Diabetes mellitus (DM) is one of the most common and
costly chronic metabolic diseases. Compared with 451 million adults
in 2017, it is estimated that there will be 693 million adults with
DM worldwide by 2045 (1).
Long-term hyperglycaemia may lead to macrovascular and
microvascular complications, such as cerebro-cardiovascular
diseases, diabetic nephropathy (DN), diabetic retinopathy (DR),
diabetic neuropathy (DNP) and diabetic cardiomyopathy (DCM),
leading to increased disability and mortality of diabetic patients
(1,2). However, early diagnosis and precisely
targeted treatment of diabetic complications may reduce the medical
burden and improve prognosis.
Circular RNAs (circRNAs) are novel noncoding RNAs
that were verified in viruses by electron microscopy. Recently,
thousands of circRNAs in eukaryotes have been identified by
high-throughput RNA sequencing (RNA-seq). Of note, they have
differentially expressed tissue patterns (3,4) and
may be detected non-invasively in blood samples (5). CircRNAs possess covalently closed
circular structures and are difficult to degrade, suggesting that
circRNAs exhibit higher stability than linear RNAs (6). In addition, circRNAs have been
demonstrated to participate in the occurrence and development of
various diseases (5,7–9),
which is well known in the field of oncology (10,11).
However, noteworthy results have become apparent from studies
investigating diabetic complications (5), according to which circRNAs may be
promising biomarkers for diabetic complications. The
circRNA_15698/microRNA (miR)-185/transforming growth factor
(TGF)-β1 pathway has been found to participate in the pathogenesis
of DN (12). In addition, circRNA
homeodomain interacting protein kinase 3 (circHIPK3) has been
demonstrated to be involved in DR and DCM both in vivo and
in vitro (13,14). Furthermore, circRNA_000203
expression is upregulated in the myocardium of diabetic mice and
caspase-1-associated circRNA (CACR) is enhanced in the serum of
patients with DM and cardiomyocytes exposed to high glucose (HG)
(15,16).
The present review summarizes recent studies about
circRNAs and diabetic complications, highlighting the unmet needs
and future research directions for diabetic complications. In doing
so, the study aims to provide novel concepts of prevention and
treatment for diabetic complications.
CircRNAs are closed circular single-stranded RNA
molecules formed by the variable cutting of pre-mRNA, without
polyadenylation [poly(A)] and capping. According to the sequence
type, circRNAs are divided into exon types, intron types and
exon-intron complexes. CircRNAs are generated by back splicing, in
which downstream splicing donor sites are covalently linked with
upstream splice-acceptor sites (Fig.
1) (17). This high stability
is attributed to covalently closed circular structures, which
protect these molecules from exonuclease-mediated degradation
compared to their linear mRNA counterparts (18). Furthermore, the high stability of
circRNAs may allow them to be detected non-invasively in bodily
fluids (10,19,20).
Thus, circRNA may be a promising biomarker for various
diseases.
There are three aspects in regulating circRNA
biogenesis: i) Regulating circRNA-producing pre-mRNA-circRNAs may
compete with linear splicing to exert gene regulation (21). ii) CircRNA degradation-circRNAs may
be degraded by endonuclease activity in vitro, such as RNase
H and RNase L (22). Furthermore,
circRNA levels may be reduced through sponging miRNAs. For
instance, circRNA circular cerebellar degeneration-related protein
1 antisense (circCDR1as) is degraded by sponging miR-671 via
protein Argonaute 2 (23). In
addition, circRNAs may be secreted into blood and urine in the form
of exosomes, which are eliminated by the liver, kidney and
reticuloendothelial system (20,24).
iii) Controlling the back-splicing machinery (cis-regulatory
elements and trans-acting factors). Usually, back-splicing events
are facilitated by flanking inverted repeated Alu pairs (25). However, it is mammalian-wide
interspersed repeats that regulate the circularization of
circCDR1as (26).
The four main biological functions of circRNAs are
as follows: Acting as miRNA sponges [competing endogenous (ceRNAs)]
(18), RNA-binding protein (RBP)
sponges, protein-coding in the cytoplasm and transcriptional
regulation of host genes in the nucleus (Fig. 1). Individual circRNAs act as miRNA
sponges or decoys, protecting target mRNAs from miRNA-dependent
degradation. Thus, the target RNAs are more actively translated and
bound by ribosomes (27) (Fig. 1). CircCDR1as and circRNA zinc
finger protein 91 are the most likely circRNAs to act as miRNA
sponges, since they contain a large number of conserved binding
sites for miRNAs. CiRS-7, containing 63 target sites for miR-7,
upregulates tumor genes through increasing miR-7 target mRNAs
(8,28). In addition, circRNA zinc finger
protein 91, with 24 binding sites for miR-23b-3p, participates in
the differentiation of keratinocytes (29). More so, circRNA coiled-coil domain
containing 66 contains a variety of miRNA binding sites, including
miR-33b and miR-93, which target the MYC gene (30). Furthermore, circCHIPK3 regulates
HG-induced human umbilical vein endothelial cell (HUVEC) apoptosis
by sponging miR-124 and regulating retinal endothelial cell (EC)
proliferation, viability, migration and tube formation in
HG-induced human retinal vascular ECs (HRVECs) by decoying
miR-30a-3p (13,31).
CircRNAs acting as ceRNAs are more commonly studied
in diabetic complications, but the other three biological functions
of circRNAs are generally used in oncology. RBP sponges would block
the process from mRNA to proteins, as RBPs are able to control the
splicing stability and the translation of mRNAs. Therefore,
circRNAs indirectly intervene in the post-transcriptional steps of
mRNAs. In addition, circSMARCA5 regulates vascular endothelial
growth factor (VEGF)A mRNA splicing in glioblastoma multiforme
through the binding of serine and arginine rich splicing factor 1
(32). Abdelmohsen et al
(33) found that RNA-binding
protein human antigen R (HuR) directly interacted with autophagy
related 16 like 1 (ATG16L1) mRNA via the 3′-UTR and enhanced
ATG16L1 translation. Thus, circRNA poly(A) binding protein nuclear
1 (circPABPN1) was able to block HuR binding to Atg16l1 mRNA and
decrease ATG16L1 protein production. The results suggest that the
interaction between HuR and circPABPN1 modulated autophagy in the
intestinal epithelium by altering ATG16L1 translation (34). As circRNAs lack the necessary
elements for translation, such as the 5′Cap and Poly(A) tail,
circRNAs may be translated either through the internal ribosomal
entry site or after m6A RNA modification at 5′-UTR (35,36).
CircRNA eukaryotic translation initiation factor 6 (EIF6) encodes a
novel peptide called EIF6-224AA, which is responsible for the
carcinogenic effect of circEIF6 in triple-negative breast cancer
via stabilizing myosin heavy chain 9 and activating the
Wnt/β-catenin pathway (37).
Likewise, circRNAs also act as transcriptional regulators of host
genes by binding to RNA polymerase II (12,38).
More so, circular intronic RNA ankyrin repeat domain 52 (ANKRD52)
was able to regulate its host gene, ANKRD52, in transcriptional
elongation by influencing polymerase II (39).
Chronic diabetic complications increase disability
and mortality in patients with DM. The availability of circRNA-seq
and bioinformatics have indicated the critical role of circRNA in
the pathogenetic mechanisms of diabetic complications and were
reviewed below.
Diabetic patients are more prone to suffering from
life-limiting and life-threatening macrovascular events, such as
cardio-cerebrovascular diseases (40). Of note, diabetic macrovascular
complications are closely related to the dysfunction of vascular
ECs and proliferation of vascular smooth muscle cells (VSMCs),
resulting in atherosclerosis under HG conditions. In two studies,
as many as 95–214 differentially expressed circRNAs were found in
HG-induced HUVECs by RNA-seq (41,42).
Pan et al (43) also found
that HG increased the expression of human circular RNA-0054633
(hsa_circ_0054633), while downregulation of hsa_circ_0054633
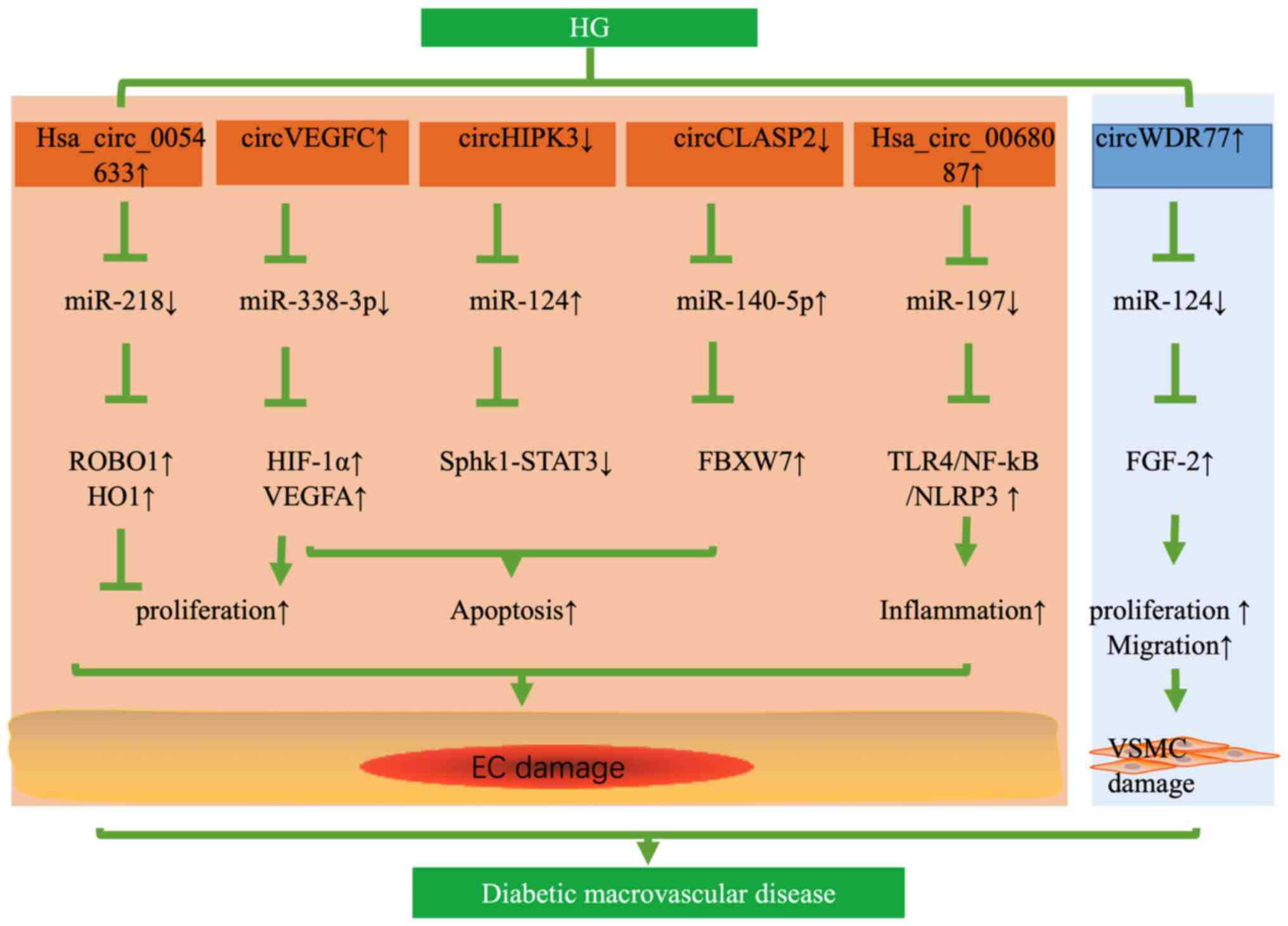
aggravated the HG-induced EC dysfunction. In addition,
hsa_circ_0054633 has a protective effect against HG-induced ECs
dysfunction through the miR-218/roundabout 1 and heme oxygenase-1
axes in vitro (43). The
expression of circHIPK3 was decreased in HG-induced HUVECs and
human aortic ECs from patients with type 2 DM (T2DM).
Overexpression of circHIPK3 inhibited HG-induced apoptosis of ECs
via the miR-124 axis (31).
Likewise, the expression of circRNA CLIP-associating protein 2
(circCLASP2) was downregulated in HG-induced HUVECs. By contrast,
upregulation of circCLASP2 inhibited apoptosis of HUVECs under HG
conditions via miR-140-5p/F-box and the WD repeat domain-containing
7 axis (44). Therefore,
upregulation of hsa_circ_0054633, circHIPK3 and circCLASP2 was able
to alleviate HG-induced EC dysfunction.
However, various circRNAs have deleterious effects
on blood vessels and may result in the dysfunction of ECs. Wei
et al (45) reported that
circVEGFC was upregulated in HG-induced HUVECs and able to sponge
miR-338-3p; the latter targeted the 3′-UTR of hypoxia-inducible
factor 1α (HIF-1α), thereby activating the transcription of VEGFA.
Downregulation of circVEGFC was able to reduce HG-induced apoptosis
of HUVECs and recover the proliferation through the
miR-338-3p/HIF-1α/VEGFA axis (45). Similarly, downregulation of
hsa_circ_0068087 inhibited HG-induced HUVEC inflammation by
suppressing the Toll-like receptor (TLR)4/NF-κB/NOD-like receptor
thermal protein domain associated protein 3 (NLRP3) pathway.
However, the effects of inhibiting inflammation disappeared through
downregulation of miR-197, suggesting that circ_0068087 functioned
as a sponge of miR-197 (46)
(Fig. 2). Chen et al
(47) determined circRNA
expression profiles in HG-induced VSMCs by circRNAs microarray
analysis and verified that circWDR77 was upregulated in HG-induced
VSMCs. Knockout of circWDR77 inhibited HG-induced VSMC
proliferation and migration targeting miR-124/FGF-2. In their
study, an RNA immunoprecipitation assay was used to validate the
interaction between circRNA WD repeat domain 77 (circWDR77) and
miR-124, suggesting that circWDR77 acted as a ceRNA of miR-124
(47) (Fig. 2). Recently, Zaiou (48) also indicated that these
differentially expressed circRNAs in HG conditions among studies
may act as vital contributors to the impairment of vascular ECs and
the proliferation of VSMCs, therefore being involved in
cardiovascular diseases. Fang et al (49) also found that circANKRD36 was
markedly upregulated in the peripheral blood leucocytes of patients
with T2DM and related to inflammatory factors, and speculated that
circANKRD36 may serve as a biomarker for the development of
inflammatory CVD among diabetic patients. Therefore, downregulation
of circVEGFC, hsa_circ_0068087, circWDR77 may alleviate HG-induced
dysfunction of ECs and VSMCs. Novel circRNAs have been found that
may serve as therapeutic targets for cardio-cerebrovascular
diseases and the prediction of inflammation in type 2 diabetes.
Microvascular complications of DM comprise DN, DR
and DNP. DN is a leading cause of end-stage renal disease
worldwide. However, the current treatment for DN is insufficient.
DR and, notably, DNP severely affect the life quality of patients
with DM. Thus, there is an urgent need to further explore ideal
therapies for diabetic microvascular complications. As such,
circRNAs can regulate the occurrence and development of diseases,
and it is reported that there is a significant relationship between
microangiopathy and circRNAs (50).
The underlying mechanisms of DN include oxidative
stress, inflammatory cell recruitment and infiltration, mesangial
cell hypertrophy, tubulointerstitial fibrosis, as well as podocyte
loss and apoptosis (12,51–57).
Recently, the roles of circRNAs in the pathogenesis of DN have
received increasing attention. Evidence for the roles of circRNAs
in DN is mainly derived from mesangial cells (MCs), tubular
epithelial cells (TECs) and podocytes (Fig. 3). Hu et al (12) revealed that circRNA_15698, a sponge
of miR-185, was upregulated in the kidney cortex of db/db mice and
HG-treated MCs. Likewise, knockdown of circRNA_15698 alleviated
extracellular matrix (ECM) accumulation by inhibiting the
expression of transforming growth factor-β 1 (TGF-β1) protein in
vivo and in vitro. The results suggested that the
CircRNA_15698/miR-185/TGF-β1 axis has a role in diabetic renal
fibrosis (12). Mou et al
(58) confirmed 18 upregulated
circRNAs and 22 downregulated circRNAs in the DN kidney from db/db
mice using circRNA-seq. Furthermore, circ_0000491 levels were
significantly augmented in DN mice and HG-induced mouse MCs. In
addition, circ-0000491 sponged miR-101b and activated TGFβR1,
leading to ECM accumulation (58).
Chen et al (52) also
demonstrated that circRNA LDL receptor related protein 6 (circLRP6)
was increased in HG-treated mouse mesangial SV40-Mes13 cells.
Knockdown of circLRP6 mitigated HG-induced proliferation, oxidative
stress, inflammation and ECM in MCs via upregulating miR-205 and
repressing the high mobility group box 1 and TLR4/NF-κB pathway.
Similarly, Ge et al (54)
found that circ_0000064 was upregulated in HG-induced mouse MCs and
knockdown of circ_0000064 inhibited cell proliferation and fibrosis
through upregulating miR-143.
Accumulating research has demonstrated that circRNAs
are associated with tubular epithelial cell damage in DN (Fig. 3). Wen et al (59) explored the circRNA expression
profiles and found that circRNA circular RNA actin related protein
2 (circACTR2) was upregulated in glucose-stressed HK-2 cells and
mediated inflammation and pyroptosis. Knockdown of circACTR2
prevented HG-induced pyroptosis, inflammation and fibrosis of TECs,
suggesting that circACTR2 has a vital role in the pathogenesis of
DN (59). Analogously, circRNA
Williams-Beuren syndrome chromosome region 17 (circ_0080425) was
increased in kidney tissues from streptozotocin (STZ)-induced
diabetic mice and HG-induced human kidney tubular cells and
aggravated inflammatory responses and kidney fibrosis by targeting
the miR-185-5p/transcription factor SOX 6 axis (56). CircRNA eukaryotic translation
initiation factor 4 gamma 2 (circEIF4G2) was increased in db/db
mice and HG-induced NRK-52E cells. By contrast, the downregulation
of circEIF4G2 mitigated renal fibrosis in DN by sponging miR-218
(60). Recent evidence also
suggested that circRNAs are involved in the pathological processes
of podocytes injury and apoptosis in DN (Fig. 3). Yao et al (55) found that circ_0000285 was increased
in kidney tissues of mouse models of DN and podocytes exposed to
HG, leading to inflammation and podocyte injury through sponging
miR-654-3p/mitogen-activated protein kinase 6.
Although most circRNAs have negative effects on DN,
certain circRNAs have protective effects against DN. CircHIPK3 was
decreased in HG-induced HK-2 cells, while upregulation of circHIPK3
could reverse the HG-induced HK-2 cell proliferation and apoptosis
via miR-326/miR-487a-3p/sirtuin 1 (61). However, further in vivo
experiments on circHIPK3 and DN are still needed. Research has also
found that circRNA-010383 was downregulated in the diabetic
kidneys, HG-induced MCs and TECs. Overexpression of circRNA_010383
in the kidney inhibited renal fibrosis and proteinuria in db/db
mice via miRNA-135a/transient receptor potential cation channel
subfamily C member 1 (51).
Mmu_circRNA_0000309 showed a sharp decrease in DN mice and a
remarkable recovery in Germacrone-challenged DN mice. Furthermore,
mmu_circRNA_0000309 knockdown inhibited the protective effect of
Germacrone via ferroptosis-depended mitochondrial damage and
podocyte apoptosis by regulating miR-188-3p/glutathione peroxidase
4 axes both in vivo and in vitro (62). Therefore, upregulation of
circHIPK3, circRNA-010383 and mmu_circRNA_0000309 could alleviate
the dysfunction of TECs, MCs and podocytes, respectively.
CircRNAs in patients with DN are increasingly
becoming the subject of ongoing research. Wang et al
(53) reported that circ_0123996
was consistently increased in kidney tissues from patients with DN
and mouse models of DN, as well as in HG-exposed MCs. Inhibition of
circ_0123996 reduced cell proliferation and fibrosis in MCs via
sponging miR-149-5p and inducing Bach1 expression. A recent study
also indicated that hsa_circ_0003928 was upregulated, while
miR-151-3p was downregulated in the serum from patients with DN and
HG-induced HK-2 cells. Furthermore, downregulation of
hsa_circ_0003928 repressed HG-induced cell apoptosis and
inflammation via miR-151-3p/annexin A2 in vitro (57). Therefore, these studies hint at
circRNAs having an essential role in the pathogenesis of DN by
acting as a sponge for miRNAs and may be a novel therapeutic target
for DN.
Proliferative DR is a major cause of sustained
blindness, which greatly impacts the life of patients with DM.
Consequently, changes in the interaction among retinal cells due to
diabetes result in severe vascular damage, loss of the
blood-retinal barrier and impaired neuronal function (63). Of note, circRNAs regulate certain
important physiological and pathological processes. Numerous
studies have demonstrated differentially expressed circRNAs in
serum, vitreous humour and retinas from patients with DR or in the
retinas of db/db mice and human retinal pericytes induced by HG
(64–68). Representative circRNAs in DR are
listed in Table I.
The crosstalk between pericytes and ECs is vital for
microvascular homeostasis and remodeling. First, with regard to
HG-induced human retinal pericytes, Liu et al (65) found that circRNA PWWP domain
containing 2A (cPWWP2A, circ_0000254) has high homology in gene
sequence with humans. Likewise, cPWWP2A was upregulated in the
retinas of STZ-induced diabetic mice in vivo and HG-induced
human retinal pericytes in vitro, and pericyte-derived
cPWWP2A was able to affect pericyte coverage and vascular
integrality by acting on miR-579 and its target genes. This study
revealed that cPWWP2A-mediated signaling has a vital role in
retinal microvascular dysfunction (65). In addition, overexpression of
circRNA zinc finger protein (ZNF)532 was verified in human retinal
pericytes treated with HG and in the vitreous of diabetic patients
with macular edema, proliferative DR or neovascularization.
Knockdown of cZNF532 exacerbated pericyte damage and retinal
vascular dysfunction via the miR-29a-3p/neuron glial antigen
2-lysyl oxidase like 2-cyclin-dependent protein kinase 2 network
(66). In another study, Wu et
al (68) proved that
hsa_circ_0001953 was upregulated in the serum of patients with
proliferative DR and revealed that hsa_circ_0001953 was a potential
diagnostic biomarker for proliferative DR. As such, there were
consistent changes in the expression of certain circRNAs in serum
of patients with DR and in the retinas of diabetic animal models,
as well as in HG-induced human retinal pericytes. Therefore, these
findings imply that serum circRNAs may be used as a biomarker to
diagnose DR.
Furthermore, more detailed studies have been
conducted on HG-induced ECs and circRNAs. A study indicated that
circHIPK3 was significantly upregulated in diabetic retinas and
retinal ECs following stressors related to DM, and circHIPK3 acted
as a ceRNA of miR-30a-3p. Knocking down circHIPK3 was able to
reduce retinal EC proliferation, viability, migration and tube
formation in vitro and ameliorate hyperglycemia-induced
retinal acellular capillaries, vascular leakage and inflammation
in vivo (13). Another
investigation suggested that circZNF609 was upregulated in
HG-treated HUVECs and retinas from STZ-induced diabetic mice, in
which silencing of circZNF609 alleviated endothelial dysfunction in
HUVECs and retinal vessel loss and pathological angiogenesis in
vivo by targeting the miR-615-5p/myocyte-specific enhancer
factor 2A axis (70). Zou et
al (71) found that circRNA
collagen type I α2 chain (circCOL1A2, hsa_circ_0081108) was
upregulated in HG-induced HRMECs. CircCOL1A2 knockdown inhibited
HG-induced migration, proliferation, angiogenesis and vascular
permeability of HRMECs in vitro and suppressed angiogenesis
in vivo through regulating the miR-29b/VEGF axis.
Endothelial tip cell specialization is vital for angiogenesis.
CircMET was increased in STZ-induced mice and DR patients' retinas.
CircMET silencing decreased the expression of tip cell-enriched
genes (CXCR4, CD34 and VEGFA) in HRVECs (72). Another study found that circFTO was
upregulated in HG-treated retinal vascular ECs. In addition,
circFTO knockdown reversed angiogenesis and impaired the
blood-retinal barrier in HG-induced retinal vascular ECs via
miR-128-3p/thioredoxin (73).
However, Zhu et al (74)
demonstrated that circDNMT3B was downregulated in HRMECs under HG
conditions and retinas from STZ-induced diabetic rats. CircDNMT3B
overexpression reduced the retinal acellular capillary number and
alleviated visual damage in STZ-induced diabetic rats, hinting at
circDNMT3B regulating diabetic retinal vascular dysfunction via
miR-20b-5p/target gene. In another approach, Ye et al
(75) found that exosomal
circEhmt1 released from hypoxia-stimulated pericytes protected
endotheliocytes from HG-induced injury by downregulating the
NFIA/NLRP3 pathway. In addition to pericytes and ECs, studies on
circRNAs and HG-induced retinal pigment epithelial cells have also
appeared in recent years. Upregulated circRNA_0084043 and
hsa_circ_0041795 participate in DR via sponging miR-140-3p and
miR-646, subsequently increasing TGFα and VEGFC expression in
retinal pigment epithelial cells (76,77).
Therefore, these data suggest that these differentially expressed
circRNAs have an important role in the pathogenesis of DR and may
be potential targets to control DR.
Neuropathic pain is one of the most common diabetic
complications in the clinic, but the aetiology of DNP has remained
to be fully elucidated. Two studies suggested that circRNAs
contribute to the development of DNP (78,79).
One of the studies showed that the expression of circHIPK3 in the
serum of patients with T2DM was significantly higher than that in
the control group. Furthermore, upregulated circHIPK3 was
positively associated with grade neuropathic pain in T2DM patients.
Knockout of circHIPK3 was able to relieve neuropathic pain and
inhibit neuroinflammation in STZ-induced diabetic rats (78). Likewise, circHIPK3 interacted with
miR-124 and negatively regulated its expression (78). The study implies that intrathecal
circHIPK3 short hairpin RNA may be used to treat DNP in the future.
Zhang et al (79) also
observed the expression profile of circRNAs in dorsal root ganglia
from DM mice by high-throughput RNA-seq, in which 15 differentially
expressed circRNAs and 133 differentially expressed mRNAs were
identified. A total of 11 circRNAs and 14 mRNAs had a marked
correlation and circRNA-Atp9b was validated to be observably
upregulated in dorsal root ganglia of DM mice (79). Liu et al (80) performed circRNA sequencing on sural
nerves in patients with diabetic peripheral neuropathy, and 11
differentially expressed circRNAs were verified and circ_0002538
was downregulated. The authors found that overexpression of
circ_0002538 improved the function of the sciatic nerve in
vivo. Indeed, further studies are necessary to explore the
mechanisms by which circRNAs are involved in DNP.
DCM is a specific cardiovascular complication in
patients with DM; it first manifests as diastolic dysfunction and
eventually progresses to refractory heart failure (HF). Before the
emergence of systolic dysfunction and clinical HF, DCM is initially
characterized by cardiomyocyte apoptosis, hypertrophy, myocardial
fibrosis and remodeling. It has been reported that circRNAs
potentially act on cardioprotective processes by sponging miRNAs in
DCM (14,15,16,81,82).
Representative circRNA dysregulation mechanisms in DCM are
presented in Fig. 4.
Through microarray analysis and RNA-seq,
differentially expressed circRNAs were detected in heart tissues of
diabetic animal models, such as 45 circRNAs up-regulated by
>2-fold and 31 circRNAs downregulated by >2-fold in db/db
mice (15). Dong et al
(82) also found that 58 circRNAs
were significantly differentially expressed in the myocardium at an
early stage of DCM, including 29 upregulated circRNAs and 29
downregulated circRNAs. Of note, cardiac fibrosis is a critical
event in the pathogenesis of DCM. This study revealed that
circ_000203 was upregulated in myocardium from db/db mice and
overexpression of circRNA_000203 increased the expression of
COL1A2, COL3A1 and α-smooth muscle actin (α-SMA) in mouse cardiac
fibroblasts (CFs) via sponging miR-26b-5p, and COL1A2 and
connective tissue growth factor were the target genes of miR-26b-5p
(15). Zhou and Yu (81) found 24 upregulated circRNAs and 19
downregulated circRNAs in the myocardium of db/db mice.
Particularly, circ_010567 was markedly increased and circ-010567
silencing was able to reduce fibrosis-associated protein resection
via upregulating miR-141 and inhibiting the TGF-β1 pathway in
Angiotensin II (AngII)-treated CFs (81). The limitation of this in
vitro study was that AngII instead of HG was used to induce
CFs, which does not wholly reflect the pathogenesis of diabetic
myocardial fibrosis. Wang et al (14) also found that circHIPK3 was
upregulated in a DCM model of STZ-induced diabetic mice. CircHIPK3
knockdown was able to ameliorate myocardial fibrosis and improve
cardiac function in vivo, while decreasing the proliferation
of CFs treated with AngII via miR-29b-3p/Col1a1-Col3a1 in
vitro. Pyroptosis is a newly discovered form of programmed cell
death and has an important role in the progression of DCM alongside
apoptosis. Of note, Yang et al (16) found that hsa_circ_0076631, also
known as CACR, was increased in HG-induced AC16 cells and the serum
of diabetic patients. Knockdown of CACR alleviated myocardial
pyroptosis and inflammation via targeting miR-214-3p/caspase-1 in
HG-induced AC16 cells. Only diabetic models were used in these
studies, but it was not verified whether these mice also had DCM.
It was reported that circRNA CDR1as was upregulated in DCM hearts
in STZ-induced diabetic mice, which promoted cardiomyocyte
apoptosis through activating the MST1-Hippo pathway in vivo
and in HG-treated primary cardiomyocytes. Knocking down CDR1as was
able to inhibit cardiomyocyte apoptosis in DCM. Thus, inhibition of
CDR1as may become a potential therapeutic strategy for DCM
(83).
The roles of circRNAs in ischemic heart disease have
also been reported. CircRNA sodium/calcium exchanger 1 (circNCX1)
was increased in the myocardial ischemia-reperfusion mouse model
and silencing of circNCX1 in the heart attenuated myocardial
fibrosis via action on miR-133a-3p (84). In addition, downregulation of
circRNA actin α2 alleviated myocardial fibrosis via targeting
miR-548f-5p and suppressing α-SMA expression in human aortic smooth
muscle cells of coronary artery diseases (85). Furthermore, CircHIPK3 increased the
expression of fibrosis-associated genes, such as COL1A2, COL3A1 and
α-SMA, via sponging miR-29b-3p in AngII-induced mouse myocardium
(86). Furthermore, circRNA
nuclear factor I B (circNFIB) was decreased in cardiac fibrosis
in vivo and in vitro, and upregulation of circNFIB
attenuated cardiac fibrosis by directly targeting miR-433 in a
mouse myocardial infarction model (87). Thus, overexpression or inhibition
of circRNA may regulate gene expression and potentially become an
effective method to improve cardiac function.
Increasing evidence indicates that certain circRNAs
have crucial roles in the pathogenesis of diabetic complications
through ceRNA mechanisms, acting as miRNA sponges. Due to their
abundance and stability, circRNAs may serve as useful biomarkers or
therapeutic targets for diabetic complications. An overview of
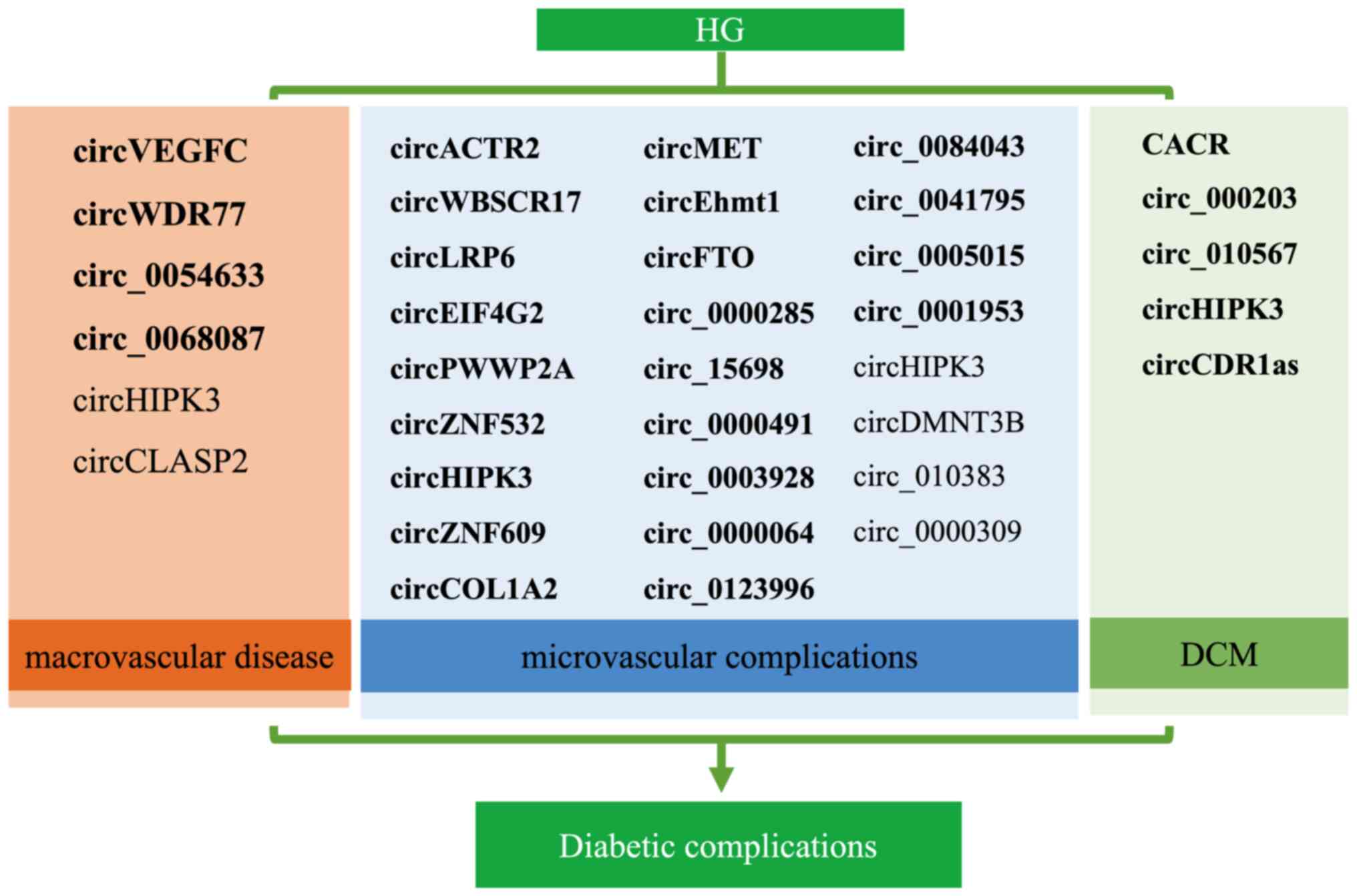
circRNAs involved in diabetic complications is provided in Fig. 5. CircHIPK3 is the most well-studied
circRNA in the field of diabetes and is most likely to become a
biological marker and therapeutic target for diabetes and its
complications. The present review indicated that the number of
upregulated circRNAs was higher than that of downregulated circRNAs
in diabetic complications both in vivo and in vitro
(Fig. 5), implying that exploring
the mechanisms of promoting DNA repair is important. Although
circRNA-miRNA-mRNA axis mechanisms have been predicted in current
studies, circRNAs as drugs may cause side effects or resistance.
Despite these limitations, circRNAs have immense potential as
therapeutic targets and stable biomarkers for diabetic
complications.
Not applicable.
Funding: No funding was received.
Not applicable.
LY and JD searched and selected the literature,
wrote the original draft, and contributed equally to this work. HZ
conceived the study, and reviewed and edited the manuscript. Data
authentication is not applicable. All authors have read and
approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Cho NH, Shaw JE, Karuranga S, Huang Y, da
Rocha Fernandes JD, Ohlrogge AW and Malanda B: IDF Diabetes Atlas:
Global estimates of diabetes prevalence for 2017 and projections
for 2045. Diabetes Res Clin Pract. 138:271–281. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morrish NJ, Wang SL, Stevens LK, Fuller JH
and Keen H: Mortality and causes of death in the WHO multinational
study of vascular disease in diabetes. Diabetologia. 44 (Suppl
2):S14–S21. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang PL, Bao Y, Yee MC, Barrett SP, Hogan
GJ, Olsen MN, Dinneny JR, Brown PO and Salzman J: Circular RNA is
expressed across the eukaryotic tree of life. PLoS One.
9:e908592014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang HD, Jiang LH, Sun DW, Hou JC and Ji
ZL: CircRNA: A novel type of biomarker for cancer. Breast Cancer.
25:1–7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang G, Ma Y, An T, Pan Y, Mo F, Zhao D,
Liu Y, Miao JN, Gu YJ, Wang Y and Gao SH: Relationships of circular
RNA with diabetes and depression. Sci Rep. 7:72852017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fan X, Weng X, Zhao Y, Chen W, Gan T and
Xu D: Circular RNAs in cardiovascular disease: An overview. Biomed
Res Int. 2017:51357812017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smid M, Wilting SM, Uhr K,
Rodríguez-González FG, de Weerd V, Prager-Van der Smissen WJC, van
der Vlugt-Daane M, van Galen A, Nik-Zainal S, Butler A, et al: The
circular RNome of primary breast cancer. Genome Res. 29:356–366.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hu W, Han Q, Zhao L and Wang L: Circular
RNA circRNA_15698 aggravates the extracellular matrix of diabetic
nephropathy mesangial cells via miR-185/TGF-β1. J Cell Physiol.
234:1469–1476. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Shan K, Liu C, Liu BH, Chen X, Dong R, Liu
X, Zhang YY, Liu B, Zhang SJ, Wang JJ, et al: Circular noncoding
RNA HIPK3 mediates retinal vascular dysfunction in diabetes
mellitus. Circulation. 136:1629–1642. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang W, Zhang S, Xu L, Feng Y, Wu X, Zhang
M, Yu Z and Zhou X: Involvement of circHIPK3 in the pathogenesis of
diabetic cardiomyopathy in mice. Diabetologia. 64:681–692. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN,
Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, et al: CircRNA_000203
enhances the expression of fibrosis-associated genes by
derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac
fibroblasts. Sci Rep. 7:403422017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang F, Li A, Qin Y, Che H, Wang Y, Lv J,
Li Y, Li H, Yue E, Ding X, et al: A novel circular RNA mediates
pyroptosis of diabetic cardiomyopathy by functioning as a competing
endogenous RNA. Mol Ther Nucleic Acids. 17:636–643. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs As
a new class of putative biomarkers in human blood. PLoS One.
10:e01412142015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashwal-Fluss R, Meyer M, Pamudurti N,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu CX, Li X, Nan F, Jiang S, Gao X, Guo
SK, Xue W, Cui Y, Dong K, Ding H, et al: Structure and degradation
of circular RNAs regulate PKR activation in innate immunity. Cell.
177:865–880.e21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang
W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs:
Biogenesis, effect and application in human diseases. Mol Cancer.
18:1162019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yoshimoto R, Rahimi K, Hansen TB, Kjems J
and Mayeda A: Biosynthesis of Circular RNA ciRS-7/CDR1as is
mediated by mammalian-wide interspersed repeats. iScience.
23:1013452020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Piwecka M, Glažar P, Hernandez-Miranda LR,
Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda
Jara CA, Fenske P, et al: Loss of a mammalian circular RNA locus
causes miRNA deregulation and affects brain function. Science.
357:eaam85262017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kristensen LS, Okholm TLH, Venø MT and
Kjems J: Circular RNAs are abundantly expressed and upregulated
during human epidermal stem cell differentiation. RNA Biol.
15:280–291. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hsiao KY, Lin YC, Gupta SK, Chang N, Yen
L, Sun HS and Tsai SJ: Noncoding effects of circular RNA CCDC66
promote colon cancer growth and metastasis. Cancer Res.
77:2339–2350. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cao Y, Yuan G, Zhang Y and Lu R: High
glucose-induced circHIPK3 downregulation mediates endothelial cell
injury. Biochem Biophys Res Commun. 507:362–368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Barbagallo D, Caponnetto A, Brex D,
Mirabella F, Barbagallo C, Lauretta G, Morrone A, Certo F, Broggi
G, Caltabiano R, et al: CircSMARCA5 Regulates VEGFA mRNA splicing
and angiogenesis in glioblastoma multiforme through the binding of
SRSF1. Cancers (Basel). 11:1942019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Abdelmohsen K, Panda AC, Munk R,
Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM,
Martindale JL and Gorospe M: Identification of HuR target circular
RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA
Biol. 14:361–369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li XX, Xiao L, Chung HK, Ma XX, Liu X,
Song JL, Jin CZ, Rao JN, Gorospe M and Wang JY: Interaction between
HuR and circPABPN1 Modulates autophagy in the intestinal epithelium
by altering ATG16L1 translation. Mol Cell Biol. 40:e004922020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Abe N, Matsumoto K, Nishihara M, Nakano Y,
Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y and Abe
H: Rolling circle translation of circular RNA in living human
cells. Sci Rep. 5:164352015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Meyer KD, Patil DP, Zhou J, Zinoviev A,
Skabkin MA, Elemento O, Pestova TV, Qian SB and Jaffrey SR: 5′ UTR
m(6)A Promotes Cap-Independent Translation. Cell. 163:999–1010.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li Y, Wang Z, Su P, Liang Y, Li Z, Zhang
H, Song X, Han D, Wang X, Liu Y, et al: Circ-EIF6 encodes
EIF6-224aa to promote TNBC progression via stabilizing MYH9 and
activating Wnt/beta-catenin pathway. Mol Ther. 30:415–430. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Han YN, Xia SQ, Zhang YY, Zheng JH and Li
W: Circular RNAs: A novel type of biomarker and genetic tools in
cancer. Oncotarget. 8:64551–64563. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rines AK, Sharabi K, Tavares CD and
Puigserver P: Targeting hepatic glucose metabolism in the treatment
of type 2 diabetes. Nat Rev Drug Discov. 15:786–804. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shang FF, Luo S, Liang X and Xia Y:
Alterations of circular RNAs in hyperglycemic human endothelial
cells. Biochem Biophys Res Commun. 499:551–555. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jin G, Wang Q, Hu X, Li X, Pei X, Xu E and
Li M: Profiling and functional analysis of differentially expressed
circular RNAs in high glucose-induced human umbilical vein
endothelial cells. FEBS Open Bio. 9:1640–1651. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pan L, Lian W, Zhang X, Han S, Cao C, Li X
and Li M: Human circular RNA-0054633 regulates high glucose-induced
vascular endothelial cell dysfunction through the
microRNA-218/roundabout 1 and microRNA-218/heme oxygenase-1 axes.
Int J Mol Med. 42:597–606. 2018.PubMed/NCBI
|
|
44
|
Zhang Q, Long J, Li N, Ma X and Zheng L:
Circ_CLASP2 regulates high glucose-induced dysfunction of human
endothelial cells through targeting miR-140-5p/FBXW7 Axis. Front
Pharmacol. 12:5947932021. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wei H, Cao C, Wei X, Meng M, Wu B, Meng L,
Wei X, Gu S and Li H: Circular RNA circVEGFC accelerates high
glucose-induced vascular endothelial cells apoptosis through
miR-338-3p/HIF-1α/VEGFA axis. Aging (Albany NY). 12:14365–14375.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cheng J, Liu Q, Hu N, Zheng F, Zhang X, Ni
Y and Liu J: Downregulation of hsa_circ_0068087 ameliorates
TLR4/NF-κB/NLRP3 inflammasome-mediated inflammation and endothelial
cell dysfunction in high glucose conditioned by sponging miR-197.
Gene. 709:1–7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen J, Cui L, Yuan J, Zhang Y and Sang H:
Circular RNA WDR77 target FGF-2 to regulate vascular smooth muscle
cells proliferation and migration by sponging miR-124. Biochem
Biophys Res Commun. 494:126–132. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zaiou M: circRNAs signature as potential
diagnostic and prognostic biomarker for diabetes mellitus and
related cardiovascular complications. Cells. 9:6592020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Fang Y, Wang X, Li W, Han J, Jin J, Su F,
Zhang J, Huang W, Xiao F, Pan Q and Zou L: Screening of circular
RNAs and validation of circANKRD36 associated with inflammation in
patients with type 2 diabetes mellitus. Int J Mol Med.
42:1865–1874. 2018.PubMed/NCBI
|
|
50
|
An Y, Furber KL and Ji S: Pseudogenes
regulate parental gene expression via ceRNA network. J Cell Mol
Med. 21:185–192. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Peng F, Gong W, Li S, Yin B, Zhao C, Liu
W, Chen X, Luo C, Huang Q, Chen T, et al: circRNA_010383 Acts as a
Sponge for miR-135a, and its downregulated expression contributes
to renal fibrosis in diabetic nephropathy. Diabetes. 70:603–615.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chen B, Li Y, Liu Y and Xu Z: circLRP6
regulates high glucose-induced proliferation, oxidative stress, ECM
accumulation, and inflammation in mesangial cells. J Cell Physiol.
234:21249–21259. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang W, Feng J, Zhou H and Li Q:
Circ_0123996 promotes cell proliferation and fibrosisin mouse
mesangial cells through sponging miR-149-5p and inducing Bach1
expression. Gene. 761:1449712020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ge X, Xi L, Wang Q, Li H, Xia L, Cang Z,
Peng W and Huang S: Circular RNA Circ_0000064 promotes the
proliferation and fibrosis of mesangial cells via miR-143 in
diabetic nephropathy. Gene. 758:1449522020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yao T, Zha D, Hu C and Wu X: Circ_0000285
promotes podocyte injury through sponging miR-654-3p and activating
MAPK6 in diabetic nephropathy. Gene. 747:1446612020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li G, Qin Y, Qin S, Zhou X, Zhao W and
Zhang D: Circ_WBSCR17 aggravates inflammatory responses and
fibrosis by targeting miR-185-5p/SOX6 regulatory axis in high
glucose-induced human kidney tubular cells. Life Sci.
259:1182692020. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
An L, Ji D, Hu W, Wang J, Jin X, Qu Y and
Zhang N: Interference of Hsa_circ_0003928 alleviates high
glucose-induced cell apoptosis and inflammation in HK-2 Cells via
miR-151-3p/Anxa2. Diabetes Metab Syndr Obes. 13:3157–3168. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mou X, Chenv JW, Zhou DY, Liu K, Chen LJ,
Zhou D and Hu YB: A novel identified circular RNA, circ_0000491,
aggravates the extracellular matrix of diabetic nephropathy
glomerular mesangial cells through suppressing miR-101b by
targeting TGFβRI. Mol Med Rep. 22:3785–3794. 2020.PubMed/NCBI
|
|
59
|
Wen S, Li S, Li L and Fan Q: circACTR2: A
novel mechanism regulating high glucose-induced fibrosis in renal
tubular cells via pyroptosis. Biol Pharm Bull. 43:558–564. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Xu B, Wang Q, Li W, Xia L, Ge X, Shen L,
Cang Z, Peng W, Shao K and Huang S: Circular RNA circEIF4G2
aggravates renal fibrosis in diabetic nephropathy by sponging
miR-218. J Cell Mol Med. 26:1799–1805. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhuang L, Wang Z, Hu X, Yang Q, Pei X and
Jin G: CircHIPK3 alleviates high glucose toxicity to human renal
tubular epithelial HK-2 cells through regulation of
miR-326/miR-487a-3p/SIRT1. Diabetes Metab Syndr Obes. 14:729–740.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Jin J, Wang Y, Zheng D, Liang M and He Q:
A novel identified circular RNA, mmu_mmu_circRNA_0000309, involves
in germacrone-mediated improvement of diabetic nephropathy through
regulating ferroptosis by targeting miR-188-3p/GPX4 signaling axis.
Antioxid Redox Signal. 36:740–759. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Antonetti DA, Silva PS and Stitt AW:
Current understanding of the molecular and cellular pathology of
diabetic retinopathy. Nat Rev Endocrinol. 17:195–206. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Gu Y, Ke G, Wang L, Zhou E, Zhu K and Wei
Y: Altered expression profile of circular RNAs in the serum of
patients with diabetic retinopathy revealed by microarray.
Ophthalmic Res. 58:176–184. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Liu C, Ge HM, Liu BM, Dong R, Shan K, Chen
X, Yao MD, Li XM, Yao J, Zhou RM, et al: Targeting
pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A
inhibition aggravates diabetes-induced microvascular dysfunction.
Proc Natl Acad Sci USA. 116:7455–7464. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jiang Q, Liu C, Li CP, Xu SS, Yao MD, Ge
HM, Sun YN, Li XM, Zhang SJ, Shan K, et al: Circular RNA-ZNF532
regulates diabetes-induced retinal pericyte degeneration and
vascular dysfunction. J Clin Invest. 130:3833–3847. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
He M, Wang W, Yu H, Wang D, Cao D, Zeng Y,
Wu Q, Zhong P, Cheng Z, Hu Y and Zhang L: Comparison of expression
profiling of circular RNAs in vitreous humour between diabetic
retinopathy and non-diabetes mellitus patients. Acta Diabetol.
57:479–489. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wu Z, Liu B, Ma Y, Chen H, Wu J and Wang
J: Discovery and validation of hsa_circ_0001953 as a potential
biomarker for proliferative diabetic retinopathy in human blood.
Acta Ophthalmol. 99:306–313. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zhang SJ, Chen X, Li CP, Li XM, Liu C, Liu
BH, Shan K, Jiang Q, Zhao C and Yan B: Identification and
characterization of circular RNAs as a new class of putative
biomarkers in diabetes retinopathy. Invest Ophthalmol Vis Sci.
58:6500–6509. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Liu C, Yao MD, Li CP, Shan K, Yang H, Wang
JJ, Liu B, Li XM, Yao J, Jiang Q and Yan B: Silencing of circular
RNA-ZNF609 ameliorates vascular endothelial dysfunction.
Theranostics. 7:2863–2877. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zou J, Liu KC, Wang WP and Xu Y: Circular
RNA COL1A2 promotes angiogenesis via regulating miR-29b/VEGF axis
in diabetic retinopathy. Life Sci. 256:1178882020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yao MD, Jiang Q, Ma Y, Zhu Y, Zhang QY,
Shi ZH, Zhao C and Yan B: Targeting circular RNA-MET for
anti-angiogenesis treatment via inhibiting endothelial tip cell
specialization. Mol Ther. 30:1252–1264. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Guo J, Xiao F, Ren W, Zhu Y, Du Q, Li Q
and Li X: ViaCircular Ribonucleic Acid circFTO promotes
angiogenesis and impairs blood-retinal barrier targeting the
miR-128-3p/Thioredoxin interacting protein axis in diabetic
retinopathy. Front Mol Biosci. 8:6854662021. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhu K, Hu X, Chen H, Li F, Yin N, Liu AL,
Shan K, Qin YW, Huang X, Chang Q, et al: Downregulation of circRNA
DMNT3B contributes to diabetic retinal vascular dysfunction through
targeting miR-20b-5p and BAMBI. EBioMedicine. 49:341–353. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ye L, Guo H, Wang Y, Peng Y, Zhang Y, Li
S, Yang M and Wang L: Exosomal circEhmt1 released from
hypoxia-pretreated pericytes regulates high glucose-induced
microvascular dysfunction via the NFIA/NLRP3 pathway. Oxid Med Cell
Longev. 2021:88330982021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Li Y, Cheng T, Wan C and Cang Y:
circRNA_0084043 contributes to the progression of diabetic
retinopathy via sponging miR-140-3p and inducing TGFA gene
expression in retinal pigment epithelial cells. Gene.
747:1446532020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Sun H and Kang X: hsa_circ_0041795
contributes to human retinal pigment epithelial cells (ARPE 19)
injury induced by high glucose via sponging miR-646 and activating
VEGFC. Gene. 747:1446542020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang L, Luo T, Bao Z, Li Y and Bu W:
Intrathecal circHIPK3 shRNA alleviates neuropathic pain in diabetic
rats. Biochem Biophys Res Commun. 505:644–650. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Zhang HH, Zhang Y, Wang X, Yang P, Zhang
BY, Hu S, Xu GY and Hu J: Circular RNA profile in diabetic
peripheral neuropathy: Analysis of coexpression networks of
circular RNAs and mRNAs. Epigenomics. 12:843–857. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu YT, Xu Z, Liu W, Ren S, Xiong HW,
Jiang T, Chen J, Kang Y, Li QY, Wu ZH, et al: The
circ_0002538/miR-138-5p/plasmolipin axis regulates Schwann cell
migration and myelination in diabetic peripheral neuropathy. Neural
Regen Res. 18:1591–1600. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Zhou B and Yu JW: A novel identified
circular RNA, circRNA_010567, promotes myocardial fibrosis via
suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res
Commun. 487:769–775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Dong S, Tu C, Ye X, Li L, Zhang M, Xue A,
Chen S, Zhao Z, Cong B, Lin J and Shen Y: Expression profiling of
circular RNAs and their potential role in early-stage diabetic
cardiomyopathy. Mol Med Rep. 22:1958–1968. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Shao Y, Li M, Yu Q, Gong M, Wang Y, Yang
X, Liu L, Liu D, Tan Z, Zhang Y, et al: CircRNA CDR1as promotes
cardiomyocyte apoptosis through activating hippo signaling pathway
in diabetic cardiomyopathy. Eur J Pharmacol. 922:1749152022.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li M, Ding W, Tariq MA, Chang W, Zhang X,
Xu W, Hou L, Wang Y and Wang J: A circular transcript of ncx1 gene
mediates ischemic myocardial injury by targeting miR-133a-3p.
Theranostics. 8:5855–5869. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Sun Y, Yang Z, Zheng B, Zhang XH, Zhang
ML, Zhao XS, Zhao HY, Suzuki T and Wen JK: A novel regulatory
mechanism of smooth muscle α-actin expression by
NRG-1/circACTA2/miR-548f-5p axis. Circ Res. 121:628–635. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Ni H, Li W, Zhuge Y, Xu S, Wang Y, Chen Y,
Shen G and Wang F: Inhibition of circHIPK3 prevents angiotensin
II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol.
292:188–196. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zhu Y, Pan W, Yang T, Meng X, Jiang Z, Tao
L and Wang L: Upregulation of circular RNA CircNFIB attenuates
cardiac fibrosis by sponging miR-433. Front Genet. 10:5642019.
View Article : Google Scholar : PubMed/NCBI
|