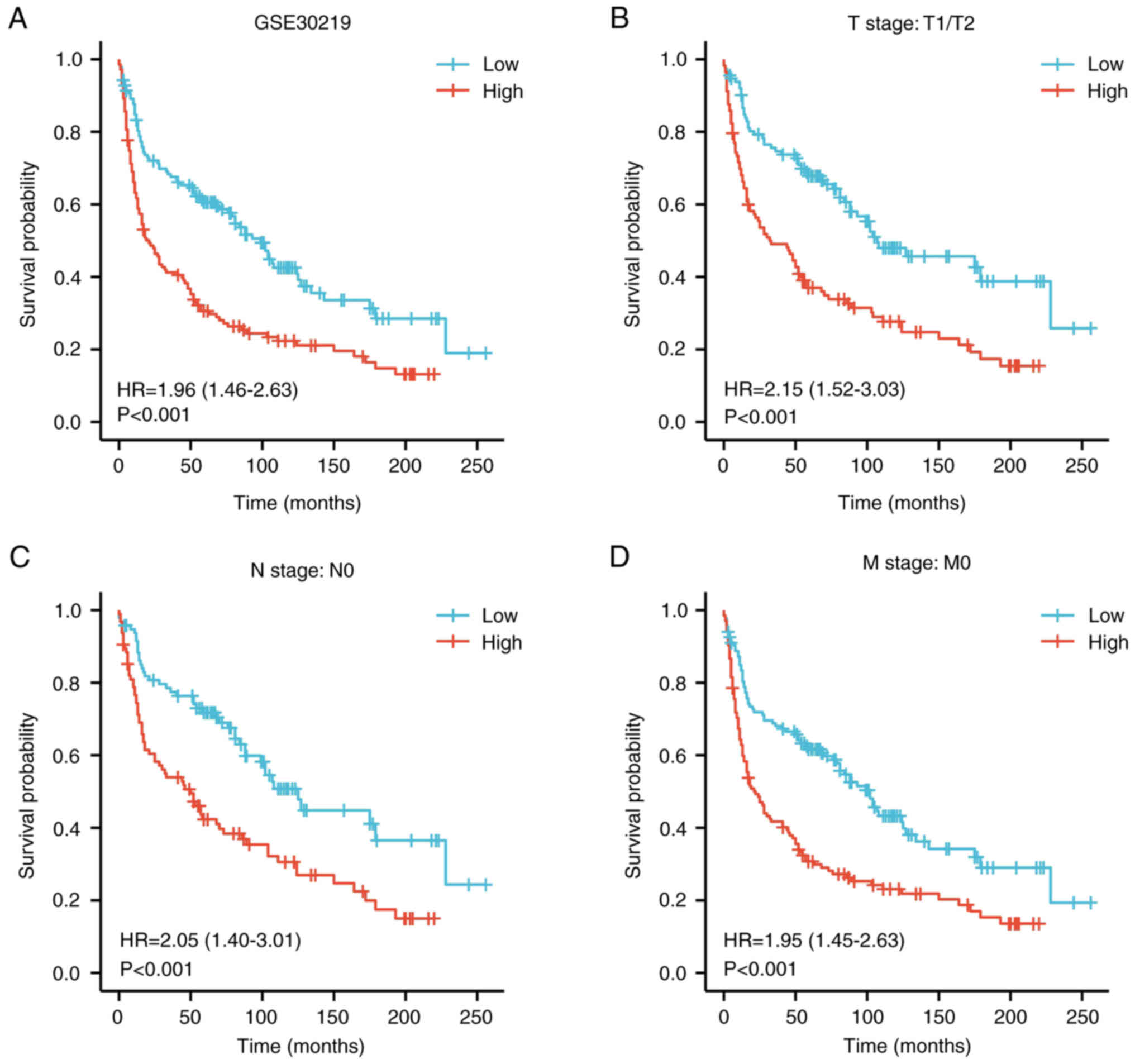
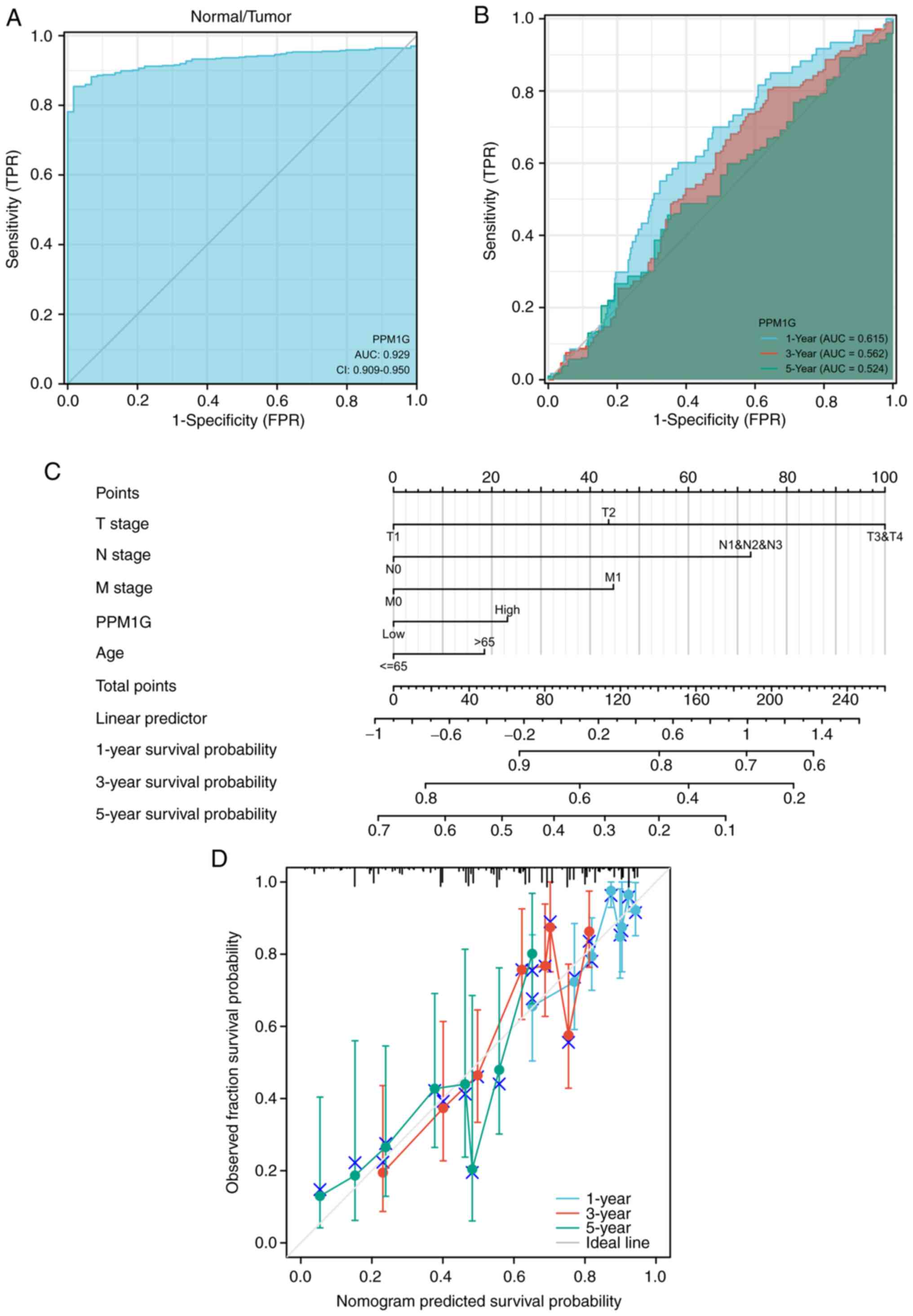
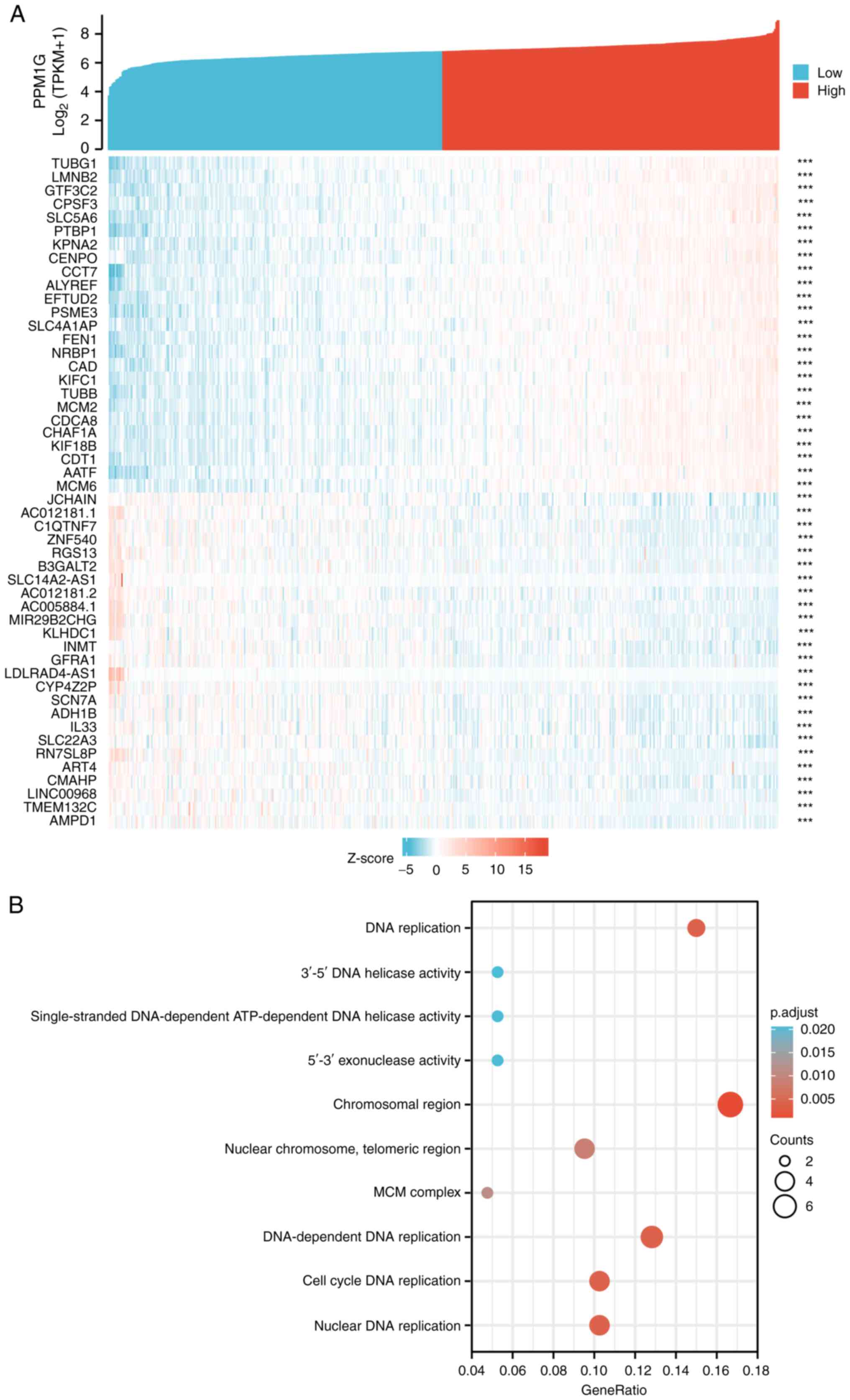
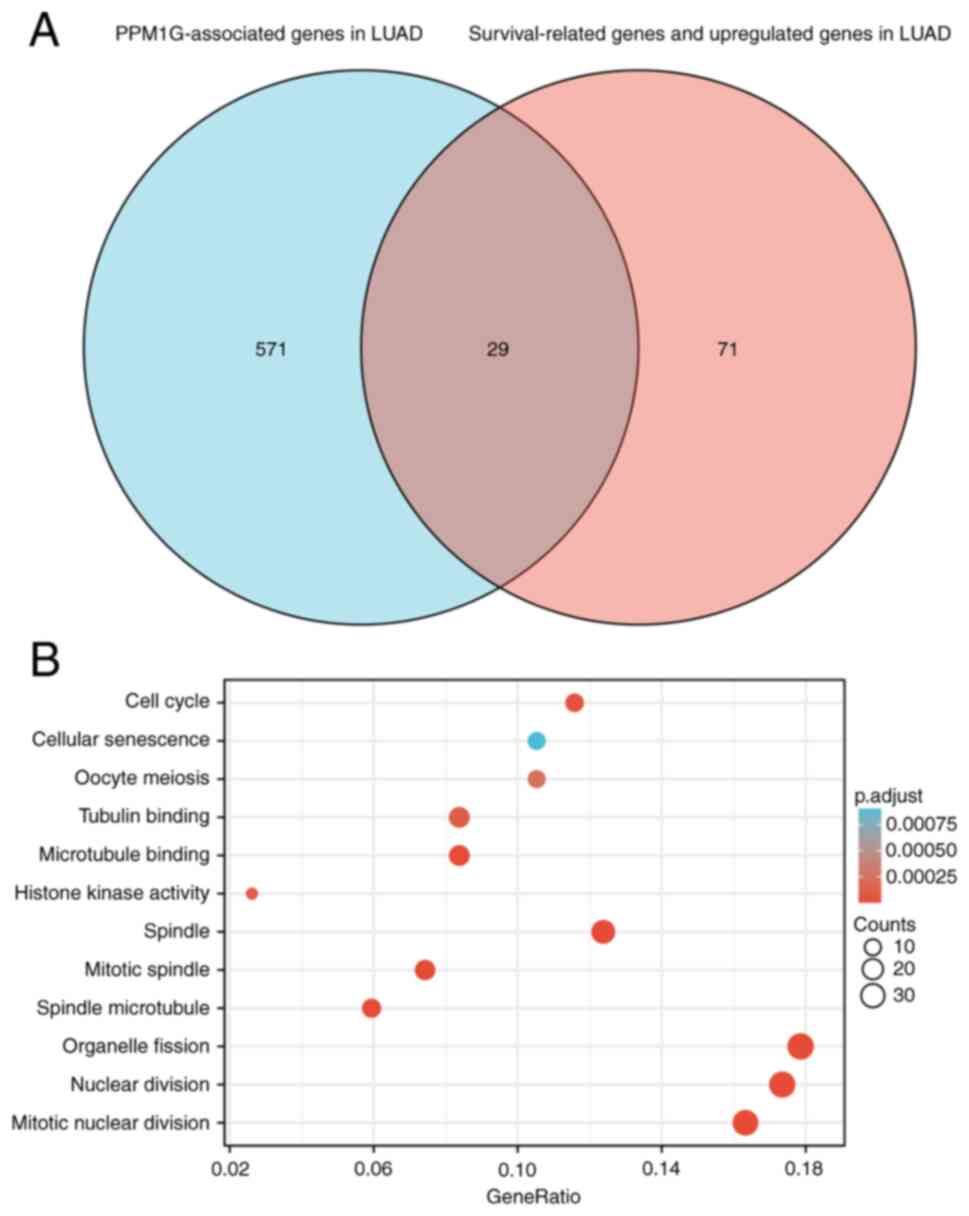
|
1
|
Wang X, Chen X and Liu H: Expression and
bioinformatics-based functional analysis of UAP1 in lung
adenocarcinoma. Cancer Manag Res. 12:12111–12121. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cho JH: Immunotherapy for non-small-cell
lung cancer: Current status and future obstacles. Immune Netw.
17:378–391. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
O'Brien TD, Jia P, Caporaso NE, Landi MT
and Zhao Z: Weak sharing of genetic association signals in three
lung cancer subtypes: Evidence at the SNP, gene, regulation, and
pathway levels. Genome Med. 10:162018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Friedlaender A, Banna G, Malapelle U,
Pisapia P and Addeo A: Next generation sequencing and genetic
alterations in squamous cell lung carcinoma: Where are we today?
Front Oncol. 9:1662019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
AbdulJabbar K, Raza SEA, Rosenthal R,
Jamal-Hanjani M, Veeriah S, Akarca A, Lund T, Moore DA, Salgado R,
Al Bakir M, et al: Geospatial immune variability illuminates
differential evolution of lung adenocarcinoma. Nat Med.
26:1054–1062. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Herbst RS, Morgensztern D and Boshoff C:
The biology and management of non-small cell lung cancer. Nature.
553:446–454. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Avancini A, Sartori G, Gkountakos A,
Casali M, Trestini I, Tregnago D, Bria E, Jones LW, Milella M,
Lanza M and Pilotto S: Physical activity and exercise in lung
cancer care: Will promises be fulfilled? Oncologist. 25:e555–e569.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kamada R, Kudoh F, Ito S, Tani I, Janairo
JIB, Omichinski JG and Sakaguchi K: Metal-dependent Ser/Thr protein
phosphatase PPM family: Evolution, structures, diseases and
inhibitors. Pharmacol Ther. 215:1076222020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin YR, Yang WJ and Yang GW: Prognostic
and immunological potential of PPM1G in hepatocellular carcinoma.
Aging (Albany NY). 13:12929–12954. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang M, Xu E, Zhang J and Chen X: PPM1D
phosphatase, a target of p53 and RBM38 RNA-binding protein,
inhibits p53 mRNA translation via dephosphorylation of RBM38.
Oncogene. 34:5900–5911. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peng TS, He YH, Nie T, Hu XD, Lu HY, Yi J,
Shuai YF and Luo M: PPM1D is a prognostic marker and therapeutic
target in colorectal cancer. Exp Ther Med. 8:430–434. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li K, Liu Y, Xu S and Wang J: PPM1D
functions as oncogene and is associated with poor prognosis in
esophageal squamous cell carcinoma. Pathol Oncol Res. 26:387–395.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang C, Chen Y, Wang M, Chen X, Li Y,
Song E, Liu X, Kim S and Peng H: PPM1D silencing by RNA
interference inhibits the proliferation of lung cancer cells. World
J Surg Oncol. 12:2582014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang H, Gao XY, Li P and Jiang TS: PPM1D
overexpression predicts poor prognosis in non-small cell lung
cancer. Tumour Biol. 36:2179–2184. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun C, Wang G, Wrighton KH, Lin H,
Songyang Z, Feng XH and Lin X: Regulation of p27Kip1
phosphorylation and G1 cell cycle progression by protein
phosphatase PPM1G. Am J Cancer Res. 6:2207–2220. 2016.PubMed/NCBI
|
|
17
|
Chen D, Zhao Z, Chen L, Li Q, Zou J and
Liu S: PPM1G promotes the progression of hepatocellular carcinoma
via phosphorylation regulation of alternative splicing protein
SRSF3. Cell Death Dis. 12:7222021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Di C, Syafrizayanti, Zhang Q, Chen Y, Wang
Y, Zhang X, Liu Y, Sun C, Zhang H and Hoheisel JD: Function,
clinical application, and strategies of Pre-mRNA splicing in
cancer. Cell Death Differ. 26:1181–1194. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46(D1): D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pachter L: Models for transcript
quantification from RNA-Seq. arXiv preprint arXiv:11043889.
2011.
|
|
21
|
Rousseaux S, Debernardi A, Jacquiau B,
Vitte AL, Vesin A, Nagy-Mignotte H, Moro-Sibilot D, Brichon PY,
Lantuejoul S, Hainaut P, et al: Ectopic activation of germline and
placental genes identifies aggressive metastasis-prone lung
cancers. Sci Transl Med. 5:186ra1662013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Moreno Leon L, Gautier M, Allan R, Ilié M,
Nottet N, Pons N, Paquet A, Lebrigand K, Truchi M, Fassy J, et al:
The nuclear hypoxia-regulated NLUCAT1 long non-coding RNA
contributes to an aggressive phenotype in lung adenocarcinoma
through regulation of oxidative stress. Oncogene. 38:7146–7165.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Landi MT, Dracheva T, Rotunno M, Figueroa
JD, Liu H, Dasgupta A, Mann FE, Fukuoka J, Hames M, Bergen AW, et
al: Gene expression signature of cigarette smoking and its role in
lung adenocarcinoma development and survival. PLoS One.
3:e16512008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu
JS, Li B and Liu XS: TIMER: A web server for comprehensive analysis
of tumor-infiltrating immune cells. Cancer Res. 77:e108–e110. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stockhausen K: The declaration of
Helsinki: Revising ethical research guidelines for the 21st
century. Med J Aust. 172:252–253. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lánczky A, Nagy Á, Bottai G, Munkácsy G,
Szabó A, Santarpia L and Győrffy B: miRpower: A web-tool to
validate survival-associated miRNAs utilizing expression data from
2178 breast cancer patients. Breast Cancer Res. 160:439–446. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45(D1): D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Liu W, Li JC, Li M, Li B and Zhu
R: Hepcidin downregulation correlates with disease aggressiveness
and immune infiltration in liver cancers. Front Oncol.
11:7147562021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43((Database Issue)): D447–D452. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Prates L, Lemes RB, Hünemeier T and
Leonardi F: Population-based change-point detection for the
identification of homozygosity islands. Bioinformatics.
39:btad1702023. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wickham H: ggplot2: Elegant graphics for
data analysis. New York, NY: Springer; 2009, View Article : Google Scholar
|
|
35
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Darvin P, Toor SM, Sasidharan Nair V and
Elkord E: Immune checkpoint inhibitors: Recent progress and
potential biomarkers. Exp Mol Med. 50:1–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ge MX, Liu HT, Zhang N, Niu WX, Lu ZN, Bao
YY, Huang R, Yu DK, Shao RG and He HW: Costunolide represses
hepatic fibrosis through WW domain-containing protein 2-mediated
Notch3 degradation. Br J Pharmacol. 177:372–387. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pan C, Liu HD, Gong Z, Yu X, Hou XB, Xie
DD, Zhu XB, Li HW, Tang JY, Xu YF, et al: Cadmium is a potent
inhibitor of PPM phosphatases and targets the M1 binding site. Sci
Rep. 3:23332013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chaudhary N and Maddika S: WWP2-WWP1
ubiquitin ligase complex coordinated by PPM1G maintains the balance
between cellular p73 and ΔNp73 levels. Mol Cell Biol. 34:3754–3764.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xiao Q, Cheng Z, Kuang W, Wu H, Luo X and
Wang R: Clinical value of PPM1G gene in survival prognosis and
immune infiltration of hepatocellular carcinoma. Appl Bionics
Biomech. 2022:89262212022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen J, Li J, Sun H, Hu T, Wang Y, Kang G,
Cao M and Li X: PPM1G promotes the progression of lung
adenocarcinoma by inhibiting p38 activation via dephosphorylation
of MEK6. Carcinogenesis. 44:93–104. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gudipaty SA, McNamara RP, Morton EL and
D'Orso I: PPM1G binds 7SK RNA and hexim1 To block P-TEFb assembly
into the 7SK snRNP and sustain transcription elongation. Mol Cell
Biol. 35:3810–3828. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kumar P, Tathe P, Chaudhary N and Maddika
S: PPM1G forms a PPP-type phosphatase holoenzyme with B56δ that
maintains adherens junction integrity. EMBO Rep. 20:e469652019.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dyer DP, Medina-Ruiz L, Bartolini R,
Schuette F, Hughes CE, Pallas K, Vidler F, Macleod MKL, Kelly CJ,
Lee KM, et al: Chemokine receptor redundancy and specificity are
context dependent. Immunity. 50:378–389.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang M, Chang M, Li C, Chen Q, Hou Z, Xing
B and Lin J: Tumor-microenvironment-activated reactive oxygen
species amplifier for enzymatic cascade cancer
starvation/chemodynamic /immunotherapy. Adv Mater. 34:e21060102022.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Stankovic B, Bjørhovde HAK, Skarshaug R,
Aamodt H, Frafjord A, Müller E, Hammarström C, Beraki K, Bækkevold
ES, Woldbæk PR, et al: Immune cell composition in human non-small
cell lung cancer. Front Immunol. 9:31012019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Welsh TJ, Green RH, Richardson D, Waller
DA, O'Byrne KJ and Bradding P: Macrophage and mast-cell invasion of
tumor cell islets confers a marked survival advantage in
non-small-cell lung cancer. J Clin Oncol. 23:8959–8967. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kilic A, Landreneau RJ, Luketich JD,
Pennathur A and Schuchert MJ: Density of tumor-infiltrating
lymphocytes correlates with disease recurrence and survival in
patients with large non-small-cell lung cancer tumors. J Surg Res.
167:207–210. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen X, Wan J, Liu J, Xie W, Diao X, Xu J,
Zhu B and Chen Z: Increased IL-17-producing cells correlate with
poor survival and lymphangiogenesis in NSCLC patients. Lung Cancer.
69:348–354. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Horne ZD, Jack R, Gray ZT, Siegfried JM,
Wilson DO, Yousem SA, Nason KS, Landreneau RJ, Luketich JD and
Schuchert MJ: Increased levels of tumor-infiltrating lymphocytes
are associated with improved recurrence-free survival in stage 1A
non-small-cell lung cancer. J Surg Res. 171:1–5. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Al-Shibli KI, Donnem T, Al-Saad S, Persson
M, Bremnes RM and Busund LT: Prognostic effect of epithelial and
stromal lymphocyte infiltration in non-small cell lung cancer. Clin
Cancer Res. 14:5220–5227. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zuazo M, Arasanz H, Fernández-Hinojal G,
García-Granda MJ, Gato M, Bocanegra A, Martínez M, Hernández B,
Teijeira L, Morilla I, et al: Functional systemic CD4 immunity is
required for clinical responses to PD-L1/PD-1 blockade therapy.
EMBO Mol Med. 11:e102932019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Frafjord A, Buer L, Hammarström C, Aamodt
H, Woldbæk PR, Brustugun OT, Helland Å, Øynebråten I and Corthay A:
The immune landscape of human primary lung tumors Is Th2 skewed.
Front Immunol. 12:7645962021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Phua SC, Chiba S, Suzuki M, Su E, Roberson
EC, Pusapati GV, Schurmans S, Setou M, Rohatgi R, Reiter JF, et al:
Dynamic remodeling of membrane composition drives cell cycle
through primary cilia excision. Cell. 178:2612019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kanemitsu N, Ebisuno Y, Tanaka T, Otani K,
Hayasaka H, Kaisho T, Akira S, Katagiri K, Kinashi T, Fujita N, et
al: CXCL13 is an arrest chemokine for B cells in high endothelial
venules. Blood. 106:2613–2618. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Pallandre JR, Krzewski K, Bedel R, Ryffel
B, Caignard A, Rohrlich PS, Pivot X, Tiberghien P, Zitvogel L,
Strominger JL and Borg C: Dendritic cell and natural killer cell
cross-talk: A pivotal role of CX3CL1 in NK cytoskeleton
organization and activation. Blood. 112:4420–4424. 2008. View Article : Google Scholar : PubMed/NCBI
|