|
1
|
Zhu L, Ling J, Zhu Z, Tian T, Song Y and
Yang C: Selection and applications of functional nucleic acids for
infectious disease detection and prevention. Anal Bioanal Chem.
413:4563–4579. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ling Z, Xiao H and Chen W: Gut microbiome:
The cornerstone of life and health. Adv Gut Microbiome Res.
2022:1–3. 2022. View Article : Google Scholar
|
|
3
|
Vengesai A, Kasambala M, Mutandadzi H,
Mduluza-Jokonya TL, Mduluza T and Naicker T: Scoping review of the
applications of peptide microarrays on the fight against human
infections. PLoS One. 17:e02486662022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Casanova JL and Abel L: Lethal Infectious
diseases as inborn errors of immunity: Toward a synthesis of the
germ and genetic theories. Annu Rev Pathol. 16:23–50. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang L, Jianying L and Pei-Yong S:
SARS-CoV-2 variants and vaccination. Zoonoses (Burlingt).
2:62022.
|
|
6
|
Micoli F, Bagnoli F, Rappuoli R and
Serruto D: The role of vaccines in combatting antimicrobial
resistance. Nat Rev Microbiol. 19:287–302. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mercer A: Protection against severe
infectious disease in the past. Pathog Glob Health. 115:151–167.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu L and Moore MD: A survey of analytical
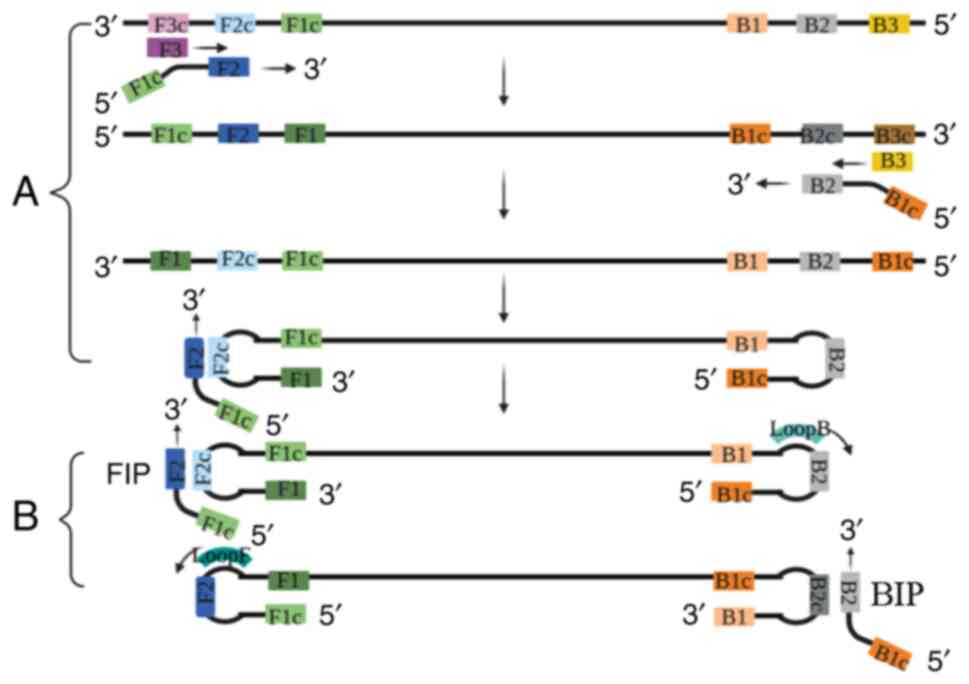
techniques for noroviruses. Foods. 9:3182020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiang Z, Jiang B, Li W, Zhai G, Zhou H,
Wang Y and Wu J: The diagnostic and prognostic value of serum
exosome-derived carbamoyl phosphate synthase 1 in HEV-related acute
liver failure patients. J Med Virol. 94:5015–5025. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang HS, Tsai CL, Chang J, Hsu TC, Lin S
and Lee CC: Multiplex PCR system for the rapid diagnosis of
respiratory virus infection: Systematic review and meta-analysis.
Clin Microbiol Infect. 24:1055–1063. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
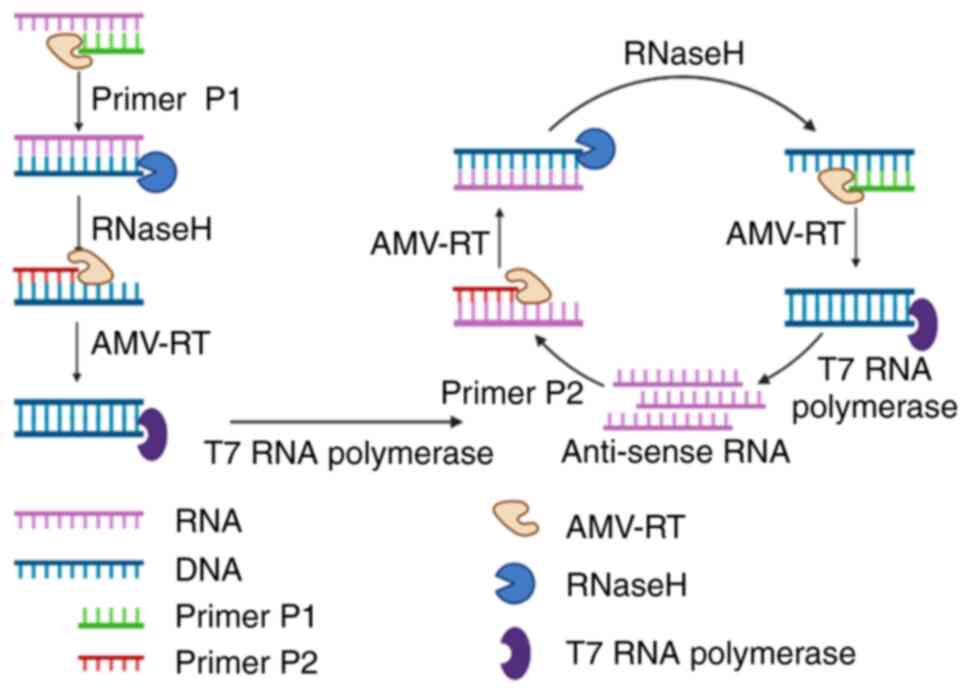
11
|
Wu J, Bortolanza M, Zhai G, Shang A, Ling
Z, Jiang B, Shen X, Yao Y, Yu J, Li L and Cao H: Gut microbiota
dysbiosis associated with plasma levels of Interferon-γ and viral
load in patients with acute hepatitis E infection. J Med Virol.
94:692–702. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu J, Xu Y, Cui Y, Bortolanza M, Wang M,
Jiang B, Yan M, Liang W, Yao Y, Pan Q, et al: Dynamic changes of
serum metabolites associated with infection and severity of
patients with acute hepatitis E infection. J Med Virol.
94:2714–2726. 2022. View Article : Google Scholar : PubMed/NCBI
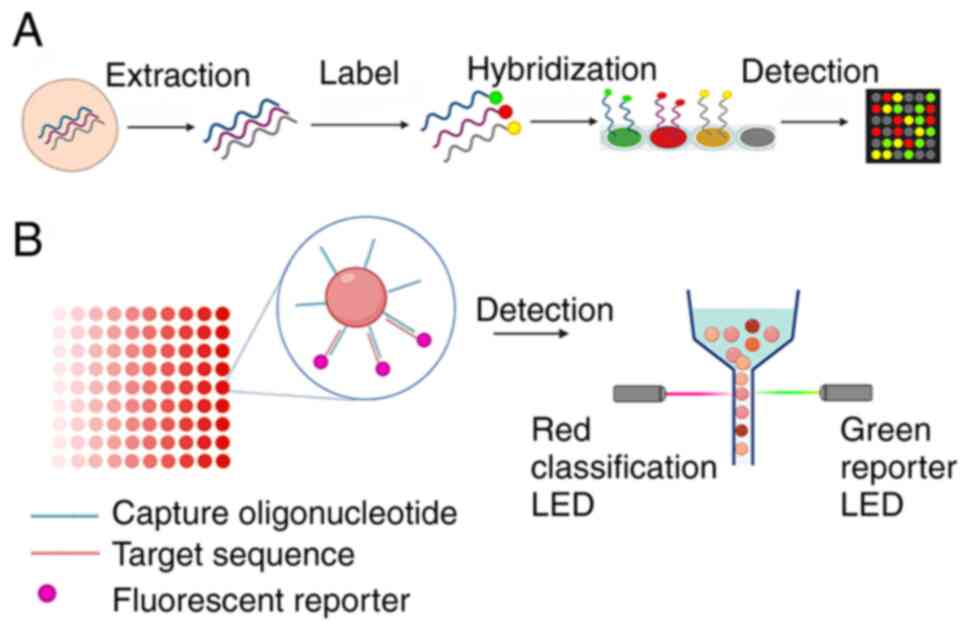
|
|
13
|
Zhang B, Zhou J, Li M, Wei Y, Wang J, Wang
Y, Shi P, Li X, Huang Z, Tang H and Song Z: Evaluation of
CRISPR/Cas9 site-specific function and validation of sgRNA sequence
by a Cas9/sgRNA-assisted reverse PCR technique. Anal Bioanal Chem.
413:2447–2456. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sidstedt M, Rådström P and Hedman J: PCR
inhibition in qPCR, dPCR and MPS-mechanisms and solutions. Anal
Bioanal Chem. 412:2009–2023. 2020. View Article : Google Scholar : PubMed/NCBI
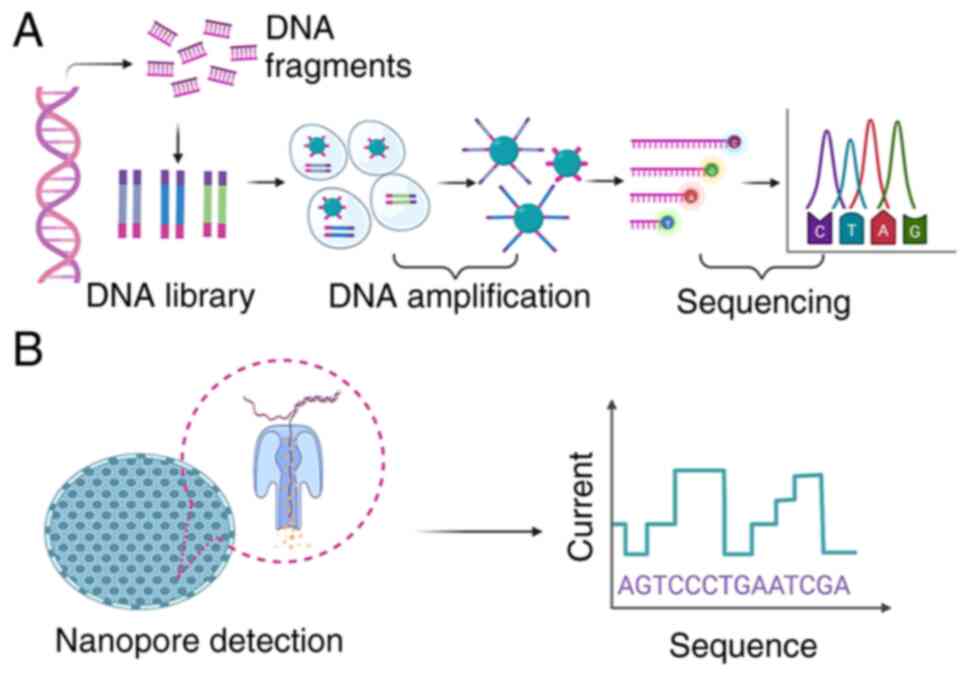
|
|
15
|
García-Bernalt Diego J, Fernández-Soto P,
Crego-Vicente B, Alonso-Castrillejo S, Febrer-Sendra B,
Gómez-Sánchez A, Vicente B, López-Abán J and Muro A: Progress in
loop-mediated isothermal amplification assay for detection of
Schistosoma mansoni DNA: Towards a ready-to-use test. Sci Rep.
9:147442019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang W, Ji W, Zhang Y, Xie Y, Chen S, Jin
Y and Duan G: An update on detection technologies for SARS-CoV-2
variants of concern. Viruses. 14:23242022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lv C, Deng W, Wang L, Qin Z, Zhou X and Xu
J: Molecular techniques as alternatives of diagnostic tools in
china as schistosomiasis moving towards elimination. Pathogens.
11:2872022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mackay IM, Arden KE and Nitsche A:
Real-time PCR in virology. Nucleic Acids Re. 30:1292–1305. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Castelli G, Bruno F, Reale S, Catanzaro S,
Valenza V and Vitale F: Molecular diagnosis of leishmaniasis:
Quantification of parasite load by a Real-Time PCR assay with high
sensitivity. Pathogens. 10:8652021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vidanapathirana G, Angulmaduwa ALSK,
Munasinghe TS, Ekanayake EWMA, Harasgama P, Kudagammana ST,
Dissanayake BN and Liyanapathirana LVC: Comparison of pneumococcal
colonization density among healthy children and children with
respiratory symptoms using real time PCR (RT-PCR). BMC Microbiol.
22:312022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ingalagi P, Bhat KG, Kulkarni RD,
Kotrashetti VS, Kumbar V and Kugaji M: Detection and comparison of
prevalence of Porphyromonas gingivalis through culture and
Real Time-polymerase chain reaction in subgingival plaque samples
of chronic periodontitis and healthy individuals. J Oral Maxillofac
Pathol. 26:2882022.PubMed/NCBI
|
|
22
|
Marrero Rolon R, Cunningham SA, Mandrekar
JN, Polo ET and Patel R: Erratum for Marrero Rolon et al.,
‘Clinical evaluation of a real-time pCR assay for simultaneous
detection of helicobacter pylori and genotypic markers of
clarithromycin resistance directly from stool’. J Clin Microbiol.
60:e02452212022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bennett S and Gunson RN: The development
of a multiplex real-time RT-PCR for the detection of adenovirus,
astrovirus, rotavirus and sapovirus from stool samples. J Virol
Methods. 242:30–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang XW, Huang TS, Xie L, Chen SZ, Wang
SD, Huang ZW, Li XY and Ling WP: Development of a diagnostic assay
by three-tube multiplex real-time PCR for simultaneous detection of
nine microorganisms causing acute respiratory infections. Sci Rep.
12:133062022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu L, Zhang Y, Cui P, Wang C, Zeng X,
Deng G and Wang X: Development of a duplex TaqMan real-time RT-PCR
assay for simultaneous detection of newly emerged H5N6 influenza
viruses. Virol J. 16:1192019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Das Mukhopadhyay C, Sharma P, Sinha K and
Rajarshi K: Recent trends in analytical and digital techniques for
the detection of the SARS-Cov-2. Biophys Chem. 270:1065382021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu CY, Chan KG, Yean CY and Ang GY:
Nucleic acid-based diagnostic tests for the detection SARS-CoV-2:
An Update. Diagnostics (Basel). 11:532021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li H, Bai R, Zhao Z, Tao L, Ma M, Ji Z,
Jian M, Ding Z, Dai X, Bao F and Liu A: Application of droplet
digital PCR to detect the pathogens of infectious diseases. Biosci
Rep. 38:BSR201811702018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lei S, Chen S and Zhong Q: Digital PCR for
accurate quantification of pathogens: Principles, applications,
challenges and future prospects. Int J Biol Macromol. 184:750–759.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Das S, Hammond-McKibben D, Guralski D,
Lobo S and Fiedler PN: Development of a sensitive molecular
diagnostic assay for detecting Borrelia burgdorferi DNA from the
blood of Lyme disease patients by digital PCR. PLoS One.
15:e02353722020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cao Y, Yu M, Dong G, Chen B and Zhang B:
Digital PCR as an emerging tool for monitoring of microbial
biodegradation. Molecules. 25:7062020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang L, Parvin R, Fan Q and Ye F:
Emerging digital PCR technology in precision medicine. Biosens
Bioelectron. 211:1143442020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Košir AB, Spilsberg B, Holst-Jensen A, Žel
J and Dobnik D: Development and inter-laboratory assessment of
droplet digital PCR assays for multiplex quantification of 15
genetically modified soybean lines. Sci Rep. 9:37352019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu L, Qu H, Alonso DG, Yu Z, Yu Y, Shi Y,
Hu C, Zhu T, Wu N and Shen F: Portable integrated digital PCR
system for the point-of-care quantification of BK virus from urine
samples. Biosens Bioelectron. 175:1129082021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sedlak RH, Nguyen T, Palileo I, Jerome KR
and Kuypers J: Superiority of Digital Reverse Transcription-PCR
(RT-PCR) over Real-Time RT-PCR for Quantitation of Highly Divergent
Human Rhinoviruses. J Clin Microbiol. 55:442–449. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
van Snippenberg W, Gleerup D, Rutsaert S,
Vandekerckhove L, De Spiegelaere W and Trypsteen W: Triplex digital
PCR assays for the quantification of intact proviral HIV-1 DNA.
Methods. 201:41–48. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bønløkke S, Stougaard M, Sorensen BS,
Booth BB, Høgdall E, Nyvang GB, Lindegaard JC, Blaakær J, Bertelsen
J, Fuglsang K, et al: The diagnostic value of circulating Cell-Free
HPV DNA in plasma from cervical cancer patients. Cells.
11:21702022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lyu L, Li Z, Pan L, Jia H, Sun Q, Liu Q
and Zhang Z: Evaluation of digital PCR assay in detection of M.
tuberculosis IS6110 and IS1081 in tuberculosis patients plasma.
BMC Infect Dis. 20:6572020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Salipante SJ and Jerome KR: Digital PCR-An
emerging technology with broad applications in microbiology. Clin
Chem. 66:117–123. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rutsaert S, Bosman K, Trypsteen W, Nijhuis
M and Vandekerckhove L: Digital PCR as a tool to measure HIV
persistence. Retrovirology. 15:162018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kojabad AA, Farzanehpour M, Galeh HEG,
Dorostkar R, Jafarpour A, Bolandian M and Nodooshan MM: Droplet
digital PCR of viral DNA/RNA, current progress, challenges, and
future perspectives. J Med Virol. 93:4182–4197. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dingle TC, Sedlak RH, Cook L and Jerome
KR: Tolerance of droplet-digital PCR vs real-time quantitative PCR
to inhibitory substances. Clin Chem. 59:1670–1672. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pan SW, Su WJ, Chan YJ, Chuang FY, Feng JY
and Chen YM: Mycobacterium tuberculosis-derived circulating
cell-free DNA in patients with pulmonary tuberculosis and persons
with latent tuberculosis infection. PLoS One. 16:e02538792021.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang D, Liu E, Liu H, Jin X, Niu C, Gao Y
and Su X: A droplet digital PCR assay for detection and
quantification of Verticillium nonalfalfae and V.
albo-atrum. Front Cell Infect Microbiol. 12:11106842023.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gundry CN, Vandersteen JG, Reed GH, Pryor
RJ, Chen J and Wittwer CT: Amplicon melting analysis with labeled
primers: A closed-tube method for differentiating homozygotes and
heterozygotes. Clin Chem. 49:396–406. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tamburro M and Ripabelli G: High
Resolution Melting as a rapid, reliable, accurate and
cost-effective emerging tool for genotyping pathogenic bacteria and
enhancing molecular epidemiological surveillance: A comprehensive
review of the literature. Ann Ig. 29:293–316. 2017.PubMed/NCBI
|
|
47
|
Hu M, Yang D, Wu X, Luo M and Xu F: A
novel high-resolution melting analysis-based method for Salmonella
genotyping. J Microbiol Methods. 172:1058062020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wen X, Chen Q, Yin H, Wu S and Wang X:
Rapid identification of clinical common invasive fungi via a
multi-channel real-time fluorescent polymerase chain reaction
melting curve analysis. Medicine (Baltimore). 99:e191942020.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Banowary B, Dang VT, Sarker S, Connolly
JH, Chenu J, Groves P, Ayton M, Raidal S, Devi A, Vanniasinkam T
and Ghorashi SA: Differentiation of Campylobacter jejuni and
campylobacter coli using Multiplex-PCR and high resolution melt
curve analysis. PLoS One. 10:e01388082015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tong SY, Dakh F, Hurt AC, Deng YM, Freeman
K, Fagan PK, Barr IG and Giffard PM: Rapid detection of the H275Y
oseltamivir resistance mutation in influenza A/H1N1 2009 by single
base pair RT-PCR and high-resolution melting. PLoS One.
6:e214462020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kafi H, Emaneini M, Halimi S, Rahdar HA,
Jabalameli F and Beigverdi R: Multiplex high-resolution melting
assay for simultaneous detection of five key bacterial pathogens in
urinary tract infections: A pilot study. Front Microbiol.
13:10491782022. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tong SY and Giffard PM: Microbiological
applications of high-resolution melting analysis. J Clin Microbiol.
50:3418–3421. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ghorbani J, Hashemi FB, Jabalameli F,
Emaneini M and Beigverdi R: Multiplex detection of five common
respiratory pathogens from bronchoalveolar lavages using high
resolution melting curve analysis. BMC Microbiol. 22:1412020.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zamani M, Furst AL and Klapperich CM:
Strategies for engineering affordable technologies for
point-of-Care diagnostics of infectious diseases. Acc Chem Res.
54:3772–3779. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Du J, Ma B, Li J, Wang Y, Dou T, Xu S and
Zhang M: Rapid detection and differentiation of legionella
pneumophila and Non-legionella pneumophila Species by using
recombinase polymerase amplification combined with EuNPs-based
lateral flow immunochromatography. Front Chem. 9:8151892022.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Soroka M, Wasowicz B and Rymaszewska A:
Loop-Mediated isothermal amplification (LAMP): The better sibling
of PCR? Cells. 10:19312021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Notomi T, Okayama H, Masubuchi H, Yonekawa
T, Watanabe K, Amino N and Hase T: Loop-mediated isothermal
amplification of DNA. Nucleic Acids Res. 28:E632000. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Parija SC and Poddar A: Molecular
diagnosis of infectious parasites in the post-COVID-19 era. Trop
Parasitol. 11:3–10. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Vo DT and Story MD: Facile and direct
detection of human papillomavirus (HPV) DNA in cells using
loop-mediated isothermal amplification (LAMP). Mol Cell Probes.
59:1017602021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen N, Si Y, Li G, Zong M, Zhang W, Ye Y
and Fan L: Development of a loop-mediated isothermal amplification
assay for the rapid detection of six common respiratory viruses.
Eur J Clin Microbiol Infect Dis. 40:2525–2532. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kim J, Park BG, Lim DH, Jang WS, Nam J,
Mihn DC and Lim CS: Development and evaluation of a multiplex
loop-mediated isothermal amplification (LAMP) assay for
differentiation of Mycobacterium tuberculosis and non-tuberculosis
mycobacterium in clinical samples. PLoS One. 16:e02447532021.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Phillips EA, Moehling TJ, Ejendal KFK,
Hoilett OS, Byers KM, Basing LA, Jankowski LA, Bennett JB, Lin LK,
Stanciu LA and Linnes JC: Microfluidic rapid and autonomous
analytical device (microRAAD) to detect HIV from whole blood
samples. Lab Chip. 19:3375–3386. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen X, Zhang J, Pan M, Qin Y, Zhao H, Qin
P, Yang Q, Li X, Zeng W, Xiang Z, et al: Loop-mediated isothermal
amplification (LAMP) assays targeting 18S ribosomal RNA genes for
identifying P. vivax and P. ovale species and
mitochondrial DNA for detecting the genus Plasmodium. Parasit
Vectors. 14:2782021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Trinh KTL and Lee NY: Fabrication of
wearable PDMS device for rapid detection of nucleic acids via
recombinase polymerase amplification operated by human body heat.
Biosensors (Basel). 12:722022. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Islam MN, Moriam S, Umer M, Phan HP,
Salomon C, Kline R, Nguyen NT and Shiddiky MJA: Naked-eye and
electrochemical detection of isothermally amplified HOTAIR long
non-coding RNA. Analyst. 143:3021–3028. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Mota DS, Guimarães JM, Gandarilla AMD,
Filho JCBS, Brito WR and Mariúba LAM: Recombinase polymerase
amplification in the molecular diagnosis of microbiological targets
and its applications. Can J Microbiol. 68:383–402. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li J, Macdonald J and von Stetten F:
Review: A comprehensive summary of a decade development of the
recombinase polymerase amplification. Analyst. 145:1950–1960. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Qi Y, Li W, Li X, Shen W, Zhang J, Li J,
Lv R, Lu N, Zong L, Zhuang S, et al: Development of rapid and
visual nucleic acid detection methods towards four serotypes of
human adenovirus species B based on RPA-LF test. Biomed Res Int.
2021:99577472021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mayran C, Foulongne V, Van de Perre P,
Fournier-Wirth C, Molès JP and Cantaloube JF: Rapid diagnostic test
for hepatitis B virus viral load based on recombinase polymerase
amplification combined with a lateral flow read-out. Diagnostics
(Basel). 12:6212022. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Li J, Pollak NM and Macdonald J: Multiplex
detection of nucleic acids using recombinase polymerase
amplification and a molecular colorimetric 7-Segment display. ACS
Omega. 4:11388–11396. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Munawar MA: Critical insight into
recombinase polymerase amplification technology. Expert Rev Mol
Diagn. 22:725–737. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Xu L, Duan J, Chen J, Ding S and Cheng W:
Recent advances in rolling circle amplification-based biosensing
strategies-A review. Anal Chim Acta. 1148:2381872021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Compton J: Nucleic acid sequence-based
amplification. Nature. 350:91–92. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Glökler J, Lim TS, Ida J and Frohme M:
Isothermal amplifications-a comprehensive review on current
methods. Crit Rev Biochem Mol Biol. 56:543–586. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Kia V, Tafti A, Paryan M and
Mohammadi-Yeganeh S: Evaluation of real-time NASBA assay for the
detection of SARS-CoV-2 compared with real-time PCR. Ir J Med Sci.
6:1–7. 2022.
|
|
76
|
Yrad FM, Castañares JM and Alocilja EC:
Visual detection of Dengue-1 RNA using gold nanoparticle-based
lateral flow biosensor. Diagnostics (Basel). 9:742019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Mohammadi-Yeganeh S, Paryan M, Mirab
Samiee S, Kia V and Rezvan H: Molecular beacon probes-base
multiplex NASBA Real-time for detection of HIV-1 and HCV. Iran J
Microbiol. 4:47–54. 2012.PubMed/NCBI
|
|
78
|
Gao YP, Huang KJ, Wang FT, Hou YY, Xu J
and Li G: Recent advances in biological detection with rolling
circle amplification: Design strategy, biosensing mechanism, and
practical applications. Analyst. 147:3396–3414. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wöhrle J, Krämer SD, Meyer PA, Rath C,
Hügle M, Urban GA and Roth G: Digital DNA microarray generation on
glass substrates. Sci Rep. 10:57702020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Xie C, Hu X, Liu Y and Shu C: Performance
comparison of GeneXpert MTB/RIF, gene chip technology, and modified
roche culture method in detecting mycobacterium tuberculosis and
drug susceptibility in sputum. Contrast Media Mol Imaging.
2022:29954642022. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Nasrabadi Z, Ranjbar R, Poorali F and
Sarshar M: Detection of eight foodborne bacterial pathogens by
oligonucleotide array hybridization. Electron Physician.
9:4405–4411. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ma X, Li Y, Liang Y, Liu Y, Yu L, Li C,
Liu Q and Chen L: Development of a DNA microarray assay for rapid
detection of fifteen bacterial pathogens in pneumonia. BMC
Microbiol. 20:1772020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Feng G, Han W, Shi J, Xia R and Xu J:
Analysis of the application of a gene chip method for detecting
Mycobacterium tuberculosis drug resistance in clinical specimens: A
retrospective study. Sci Rep. 11:179512021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Zhu L, Liu Q, Martinez L, Shi J, Chen C,
Shao Y, Zhong C, Song H, Li G, Ding X, et al: Diagnostic value of
GeneChip for detection of resistant Mycobacterium tuberculosis in
patients with differing treatment histories. J Clin Microbiol.
53:131–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sun B and Sun Y: Diagnostic performance of
DNA microarray for detecting rifampicin and isoniazid resistance in
Mycobacterium tuberculosis. J Thorac Dis. 13:4448–4454. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chandran S, Arjun R, Sasidharan A, Niyas
VK and Chandran S: Clinical performance of FilmArray
Meningitis/Encephalitis multiplex polymerase chain reaction panel
in central nervous system infections. Indian J Crit Care Med.
26:67–70. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Senescau A, Kempowsky T, Bernard E,
Messier S, Besse P, Fabre R and François JM: Innovative
DendrisChips® Technology for a syndromic approach of in
vitro diagnosis: Application to the respiratory infectious
diseases. Diagnostics (Basel). 8:772018. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Dien Bard J and McElvania E: Panels and
syndromic testing in clinical microbiology. Clin Lab Med.
40:393–420. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Gonsalves S, Mahony J, Rao A, Dunbar S and
Juretschko S: Multiplexed detection and identification of
respiratory pathogens using the NxTAG® respiratory
pathogen panel. Methods. 158:61–68. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Ma ZY, Deng H, Hua LD, Lei W, Zhang CB,
Dai QQ, Tao WJ and Zhang L: Suspension microarray-based comparison
of oropharyngeal swab and bronchoalveolar lavage fluid for pathogen
identification in young children hospitalized with respiratory
tract infection. BMC Infect Dis. 20:1682020. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Dunbar SA: Applications of Luminex xMAP
technology for rapid, high-throughput multiplexed nucleic acid
detection. Clin Chim Acta. 363:71–82. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Reslova N, Michna V, Kasny M, Mikel P and
Kralik P: xMAP technology: Applications in detection of pathogens.
Front Microbiol. 8:552017. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Dai Z, Li T, Li J, Han Z, Pan Y, Tang S,
Diao X and Luo M: High-throughput long paired-end sequencing of a
Fosmid library by PacBio. Plant Methods. 15:1422019. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Duan H, Li X, Mei A, Li P, Liu Y, Li X, Li
W, Wang C and Xie S: The diagnostic value of metagenomic
next-generation sequencing in infectious diseases. BMC Infect Dis.
21:622021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Grumaz S, Stevens P, Grumaz C, Decker SO,
Weigand MA, Hofer S, Brenner T, von Haeseler A and Sohn K:
Next-generation sequencing diagnostics of bacteremia in septic
patients. Genome Med. 8:732016. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lyimo BM, Popkin-Hall ZR, Giesbrecht DJ,
Mandara CI, Madebe RA, Bakari C, Pereus D, Seth MD, Ngamba RM,
Mbwambo RB, et al: Potential opportunities and challenges of
deploying next generation sequencing and CRISPR-Cas systems to
support diagnostics and surveillance towards malaria control and
elimination in africa. Front Cell Infect Microbiol. 12:7578442022.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Zhang XX, Guo LY, Liu LL, Shen A, Feng WY,
Huang WH, Hu HL, Hu B, Guo X, Chen TM, et al: The diagnostic value
of metagenomic next-generation sequencing for identifying
Streptococcus pneumoniae in paediatric bacterial meningitis.
BMC Infect Dis. 19:4952019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Huang J, Jiang E, Yang D, Wei J, Zhao M,
Feng J and Cao J: Metagenomic Next-generation sequencing versus
traditional pathogen detection in the diagnosis of peripheral
pulmonary infectious lesions. Infect Drug Resist. 13:567–576. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Dong Y, Gao Y, Chai Y and Shou S: Use of
quantitative metagenomics next-generation sequencing to confirm
fever of unknown origin and infectious disease. Front Microbio.
13:9310582022. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Gu L, Liu W, Ru M, Lin J, Yu G, Ye J, Zhu
ZA, Liu Y, Chen J, Lai G and Wen W: The application of metagenomic
next-generation sequencing in diagnosing Chlamydia psittaci
pneumonia: A report of five cases. BMC Pulm Med. 20:652020.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Jerome H, Taylor C, Sreenu VB, Klymenko T,
Filipe ADS, Jackson C, Davis C, Ashraf S, Wilson-Davies E,
Jesudason N, et al: Metagenomic next-generation sequencing aids the
diagnosis of viral infections in febrile returning travellers. J
Infect. 79:383–388. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Simner PJ, Miller S and Carroll KC:
Understanding the promises and hurdles of metagenomic
next-generation sequencing as a diagnostic tool for infectious
diseases. Clin Infect Dis. 66:778–788. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Yu X, Jiang W, Shi Y, Ye H and Lin J:
Applications of sequencing technology in clinical microbial
infection. J Cell Mol Med. 23:7143–7150. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Gu W, Miller S and Chiu CY: Clinical
metagenomic next-generation sequencing for pathogen detection. Annu
Rev Patho. 14:319–338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhang L, Chen F, Zeng Z, Xu M, Sun F, Yang
L, Bi X, Lin Y, Gao Y, Hao H, et al: Advances in metagenomics and
its application in environmental microorganisms. Front Microbiol.
12:7663642021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wang X, Liu Y, Liu H, Pan W, Ren J, Zheng
X, Tan Y, Chen Z, Deng Y, He N, et al: Recent advances and
application of whole genome amplification in molecular diagnosis
and medicine. Med Comm. 3:e1162022.
|
|
107
|
Athanasopoulou K, Boti MA, Adamopoulos PG,
Skourou PC and Scorilas A: Third-Generation sequencing: The
spearhead towards the radical transformation of modern genomics.
Life (Basel). 12:302021.PubMed/NCBI
|
|
108
|
Keller MW, Rambo-Martin BL, Wilson MM,
Ridenour CA, Shepard SS, Stark TJ, Neuhaus EB, Dugan VG, Wentworth
DE and Barnes JR: Author Correction: Direct RNA Sequencing of the
coding complete influenza A virus genome. Sci Rep. 8:157462018.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Wang M, Fu A, Hu B, Tong Y, Liu R, Liu Z,
Gu J, Xiang B, Liu J, Jiang W, et al: Nanopore targeted sequencing
for the accurate and comprehensive detection of SARS-CoV-2 and
other respiratory viruses. Small. 16:e20021692020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Mongan AE, Tuda JSB and Runtuwene LR:
Portable sequencer in the fight against infectious disease. J Hum
Genet. 65:35–40. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Holshue ML, DeBolt C, Lindquist S, Lofy
KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural
A, et al: First case of 2019 novel coronavirus in the united
states. N Engl J Med. 382:929–936. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Wongsurawat T, Jenjaroenpun P, Taylor MK,
Lee J, Tolardo AL, Parvathareddy J, Kandel S, Wadley TD, Kaewnapan
B, Athipanyasilp N, et al: Rapid Sequencing of Multiple RNA Viruses
in Their Native Form. Front Microbiol. 10:2602019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Fang Y, Changavi A, Yang M, Sun L, Zhang
A, Sun D, Sun Z, Zhang B and Xu MQ: Nanopore Whole Transcriptome
Analysis and Pathogen Surveillance by a Novel Solid-Phase Catalysis
Approach. Adv Sci (Weinh). 9:e21033732022. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Akaçin İ, Ersoy Ş, Doluca O and
Güngörmüşler M: Comparing the significance of the utilization of
next generation and third generation sequencing technologies in
microbial metagenomics. Microbiol Res. 264:1271542022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Gradisteanu Pircalabioru G, Iliescu FS,
Mihaescu G, Cucu AI, Ionescu ON, Popescu M, Simion M, Burlibasa L,
Tica M, Chifiriuc MC and Iliescu C: Advances in the rapid
diagnostic of viral respiratory tract infections. Front Cell Infect
Microbiol. 12:8072532022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Sheng L, Lu Y, Deng S, Liao X, Zhang K,
Ding T, Gao H, Liu D, Deng R and Li J: A transcription aptasensor:
Amplified, label-free and culture-independent detection of
foodborne pathogens via light-up RNA aptamers. Chem Commun (Camb).
55:10096–10099. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Andryukov BG, Lyapun IN, Matosova EV and
Somova LM: Biosensor technologies in medicine: From detection of
biochemical markers to research into molecular targets (review).
Sovrem Tekhnologii Med. 12:70–83. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Robertson KL and Vora GJ: Locked nucleic
acid flow cytometry-fluorescence in situ hybridization (LNA
flow-FISH): A method for bacterial small RNA detection. J Vis Exp.
10:e36552012.PubMed/NCBI
|
|
119
|
Freen-van Heeren JJ: Flow-FISH as a tool
for studying bacteria, fungi and viruses. BioTech (Basel).
10:212021. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Israr MZ, Bernieh D, Salzano A, Cassambai
S, Yazaki Y and Suzuki T: Matrix-assisted laser desorption
ionisation (MALDI) mass spectrometry (MS): basics and clinical
applications. Clin Chem Lab Med. 58:883–896. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Kailasa SK, Koduru JR, Park TJ, Wu HF and
Lin YC: Progress of electrospray ionization and rapid evaporative
ionization mass spectrometric techniques for the broad-range
identification of microorganisms. Analyst. 145:70722020. View Article : Google Scholar : PubMed/NCBI
|