF-1α signaling pathway
(Fig. 6). Furthermore, Tan 2A may
ameliorate brain glucose metabolism by regulating GLUT-1 mediated
glucose transport in HUVECs.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 81774076) and Clinical Superior
Discipline Development Fund of Shanghai Putuo District (grant no.
2019ysxk01).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Sequencing Read Archive
repository (https://www.ncbi.nlm.nih.gov/sra/PRJNA871126).
Authors' contributions
ZL conceived and designed experiments. YZ and HZ
drafted the manuscript. YZ, HZ, YH, SW and ZL performed the
experiments and analyzed the data. All authors have read and
approved the final manuscript. YZ and ZL confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mergenthaler P, Lindauer U, Dienel GA and
Meisel A: Sugar for the brain: The role of glucose in physiological
and pathological brain function. Trends Neurosci. 36:587–597. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhao FQ and Keating AF: Functional
properties and genomics of glucose transporters. Curr Genomics.
8:113–128. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kayano T, Fukumoto H, Eddy RL, Fan YS,
Byers MG, Shows TB and Bell GI: Evidence for a family of human
glucose transporter-like proteins. Sequence and gene localization
of a protein expressed in fetal skeletal muscle and other tissues.
J Biol Chem. 263:15245–15248. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mueckler M: Facilitative glucose
transporters. Eur J Biochem. 219:713–725. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Veys K, Fan Z, Ghobrial M, Bouche A,
Garcia-Caballero M, Vriens K, Conchinha NV, Seuwen A, Schlegel F
and Gorski T: Role of the GLUT1 glucose transporter in postnatal
CNS angiogenesis and blood-brain barrier integrity. Circ Res.
127:466–482. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nagamatsu S, Kornhauser JM, Burant CF,
Seino S, Mayo KE and Bell GI: Glucose transporter expression in
brain. cDNA sequence of mouse GLUT3, the brain facilitative glucose
transporter isoform, and identification of sites of expression by
in situ hybridization. J Biol Chem. 267:467–472. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang H, Pang W, Xu X, You B, Zhang C and
Li D: Cryptotanshinone attenuates ischemia/reperfusion-induced
apoptosis in myocardium by upregulating MAPK3. J Cardiovasc
Pharmacol. 77:370–377. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu J, Zhang P, Chen Y, Xu Y, Luan P, Zhu Y
and Zhang J: Sodium tanshinone IIA sulfonate ameliorates cerebral
ischemic injury through regulation of angiogenesis. Exp Ther Med.
22:11222021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Subedi L and Gaire BP: Tanshinone IIA: A
phytochemical as a promising drug candidate for neurodegenerative
diseases. Pharmacol Res. 169:1056612021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang Y, Li C, Meng H, Guo D, Zhang Q, Lu
W, Wang Q, Wang Y and Tu P: BYD ameliorates oxidative
stress-induced myocardial apoptosis in heart failure post-acute
myocardial infarction via the P38 MAPK-CRYAB signaling pathway.
Front Physiol. 9:5052018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tang Q, Han R, Xiao H, Shen J, Luo Q and
Li J: Neuroprotective effects of tanshinone IIA and/or
tetramethylpyrazine in cerebral ischemic injury in vivo and in
vitro. Brain Res. 1488:81–91. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu J, Xu Y, Ren G, Hu X, Wang C, Yang Z,
Li Z, Mao W and Lu D: Tanshinone IIA Sodium sulfonate regulates
antioxidant system, inflammation, and endothelial dysfunction in
atherosclerosis by downregulation of CLIC1. Eur J Pharmacol.
815:427–436. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lou G, Hu W, Wu Z, Xu H, Yao H, Wang Y,
Huang Q, Wang B, Wen L, Gong D, et al: Tanshinone II A attenuates
vascular remodeling through klf4 mediated smooth muscle cell
phenotypic switching. Sci Rep. 10:138582020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen W, Li X, Guo S, Song N, Wang J, Jia L
and Zhu A: Tanshinone IIA harmonizes the crosstalk of autophagy and
polarization in macrophages via miR-375/KLF4 pathway to attenuate
atherosclerosis. Int Immunopharmacol. 70:486–497. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie Z, Truong TL, Zhang P, Xu F, Xu X and
Li P: Dan-Qi prescription ameliorates insulin resistance through
overall corrective regulation of glucose and fat metabolism. J
Ethnopharmacol. 172:70–79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guan R, Wang J, Li Z, Ding M, Li D, Xu G,
Wang T, Chen Y, Yang Q, Long Z, et al: Sodium tanshinone IIA
sulfonate decreases cigarette smoke-induced inflammation and
oxidative stress via blocking the activation of MAPK/HIF-1alpha
signaling pathway. Front Pharmacol. 9:2632018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhou L, Sui H, Wang T, Jia R, Zhang Z, Fu
J, Feng Y, Liu N, Ji Q, Wang Y, et al: Tanshinone IIA reduces
secretion of proangiogenic factors and inhibits angiogenesis in
human colorectal cancer. Oncol Rep. 43:1159–1168. 2020.PubMed/NCBI
|
|
18
|
Zhou ZY, Zhao WR, Xiao Y, Zhang J, Tang JY
and Lee SM: Mechanism study of the protective effects of sodium
tanshinone IIA sulfonate against atorvastatin-induced cerebral
hemorrhage in zebrafish: Transcriptome analysis. Front Pharmacol.
11:5517452020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fu P, Du F, Chen W, Yao M, Lv K and Liu Y:
Tanshinone IIA blocks epithelial-mesenchymal transition through
HIF-1alpha downregulation, reversing hypoxia-induced chemotherapy
resistance in breast cancer cell lines. Oncol Rep. 31:2561–2568.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu M, Cao F, Liu L, Zhang B, Wang Y, Dong
H, Cui Y, Dong M, Xu D, Liu Y, et al: Tanshinone IIA-induced
attenuation of lung injury in endotoxemic mice is associated with
reduction of hypoxia-inducible factor 1α expression. Am J Respir
Cell Mol Biol. 45:1028–1035. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu Y, Wang SJ and Song XT: Effects of
electroacupuncture on glucose transporter-1 expression of
hippocampal microvascular endothelial cells in rats with focal
cerebral ischemia. Zhen Ci Yan Jiu. 35:118–123. 2010.(In Chinese).
PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Y, Zhang Y, Zhu Y and Zhang P:
Lipolytic inhibitor G0/G1 switch gene 2 inhibits reactive oxygen
species production and apoptosis in endothelial cells. Am J Physiol
Cell Physiol. 308:C496–C504. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baudin B, Bruneel A, Bosselut N and
Vaubourdolle M: A protocol for isolation and culture of human
umbilical vein endothelial cells. Nat Protoc. 2:481–485. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Masson N and Ratcliffe PJ: HIF prolyl and
asparaginyl hydroxylases in the biological response to
intracellular O(2) levels. J Cell Sci. 116:3041–3049. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Singh AR, Zhao Y, Du T, Huang Y,
Wan X, Mukhopadhyay D, Wang Y, Wang N and Zhang P: TRIM28 regulates
sprouting angiogenesis through VEGFR-DLL4-notch signaling circuit.
FASEB J. 34:14710–14724. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Diaz-Trelles R, Scimia MC, Bushway P, Tran
D, Monosov A, Monosov E, Peterson K, Rentschler S, Cabrales P,
Ruiz-Lozano P and Mercola M: Notch-independent RBPJ controls
angiogenesis in the adult heart. Nat Commun. 7:120882016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Benarroch EE: Brain glucose transporters:
Implications for neurologic disease. Neurology. 82:1374–1379. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Szablewski L: Brain glucose transporters:
Role in pathogenesis and potential targets for the treatment of
Alzheimer's disease. Int J Mol Sci. 22:81422021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Allen A and Messier C: Plastic changes in
the astrocyte GLUT1 glucose transporter and beta-tubulin
microtubule protein following voluntary exercise in mice. Behav
Brain Res. 240:95–102. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Choeiri C, Staines W, Miki T, Seino S and
Messier C: Glucose transporter plasticity during memory processing.
Neuroscience. 130:591–600. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Simpson IA, Carruthers A and Vannucci SJ:
Supply and demand in cerebral energy metabolism: The role of
nutrient transporters. J Cereb Blood Flow Metab. 27:1766–1791.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ullner PM, Di Nardo A, Goldman JE, Schobel
S, Yang H, Engelstad K, Wang D, Sahin M and Vivo DCD: Murine Glut-1
transporter haploinsufficiency: Postnatal deceleration of brain
weight and reactive astrocytosis. Neurobiol Dis. 36:60–69. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zlokovic BV: Neurovascular pathways to
neurodegeneration in Alzheimer's disease and other disorders. Nat
Rev Neurosci. 12:723–738. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Winkler EA, Nishida Y, Sagare AP, Rege SV,
Bell RD, Perlmutter D, Sengillo JD, Hillman S, Kong P, Nelson AR,
et al: GLUT1 reductions exacerbate Alzheimer's disease
vasculo-neuronal dysfunction and degeneration. Nat Neurosci.
18:521–530. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park JL, Heilig CW and Brosius FC III:
GLUT1-deficient mice exhibit impaired endothelium-dependent
vascular relaxation. Eur J Pharmacol. 496:213–214. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao S, Liu Z, Li H, Little PJ, Liu PA and
Xu S: Cardiovascular actions and therapeutic potential of
tanshinone IIA. Atherosclerosis. 220:3–10. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xu S and Liu P: Tanshinone II-A: New
perspectives for old remedies. Expert Opin Ther Pat. 23:149–153.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun D, Shen M, Li J, Li W, Zhang Y, Zhao
L, Zhang Z, Yuan Y, Wang H and Cao F: Cardioprotective effects of
tanshinone IIA pretreatment via kinin B2 receptor-Akt-GSK-3β
dependent pathway in experimental diabetic cardiomyopathy.
Cardiovasc Diabetol. 10:42011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pan C, Lou L, Huo Y, Singh G, Chen M,
Zhang D, Wu A, Zhao M, Wang S and Li J: Salvianolic acid B and
tanshinone IIA attenuate myocardial ischemia injury in mice by NO
production through multiple pathways. Ther Adv Cardiovasc Dis.
5:99–111. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang R, Liu A, Ma X, Li L, Su D and Liu J:
Sodium tanshinone IIA sulfonate protects cardiomyocytes against
oxidative stress-mediated apoptosis through inhibiting JNK
activation. J Cardiovasc Pharmacol. 51:396–401. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang J, Tong H, Wang X, Wang X and Wang Y:
Tanshinone IIA alleviates the damage of neurocytes by targeting
GLUT1 in ischaemia reperfusion model (in vivo and in vitro
experiments). Folia Neuropathol. 58:176–193. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nanayakkara G, Alasmari A, Mouli S,
Eldoumani H, Quindry J, McGinnis G, Fu X, Berlin A, Peters B, Zhong
J and Amin R: Cardioprotective HIF-1alpha-frataxin signaling
against ischemia-reperfusion injury. Am J Physiol Heart Circ
Physiol. 309:H867–H879. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ahn GO, Seita J, Hong BJ, Kim YE, Bok S,
Lee CJ, Kim KS, Lee JC, Leeper NJ, Cooke JP, et al: Transcriptional
activation of hypoxia-inducible factor-1 (HIF-1) in myeloid cells
promotes angiogenesis through VEGF and S100A8. Proc Natl Acad Sci
USA. 111:2698–2703. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chiche J, Ilc K, Laferriere J, Trottier E,
Dayan F, Mazure NM, Brahimi-Horn MC and Pouysségur J:
Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell
growth by counteracting acidosis through the regulation of the
intracellular pH. Cancer Res. 69:358–368. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Dungwa JV, Hunt LP and Ramani P:
Overexpression of carbonic anhydrase and HIF-1α in wilms tumours.
BMC Cancer. 11:3902011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang R, Lu Y and Liu J: Identification of
tanshinone IIA as a natural monoacylglycerol lipase inhibitor by
combined in silico and in vitro approach. MedChemComm. 5:1528–1532.
2014. View Article : Google Scholar
|
|
48
|
Zeng L, Wu Q, Wang T, Li LP, Zhao X, Chen
K, Qian J, Yuan L, Xu H and Mei WJ: Selective stabilization of
multiple promoter G-quadruplex DNA by using
2-phenyl-1H-imidazole-based tanshinone IIA derivatives and their
potential suppressing function in the metastatic breast cancer.
Bioorg Chem. 106:1044332021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang L, Zhu C, Guo Y, Wei F, Lu J, Qin J,
Banerjee S, Wang J, Shang H, Verma SC, et al: Inhibition of KAP1
enhances hypoxia-induced Kaposi's sarcoma-associated herpesvirus
reactivation through RBP-Jκ. J Virol. 88:6873–6884. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hu YY, Fu LA, Li SZ, Chen Y, Li JC, Han J,
Liang L, Li L, Ji CC, Zheng MH and Hdan H: Hif-1alpha and
Hif-2alpha differentially regulate Notch signaling through
competitive interaction with the intracellular domain of Notch
receptors in glioma stem cells. Cancer Lett. 349:67–76. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lakhan R and Rathinam CV: Deficiency of
Rbpj leads to defective stress-induced hematopoietic stem cell
functions and hif mediated activation of non-canonical notch
signaling pathways. Front Cell Dev Biol. 8:6221902020. View Article : Google Scholar : PubMed/NCBI
|