Introduction
Titanium prostheses exhibit widespread uses in the
clinic, including joint replacements and dental implants. However,
in the majority of clinical cases, patients may experience ongoing
inflammation in the absence of infection, which may lead to
peri-prosthetic osteolysis (PPO) and subsequent aseptic loosening
(1–4). The results of numerous studies have
indicated that titanium implants undergo corrosion via various
mechanisms inside the human body, including mechanical wear,
biological activity and electrochemical processes (5–7). The
aforementioned corrosion may cause gradual deterioration of the
titanium implant; thus, impacting the structural integrity. In
addition, the results of previous studies have demonstrated that
the major cause of aseptic loosening is the gradual accumulation of
wear particles around the titanium implant (8–10).
Following 10 years of follow up, this phenomenon resulted in an
implant failure rate of 32–62% following orthopedic surgery
(11,12). For dental implants, unexplained
mucosal inflammation impacts 43% of implants, and ongoing bone loss
occurs in 6–29% of cases (13,14).
Numerous studies have focused on the molecular and
cellular interactions between humans and wear particles (15,16).
Previous studies have defined aseptic loosening as an
immune-mediated biological complication (2,17–19).
However, the complex interplay among the innate or adaptive immune
response, the skeletal system and wear particles remains to be
fully understood. Previous studies have revealed that the released
particles may induce a cascade of biological effects, including
potent proinflammatory immune responses, suppression of osteoblast
function and activation of osteoclasts, leading to peri-implant
bone loss (20–25). Thus, research has focused on the
specific molecular mechanisms and potential targeted drugs for use
in the clinic. Previous studies have revealed numerous inhibitors
that exert protective effects on titanium particle-induced bone
loss, and multiple associated downstream signaling pathways, such
as the MAPK, NF-κB, GSK-3β/β-catenin and TGF-β pathways (10,22,26–31).
Notably, multiple pathways are simultaneously activated by titanium
particles. Thus, previous studies that focused on a single target
or downstream pathway were unsuccessful in uncovering the specific
mechanisms, and therapies that target single pathways to attenuate
particle-induced osteogenic inhibition or inflammatory responses
are limited (30,32,33).
N6-methyladenosine (m6A), a dynamic
methylation at the N6 site of adenosine, is the most common
post-transcriptional RNA modification. As an epigenetic regulator,
it regulates numerous biological processes through mediating RNA
metabolism, including degradation and translation (34–36).
The methyltransferase-like (Mettl)3/Mettl14 catalytic heterodimer
serves a role in the methylation of m6A. The process of
demethylation is implemented by demethylases, including ALKB
homolog 5 and fat mass and obesity-associated protein. This
specific modification is recognized and bound by m6A
reader proteins, mainly from YT521-B homology (YTH)-domain family
proteins (37–41). Previous studies have revealed the
association between m6A modifications and bone
metabolism. RNA methylation serves a significant role in regulating
bone homeostasis. Notably, Mettl3 and/or Mettl14 deficiency may
disrupt the proliferation and differentiation of stem cells, such
as bone marrow mesenchymal stem cells, causing bone disorders, such
as osteoporosis (42–46). In addition, previous studies have
demonstrated that post-transcriptional modification via changes in
expression of methyltransferases, demethylases or reader proteins
concurrently regulated multiple downstream pathways, achieving
regulation in diverse cellular reactions (46,47).
As m6A modifications are essential in
bone homeostasis (48), the
present study aimed to explore the role of m6A
modification in titanium particle-induced osteolysis. In addition,
the present study aimed to provide novel insights into potential
therapeutic targets for aseptic loosening. It was hypothesized that
titanium particles may utilize RNA methylation as a form of
upstream regulation to induce multiple subsequent pathways; thus,
causing osteogenic inhibition and inflammatory responses.
Materials and methods
Titanium particle preparation
The titanium particles used in the present study
were obtained from Alfa Aesar; Thermo Fisher Scientific, Inc. and
prepared as previously described (10,22,29).
Briefly, the particles were incubated at 180°C for 12 h, and
subsequently immersed in 75% ethanol at room temperature for 48 h
to eliminate endotoxins. Endotoxin levels in particles were
detected using a Limulus assay kit (Xiamen Bioendo Technology Co.,
Ltd.) to ensure that endotoxin levels were <0.02 EU/ml.
Cell culture and treatment
The cells used in the present study were cultured at
37°C with 5% CO2. The murine osteoblast cell line,
MC3T3-E1 (iCell Bioscience, Inc.) was cultured in α-minimum
essential medium (Gibco; Thermo Fisher Scientific, Inc.) with 10%
FBS (Gibco; Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
For osteogenic differentiation, cells were cultured in the
aforementioned growth medium with the addition of 50 µg/ml ascorbic
acid (Sigma-Aldrich; Merck KGaA) and 10 mM β-glycerophosphate
(Sigma-Aldrich; Merck KGaA). The prepared titanium particles were
used at a concentration of 100 µg/ml. For titanium particle
treatment groups, particles were cocultured with osteogenic
induction medium to figure out the effect of titanium particles on
osteoblasts in an osteogenic environment (10,22).
The RAW264.7 mouse macrophage cell line was
purchased from American Type Culture Collection and cultured in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS and 100
U/ml penicillin/streptomycin (Gibco; Thermo Fisher Scientific,
Inc.). The supernatants of osteoblast cells with different
treatments, including titanium particle treatment and the knockdown
or overexpression of the Mettl3 gene, were harvested and then
centrifuged at 500 × g at room temperature for 5 min to remove cell
debris or titanium particles. Subsequently, the supernatant was
mixed with culture medium at a ratio of 1:1 (conditioned medium)
and then RAW264.7 cells were cultured in conditioned medium for the
detection of osteoclastic differentiation. Cells cultured in growth
medium were used as the negative control (NC), while cells treated
with 50 ng/ml recombinant murine receptor activator of NF-κB ligand
(RANKL; PeproTech, Inc.) were used as the positive control. The
samples were collected at 2 and 3 days for reverse
transcription-quantitative PCR (RT-qPCR) and western blotting,
respectively.
For nucleotide binding oligomerization domain (NOD)1
pathway inhibition experiments, osteoblast cells were pretreated
with 30 µM ML130 (MedChemExpress) and 10 µM WEHI-345 (Abmole
Bioscience Inc.) at 37°C for 2 h (49,50).
Transmission electron microscopy
(TEM)
Following co-culture with titanium particles for 24
h, osteoblast cells were collected and fixed with 2.5%
glutaraldehyde in PBS at 4°C for 1 h. No stain was used as the
stain would highlight various intracellular structures. Cells were
fixed with 1% osmium tetroxide in H2O for 2 h, followed
by sequential dehydration with a graded ethanol series. Samples
were embedded in epoxy resin at 60°C for 12 h, cut into 70–90-nm
ultrathin sections and subsequently observed using a JEM-1200EX
electron microscope (JEOL, Ltd.).
RT-qPCR
Total RNA was extracted from cells using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.). cDNA was
synthesized using total RNA and HiScript III RT SuperMix (Vazyme
Biotech Co., Ltd.) at 37°C for 15 h and then 85°C for 5 sec. ChamQ
Universal SYBR qPCR Master Mix (Vazyme Biotech Co., Ltd.) and a
Light Cycler 480 (Roche Diagnostics GmbH) were employed to amplify
cDNA. The initial denaturation condition was 95°C for 10 sec. The
thermocycling (40 cycles) conditions were as follows: 95°C for 10
sec and 60°C for 30 sec. mRNA levels were quantified using the
2−ΔΔCq method and normalized to the internal reference
gene GAPDH (51). The specific
primers are listed in Table
SI.
Western blotting
Following rinsing three times with PBS, cells were
lysed using RIPA buffer (Beyotime Institute of Biotechnology)
supplemented with protease and phosphatase inhibitors (CoWin
Biosciences). Cell homogenates were obtained and centrifuged at
14,000 × g for 20 min at 4°C. The cell supernatant was obtained and
a BCA protein assay kit (CoWin Biosciences) was used to determine
the concentration of total proteins. Samples were subsequently
incubated at 95°C for 10 min in SDS-PAGE sample loading buffer
(Beyotime Institute of Biotechnology) for denaturation. 20 µg total
protein samples were separated via 4–12% SDS-polyacrylamide gel
electrophoresis and transferred to PVDF membranes (MilliporeSigma).
Membranes were blocked in 5% skimmed milk powder supplemented with
1X TBS-1‰ Tween-20 (TBS-T) at room temperature for 1 h, and
subsequently incubated with primary antibodies at 4°C for 12 h. All
primary antibodies are listed in Table SII. Following rinsing with TBS-T,
the membranes were incubated with the HRP goat anti-rabbit IgG
secondary antibody (Beijing Emarbio Science & Technology Co.,
Ltd.) at room temperature for 1 h. Protein bands were visualized
using ECL luminophore (MilliporeSigma) and an enhanced
chemiluminescence detection system (Bio-Rad Laboratories, Inc.).
Protein expression was semi-quantified using ImageJ software v1.53e
(National Institutes of Health). For the MAPK pathway detection,
vinculin (124 kDa) was used as internal reference as it has been
widely used in other studies (52,53),
to ensure there is no overlap with the molecular weight of the
experimental proteins (40–54 kDa). In all other cases, β-actin (42
kDa) was used as the internal reference.
Alizarin red staining
Cells were fixed with 4% paraformaldehyde for 15 min
at room temperature and incubated with alizarin red solution (Wuhan
Servicebio Technology Co., Ltd.) at room temperature for 10 min.
Cells were incubated in PBS to remove excess stain. Matrix calcium
depositions were imaged using a light microscope (Zeiss AG) and
subsequently dissolved in 10% cetylpyridinium chloride
(Sigma-Aldrich; Merck KGaA) at room temperature for 1 h. The
absorbance was measured at 562 nm using Gene5 CHS 3.11 software
(BioTek; Agilent Technologies, Inc.) on a microplate reader
(BioTek; Agilent Technologies, Inc.).
ELISA
Cell culture supernatants were collected from each
group and centrifuged at 500 × g for 5 min at 4°C. ELISA kits
(Beijing winter song Boye Biotechnology Co. Ltd.) were used to
measure the secreted cytokine expression of RANKL (Mouse RANKL
ELISA Kit; cat. no. DG30574M), osteoprotegerin (OPG; Mouse OPG
ELISA Kit; cat. no. DG30295M), IL-6 (Mouse IL-6 ELISA Kit; cat. no.
DG30754M), TNF-α (Mouse TNF-α ELISA Kit; cat. no. DG30048M) and
IL-1β (Mouse IL-1β ELISA Kit; cat. no. DG94767Q) according to the
manufacturer's protocol.
RNA stability
Cells were treated with 5 µg/ml actinomycin D (ACMEC
biochemical) at 37°C to inhibit mRNA transcription. Total RNA was
extracted at 0, 3 and 6 h. RT-qPCR was carried out as
aforementioned to examine mRNA expression and calculate the
degradation rate of target genes, which was presented as the
relative expression of mRNA at each time point relative to that at
the 0-h timepoint (54).
Cell transfection
Specific lentiviral vectors carrying short hairpin
RNA (shRNA/sh) or control shRNA (GeneChem, Inc.) were used for
Mettl3 knockdown. shRNA sequences are shown in Table SIII. Lentiviral vectors were used
for Mettl3 overexpression, and blank vectors were prepared by
Guangzhou IGE Biotechnology, Ltd. shRNA sequences were synthesized
and Mettl3 overexpression products were amplified. The lentivirus
transfer plasmid pLVX (Thermo Fisher Scientific, Inc.)-shRNA was
used for recombinant plasmid construction and lentivirus
production. And for Mettl3 overexpression,
pCDH-CMV-MCS-EF1-copGFP-T2A-Puro vector (Thermo Fisher Scientific,
Inc.) was employed and the Mettl3 coding region was inserted into
the MCS sites of the vector. At 18 h before transfection, HEK 293T
cells (Procell Life Science &Technology Co., Ltd.) were
inoculated in six-well cell culture plates at a density of
4×105 cells per well. Before transfection, 2 µg of the
plasmid was diluted in 100 µl Opti-MEM medium (Gibco) without serum
(tube A), and 6 µl polyethylenimine reagent was diluted in another
100 µl serum-free Opti-MEM (tube B). We then added the dilution in
tube B into tube A and mixed it gently. The mixture was incubated
at room temperature for 12 min and then added to the cell culture
medium. A 3rd generation system including four-plasmid lentivirus,
pLP/VSVG, pLP1, pLP2, and empty/recombinant pLVX vector (Thermo
Fisher Scientific, Inc.), was applied at a molar ratio of 1:1:1:2.
Cells were incubated with transfection reagent-plasmid mixture at
37°C for 6 h before replacement. At 24 h after replacement, cell
culture medium containing lentiviral particles was collected. The
above protocols were completed by company and we carried out the
subsequent transfection and selection. In brief, lentiviruses were
cultured with MC3T3-E1 cells at a MOI of 50 at 37°C for 12 h. Green
fluorescence protein labeling was used to observe transfection
using a light microscope (Zeiss AG). Cell lines with stable
expression were selected using 6 µg/ml puromycin for 3 days and
maintained using 2 µg/ml puromycin (Biosharp Life Sciences). For
YTH domain family 2 (Ythdf2) knockdown, 50 nM small interfering RNA
(si)Ythdf2 or NC (Guangzhou IGE Biotechnology, Ltd.), named siCtrl,
was employed to transfect cells with the addition of Lipofectamine®
2000 Transfection reagent (Biosharp Life Sciences) at 37°C for 24 h
(Table SIII). The transfected
cells were employed for subsequent experimentations at 24 h after
transfection. The transfection efficiency was determined using
RT-qPCR and western blot analysis and at 24 and 48 h after the
transfection, respectively.
RNA dot blot for total m6A
content analysis
Total RNA was extracted as aforementioned and
incubated at 95°C for 3 min to disrupt secondary structures.
Denatured total RNA was loaded onto nitrocellulose membranes and
subsequently crosslinked to the membrane with UV light at 254 nm
for 30 min. Unbound RNA was removed by washing with TBS-T. Western
blot analysis was carried out as aforementioned using the
anti-m6A antibody (Table
SII).
Methylated RNA immunoprecipitation
(MeRIP)-qPCR
The EpiQuik CUT&RUN m6A RNA
Enrichment kit (EpigenTek Group, Inc.) was used for the MeRIP assay
as previously described (55,56).
Briefly, total RNA was collected and randomly fragmented. RNA
fragments were immunoprecipitated with magnetic beads pre-coated
with anti-immunoglobulin (IgG), anti-m6A or anti-Ythdf2
antibodies. Targeted RNA fragments were released, purified and
eluted, followed by RT-qPCR as aforementioned. Relative enrichment
was normalized to the input sample.
Statistical analysis
All data were representative of three independent
experiments and presented as the mean ± standard deviation. Data
were plotted and analyzed using GraphPad Prism v9.0 (Dotmatics).
Comparisons between two groups were analyzed using an unpaired
Student's t-test. Comparisons among multiple groups were analyzed
using one-way ANOVA followed by Tukey's post hoc test and two-way
ANOVA followed by Bonferroni's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Titanium particles inhibit osteogenic
differentiation and mediate osteoblast-osteoclast
communication
Titanium particles engulfed into osteoblastic cells
were observed using TEM (Fig. 1A).
In the present study, RT-qPCR and western blot analysis were used
to show early trends, and alizarin red staining was used to study
later trends. The results of the present study demonstrated that
titanium particle treatment significantly inhibited the expression
of osteogenesis-associated markers and the formation of calcium
nodules compared with those in the osteogenic induction group
(Fig. 1B and C). Furthermore,
titanium particles induced the mRNA expression of proinflammatory
cytokines (Fig. 1D), and these
results were verified via ELISA. To further investigate the
regulatory role of titanium particles in osteoblast-osteoclast
communication, which is critical for bone homeostasis (57), the supernatant of titanium-treated
osteoblasts was harvested and co-cultured with preosteoclasts. As
shown in Fig. 1E, mRNA and protein
expression levels of markers associated with osteoclast
differentiation of RAW264.7 cells were increased in the titanium
particle treatment group (CM) compared with the NC group. Thus, the
expression levels of additional soluble factors released from
osteoblasts that mediate osteoblast-osteoclast communication were
investigated. Notably, RANKL expression was significantly
increased, leading to a higher ratio of RANKL/OPG (Fig. 1F).
 | Figure 1.Titanium particles inhibit osteogenic
differentiation and mediate osteoblast-osteoclast communication.
(A) Transmission electron microscopy of osteoblasts co-cultured
(A-a) without and (A-b) with titanium particles (red arrows; scale
bar, 5 µm). (B) Titanium particle treatment inhibited the formation
of mineralized nodules at 14 days. (B-a) Blank, (B-b) osteogenic
induction medium and (B-c) titanium particle treatment. Scale bar,
500 µm. (C) Titanium particle treatment inhibited the expression of
osteogenesis-associated markers. The samples were collected at 2
and 3 days for RT-qPCR and western blotting, respectively. (D)
Titanium particle treatment promoted the expression of inflammatory
cytokines. The samples were collected at 2 and 3 days for RT-qPCR
and ELISA, respectively. (E) Supernatants of titanium-treated
osteoblasts promoted the expression of osteoclast
differentiation-associated markers in preosteoclasts. (F) Titanium
treatment promoted the expression of RANKL, leading to the higher
ratio of RANKL/OPG. The samples were collected at 2 and 3 days for
RT-qPCR and ELISA, respectively. Data are representative of three
independent experiments and are presented as the mean ± standard
deviation. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. Acp5, acid phosphatase 5, tartrate resistant;
Col1a1, collagen type I α1 chain; Ctsk, cathepsin K; CM,
conditioned medium; Dcstamp, dendrocyte expressed seven
transmembrane protein; M-CSF, macrophage colony-stimulating factor;
NC, negative control; Nfatc1, nuclear factor of activated T cells
1; ns, not significant; OD, optical density; OM, osteogenic
induction medium; OPG, osteoprotegerin; PC, positive control;
RANKL, receptor activator of NF-κB ligand; RT-qPCR, reverse
transcription-quantitative PCR; Runx2, RUNX family transcription
factor 2; Ti, titanium particle. |
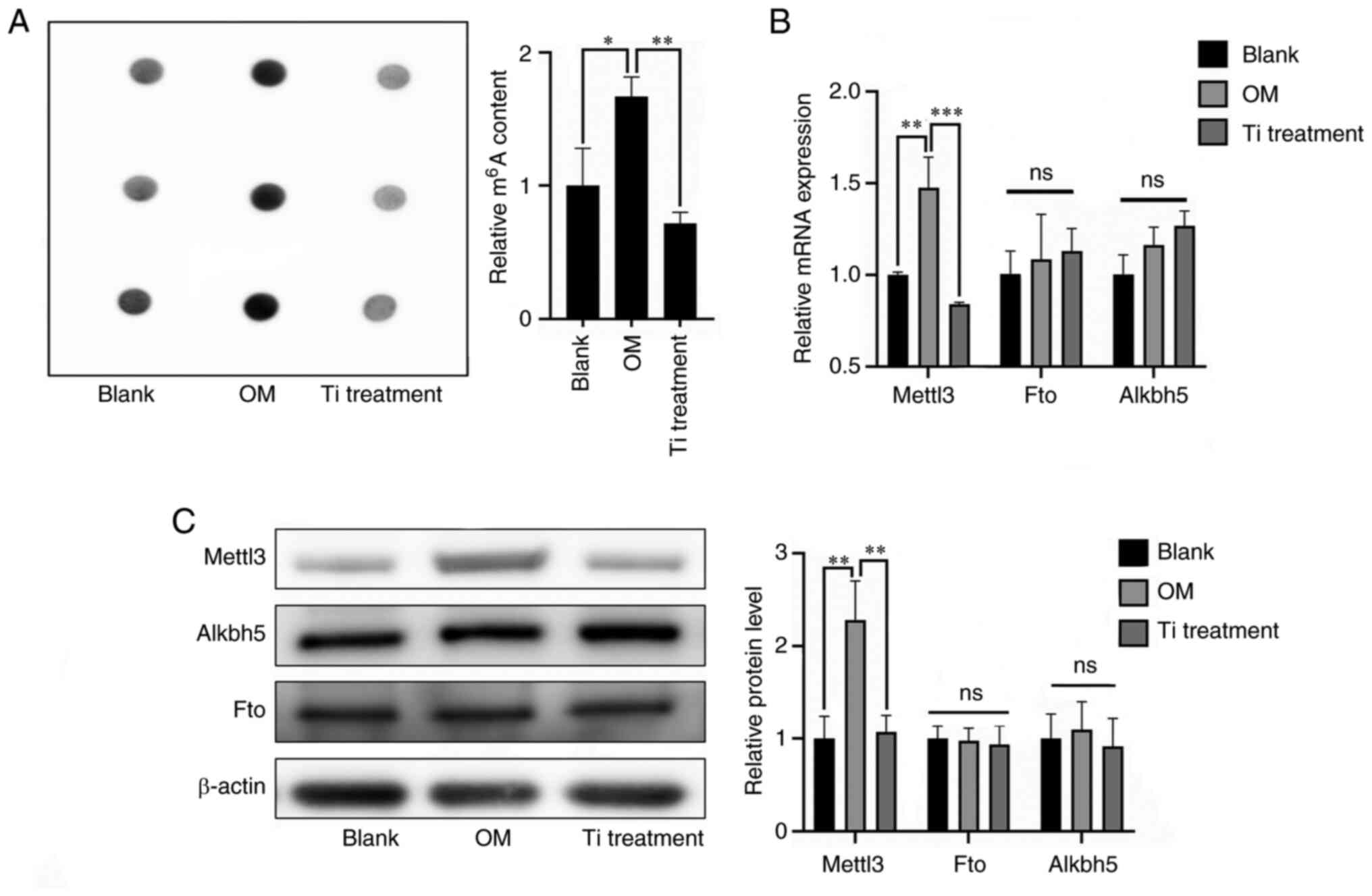
Role of m6A modification in
titanium particle treatment
To explore the role of m6A modification
in titanium particle treatment of osteoblasts, m6A
levels were evaluated. The results of the RNA dot blot analysis
demonstrated that m6A levels were reduced in the
titanium treatment group compared with the osteogenic induction
group (Fig. 2A). Subsequently,
expression levels of common methyltransferases and demethylases
were further determined via RT-qPCR and western blotting (Fig. 2B and C). The expression levels of
Mettl3, a key methyltransferase, were significantly decreased
during titanium treatment, and these results were consistent with
the measurement of m6A levels.
Mettl3 knockdown induces osteogenic
inhibition and proinflammatory responses
To explore the association between titanium
particle-induced bioactivities and the reduced expression levels of
Mettl3, Mettl3 knockdown was carried out in MC3T3-E1 cells,
referred to as shMettl3 cells (Fig.
3A). The results of the present study demonstrated that Mettl3
mRNA and protein expression was significantly reduced following
transfection compared with the NC or blank groups (Fig. 3B). The results of the RNA dot blot
analysis revealed that m6A levels were lowest in the
shMettl3 group compared with the control and titanium particle
treatment groups (Fig. 3C).
Notably, mRNA and protein expression levels of osteogenic markers
were significantly reduced in the shMettl3 group (Fig. 3D). In addition, the alizarin red
staining assay demonstrated consistent results, indicating that
Mettl3 knockdown induced osteogenic inhibition comparable with that
induced by titanium particles (Fig.
3D). Furthermore, Mettl3 knockdown increased the expression of
proinflammatory cytokines (Fig.
3E). Mettl3 knockdown increased the RANKL expression and the
ratio of RANKL/OPG compared with the control group (shCtrl
osteoblasts without titanium particle) (Fig. 3F). Additionally, the supernatant of
Mettl3-knockdown osteoblasts was harvested and co-cultured with
RAW264.7 cells. mRNA and protein expression levels of markers
associated with osteoclastogenesis were increased in RAW264.7 cells
cultured with supernatant of Mettl3-knockdown osteoblasts compared
with the control group (shCtrl osteoblasts without titanium
particle) (Fig. 3G).
 | Figure 3.Mettl3 knockdown induces osteogenic
inhibition and proinflammatory responses. (A) Construction of
Mettl3 knockdown cells (scale bar, 100 µm). (B) The verification of
transfection efficiency using reverse transcription-quantitative
PCR and western blotting. (C) Mettl3 knockdown and titanium
particle treatment were associated with reduced m6A
content in total RNA compared with the blank group (osteoblasts
without transfection). (D) Mettl3 knockdown inhibited the
expression of osteogenic markers and the formation of mineralized
nodules (scale bar, 500 µm). (E) Mettl3 knockdown promoted the
expression of inflammatory cytokines. (F) Mettl3 knockdown led to a
higher ratio of RANKL/OPG compared with that of the control group
(shCtrl cells without Ti). (G) Supernatant of Mettl3 knockdown
osteoblasts promoted the expression of osteoclast
differentiation-associated markers in preosteoclasts. Data are
representative of three independent experiments and are presented
as the mean ± standard deviation. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. Acp5, acid phosphatase 5,
tartrate resistant; Col1a1, collagen type I α1 chain; Ctrl,
control; Ctsk, cathepsin K; Dcstamp, dendrocyte expressed seven
transmembrane protein; m6A, N6-methyladenosine; Mettl3,
methyltransferase-like 3; Nfatc1, nuclear factor of activated T
cells 1; ns, not significant; OD, optical density; OPG,
osteoprotegerin; RANKL, receptor activator of NF-κB ligand; sh,
short hairpin RNA; Runx2, RUNX family transcription factor 2; Ti,
titanium particle. |
Mettl3 overexpression attenuates
titanium particle-induced inhibition of osteogenesis and
proinflammatory activities
To further verify whether titanium particles
mediated the aforementioned activities through reduced Mettl3
expression, a Mettl3-overexpression osteoblast cell line was
constructed, and a blank vector was used as the NC. RT-qPCR and
western blotting were performed to determine the transfection
efficiency following overexpression (Fig. 4A). TEM analysis demonstrated that
Mettl3 overexpression did not impact the engulfment capability of
osteoblasts (Fig. 4B). Notably,
m6A levels in the Mettl3 overexpression group were
significantly increased compared with those in the blank group
(Fig. 4C). The results of the
present study also demonstrated that Mettl3 overexpression
attenuated titanium-induced inhibition of osteogenesis (Fig. 4D). In addition, titanium-induced
proinflammatory activities were also attenuated, as well as the
ratio of RANKL/OPG (Fig. 4E and
F). The aforementioned results in Fig. 1E indicated the regulatory role of
titanium particles in osteoblast-osteoclast communication, while
this indirect regulatory effect of titanium particles on
osteoclastogenesis of RAW264.7 cells was also reversed by Mettl3
overexpression (Fig. 4G).
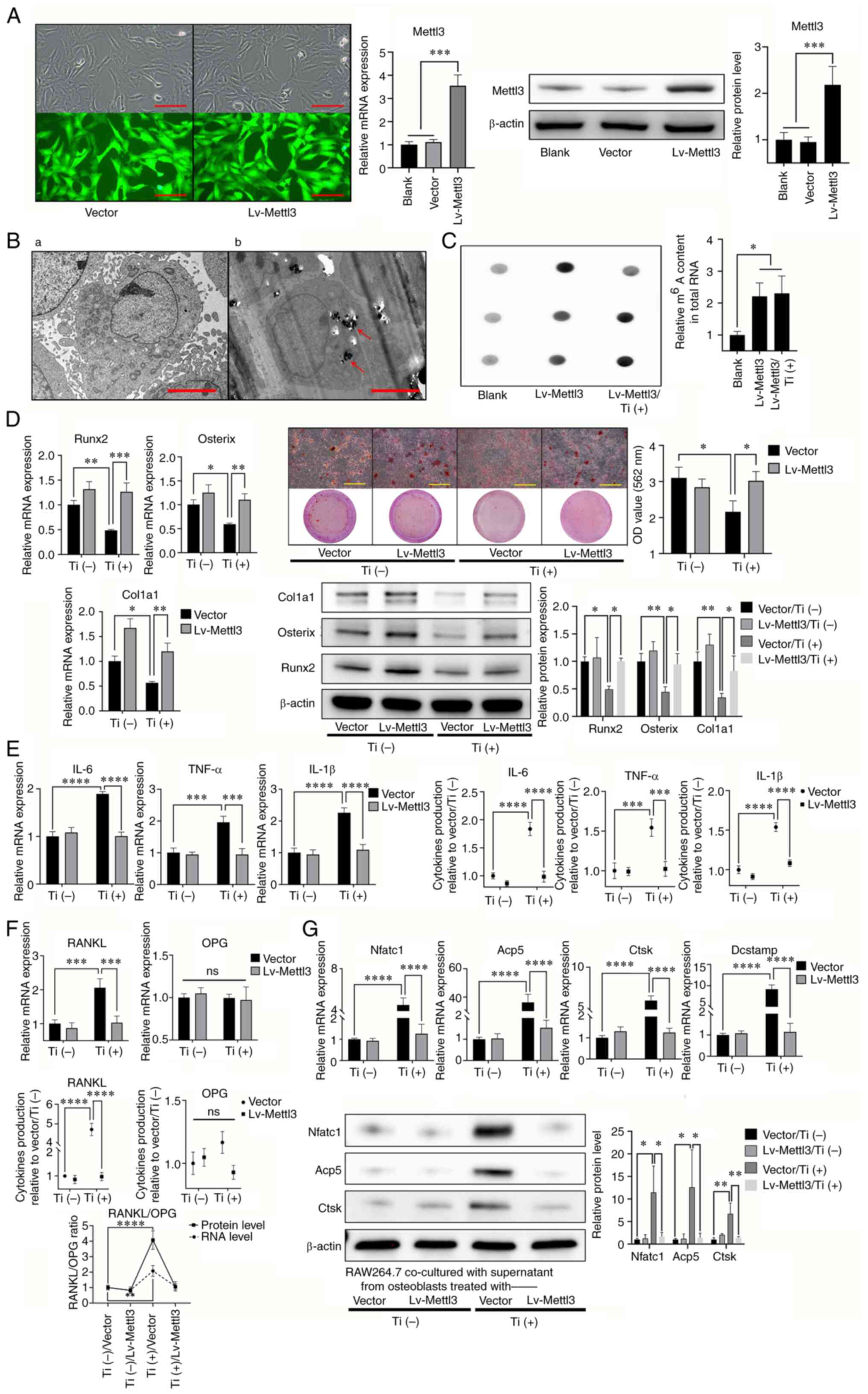 | Figure 4.Mettl3 overexpression attenuates
titanium particle-induced osteogenesis inhibition and
proinflammatory responses. (A) Construction of Mettl3
overexpression cells and the verification of transfection
efficiency using reverse transcription-quantitative PCR and western
blotting (scale bar, 100 µm). (B) Transmission electron microscopy
of Mettl3 overexpression cells co-cultured (B-a) without or (B-b)
with titanium particles (red arrows; scale bar, 5 µm). (C) Mettl3
overexpression increased the m6A content in total RNA
compared with that of the blank group (osteoblasts without
transfection). (D) Mettl3 overexpression attenuated titanium
particle-induced osteogenesis inhibition (scale bar, 500 µm). (E)
Mettl3 overexpression attenuated titanium particle-induced
proinflammatory responses. (F) Mettl3 overexpression attenuated the
titanium particle-induced increase in the RANKL/OPG ratio. (G)
Mettl3 overexpression attenuated osteoclast differentiation
promotion induced by the supernatant of Ti-treated osteoblasts.
Data are representative of three independent experiments and are
presented as the mean ± standard deviation. *P<0.05,
**P<0.01, ***P<0.001 and ****P<0.0001. Acp5, acid
phosphatase 5, tartrate resistant; Col1a1, collagen type I α1
chain; Ctsk, cathepsin K; Dcstamp, dendrocyte expressed seven
transmembrane protein; Lv, lentivirus; m6A,
N6-methyladenosine; Mettl3, methyltransferase-like 3; Nfatc1,
nuclear factor of activated T cells 1; ns, not significant; OD,
optical density; OPG, osteoprotegerin; RANKL, receptor activator of
NF-kB ligand; Runx2, RUNX family transcription factor 2; Ti,
titanium particle. |
Titanium particle treatment targets
Smad7 and SMAD specific E3 ubiquitin protein ligase 1 (Smurf1) via
Mettl3, leading to bone morphogenetic protein (BMP) signaling
inhibition
To explore the downstream signaling molecules
affected by Mettl3, the canonical BMP-Smad signaling pathway was
evaluated. The results of RT-qPCR analysis demonstrated that
titanium particle treatment increased Smad7 and Smurf1 expression
in osteoblasts, while Mettl3 overexpression attenuated the
upregulated expression of Smad7 and Smurf1 induced by titanium
particle treatment (Fig. 5A). The
results of western blotting further demonstrated that titanium
particle treatment inhibited the phosphorylation levels of
Smad1/5/9 and induced the increased expression of Smad7 and Smurf1,
and these reactions induced by titanium particle treatment were
also reversed by Mettl3 overexpression (Fig. 5B). The results of the present study
also demonstrated that both titanium particle treatment and Mettl3
knockdown increased the mRNA stability of these two genes, leading
to increased expression levels (Fig.
5C). Furthermore, MeRIP-qPCR analysis revealed that the
m6A enrichment of Smad7 and Smurf1 transcripts was
significantly reduced in the titanium particle treatment and Mettl3
knockdown groups compared with the control group (Fig. 5D).
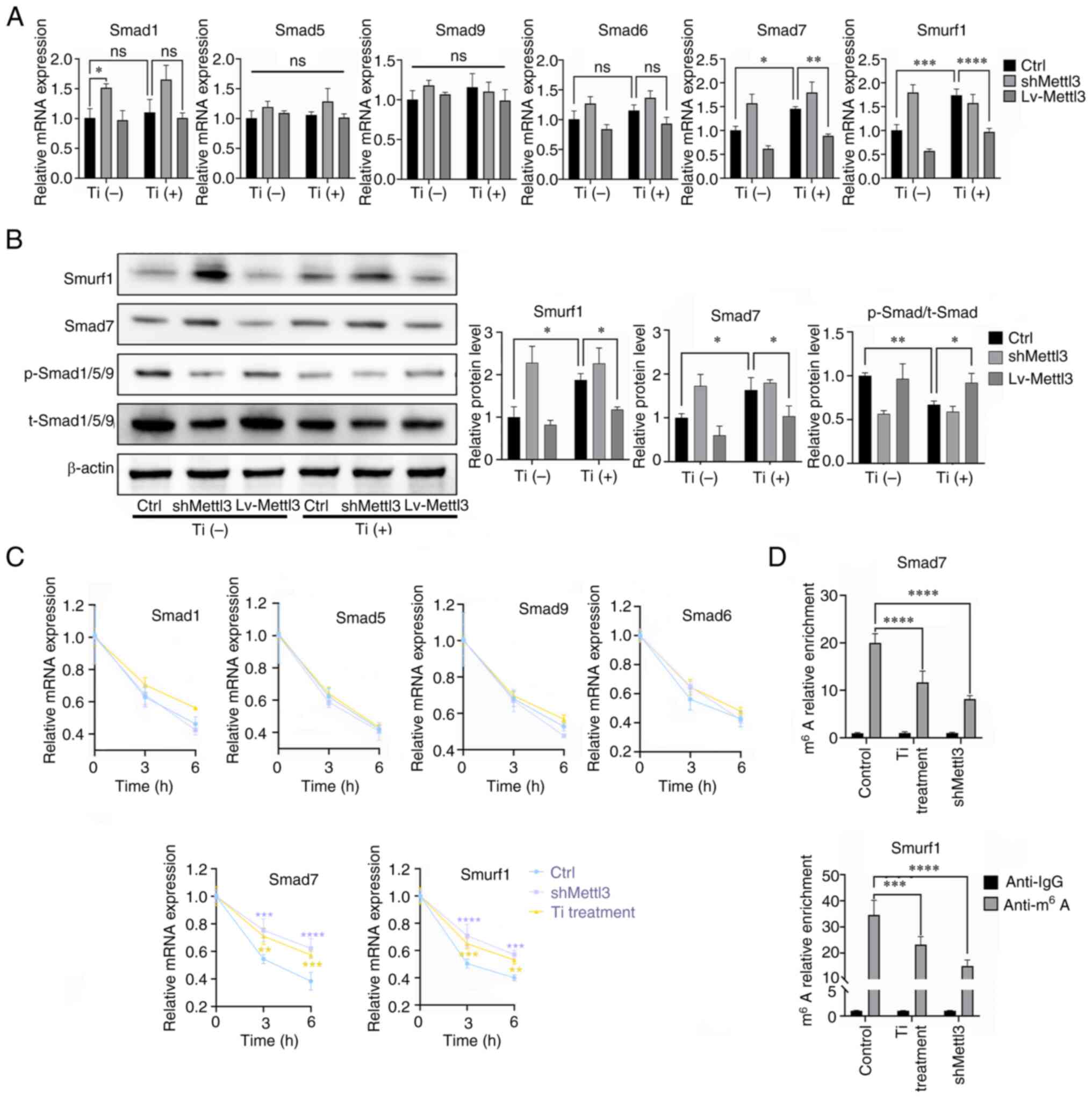 | Figure 5.Titanium particle treatment targets
Smad7 and Smurf1 via Mettl3, leading to BMP signaling inhibition.
(A) Effects of titanium particle treatment and Mettl3
knockdown/overexpression on the mRNA expression of key signaling
molecules of BMP signaling. (B) Effects of titanium particle
treatment and Mettl3 knockdown/overexpression on BMP signaling
activation. (C) Titanium particle treatment enhanced the mRNA
stability of Smad7 and Smurf1 transcripts at the selected
timepoints, following the addition of actinomycin D. (D) Titanium
particle treatment significantly decreased the m6A
modification of these two transcripts. The control group in this
figure were osteoblasts without transfection. Data are
representative of three independent experiments and are presented
as the mean ± standard deviation. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. BMP, bone morphogenetic protein;
Ctrl, control; Lv, lentivirus; m6A, N6-methyladenosine;
Mettl3, methyltransferase-like 3; ns, not significant; p-,
phosphorylated; sh, short hairpin RNA; Smurf1, SMAD specific E3
ubiquitin protein ligase 1; t-, total; Ti, titanium particle. |
Titanium particles induce NOD-like
receptors (NLRs) to exert proinflammatory activities
The results of western blotting revealed that
titanium particle treatment induced the activation of the MAPK and
NF-κB pathways, while Mettl3 overexpression attenuated this
activation. For p38, it seemed that the titanium particle induced
its activation while the Mettl3 overexpression failed to attenuated
this activation. (Fig. 6A). The
expression changes of key components of the NLR signaling pathway
were also evaluated. The results of the present study demonstrated
that titanium particle treatment increased the expression and the
mRNA half-life of NOD1 and receptor interacting serine/threonine
kinase 2 (RIPK2) in osteoblasts. However, the mRNA stability and
expression of NOD2 were not enhanced (Fig. 6B and C). MeRIP-qPCR analysis
demonstrated that NOD1 and RIPK2 transcripts functioned as the
target of Mettl3, as titanium particle treatment and Mettl3
knockdown both significantly reduced the m6A enrichment
of these two transcripts compared with the control group (Fig. 6D).
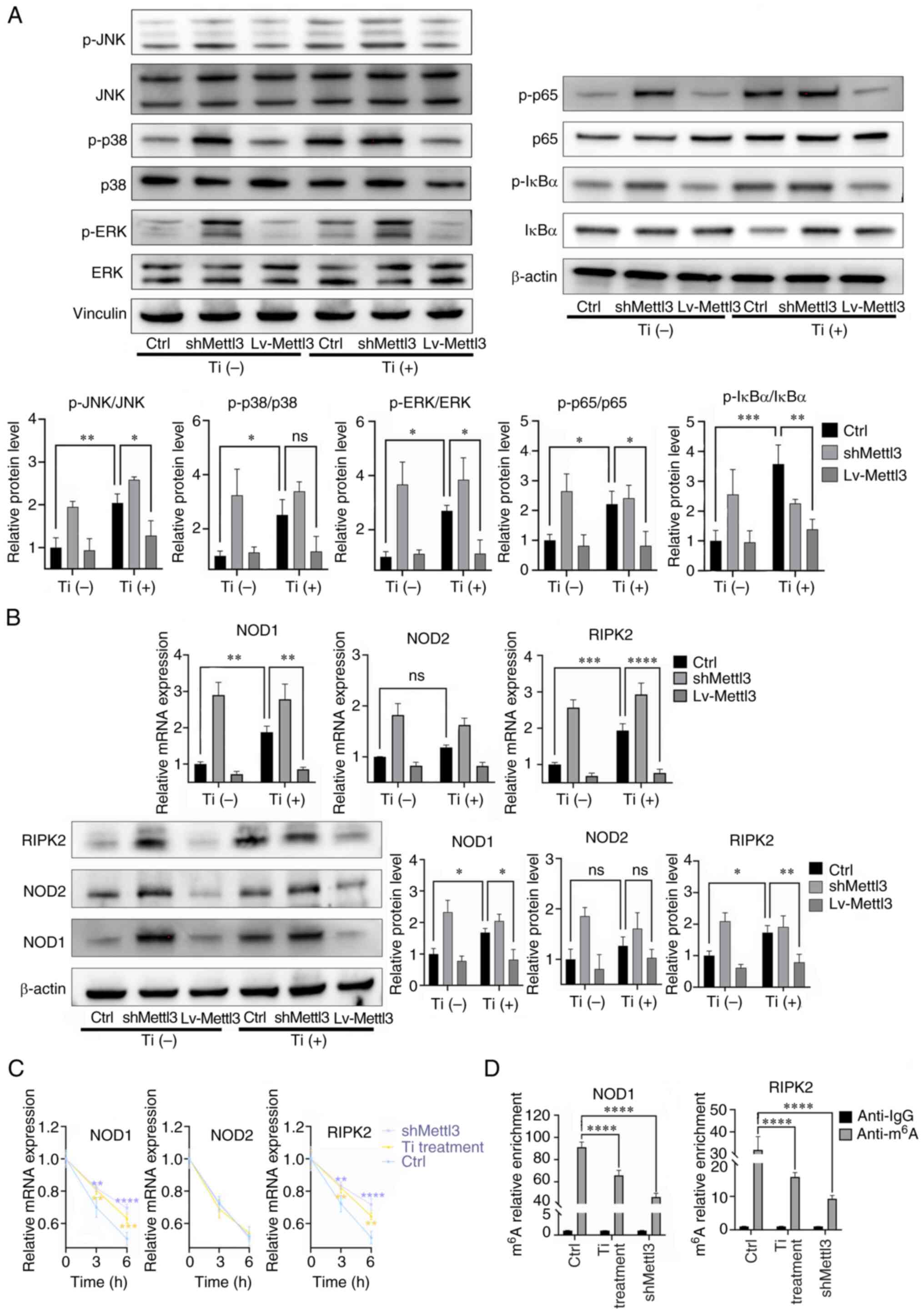 | Figure 6.Titanium particle treatment induces
NOD-like receptors to exert proinflammatory responses. (A) Titanium
particle treatment induced the activation of the MAPK and NF-κB
signaling pathways. (B) Titanium particle treatment induced the
activation of the NOD-like receptor pathway, while Mettl3
overexpression attenuated these effects. (C) Titanium particle
treatment and Mettl3 knockdown enhanced the mRNA stabilities of
NOD1 and RIPK2 following the addition of actinomycin D. (D)
Titanium particle treatment and Mettl3 knockdown decreased the
m6A modification of NOD1 and RIPK2. The control group in
this figure were osteoblasts without transfection. Data are
representative of three independent experiments and are presented
as the mean ± standard deviation. *P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001. Ctrl, control; Lv, lentivirus;
m6A, N6-methyladenosine; Mettl3, methyltransferase-like
3; NOD, nucleotide binding oligomerization domain; ns, not
significant; p-, phosphorylated; RIPK2, receptor interacting
serine/threonine kinase 2; sh, short hairpin RNA; Ti, titanium
particle. |
To further verify the role of the NLR1 signaling
pathway in the titanium particle-induced inflammatory response,
cells were pretreated with ML130 (NOD1 inhibitor) and WEHI-345
(RIPK2 inhibitor) to block the NOD1 pathway since titanium
particles can act on both NOD1 and RIPK2 mRNA based on the
aforementioned results. In Fig.
7A, without the addition of inhibitors, titanium particle
induced the activation of the MAPK and NF-κB signaling pathways,
and the activation of these two pathways was significantly
inhibited following treatment with both inhibitors from the
comparison between titanium particle treatment without inhibitors
group and titanium particle treatment with inhibitors group
(Fig. 7A). In addition, inhibition
of the NOD1 pathway attenuated the titanium particle-induced
increased expression of IL-6, TNF-α, IL-1β and RANKL (Fig. 7B). Collectively, these data
suggested that titanium particles may target the NOD1 signaling
pathway to regulate subsequent inflammatory responses.
Ythdf2 participates in the
Mettl3-mediated osteogenic inhibition and proinflammatory
activities in titanium particle treatment
m6A readers recognize m6A
modifications and then mediate regulatory effects, including RNA
translation, decay and splicing (41). The aforementioned results (Figs. 5D and 6D) revealed that downregulated Mettl3
levels led to the enhancement of mRNA stabilities of Smad7, Smurf1,
NOD1 and RIPK2, and thus, it was hypothesized that it was Ythdf2
that exerted the effects. Firstly, siYthdf2 was used to knockdown
its expression (Fig. 8A). The mRNA
and protein expression levels of Smad7, Smurf1, NOD1 and RIPK2 were
increased in the Ythdf2 knockdown group compared with the control
group (Fig. 8B). The mRNA
stabilities of these four mRNAs were also enhanced in Ythdf2
knockdown cells at 3 and 6 h, while the stability of NOD at 6 h
didn't show statistical difference (Fig. 8C), suggesting that Ythdf2 could
affect the expression of these four mRNAs by regulating their
stabilities. In Fig. 8D, we
measured the relative expressions of these four mRNAs precipitated
by Ythdf2, which were shown in anti-Ythdf2 column. Notably,
titanium particle treatment reduced the expression levels of these
mRNAs compared with control group, indicating that the binding
between Ythdf2 and mRNAs was significantly decreased following
titanium particle treatment (Fig.
8D).
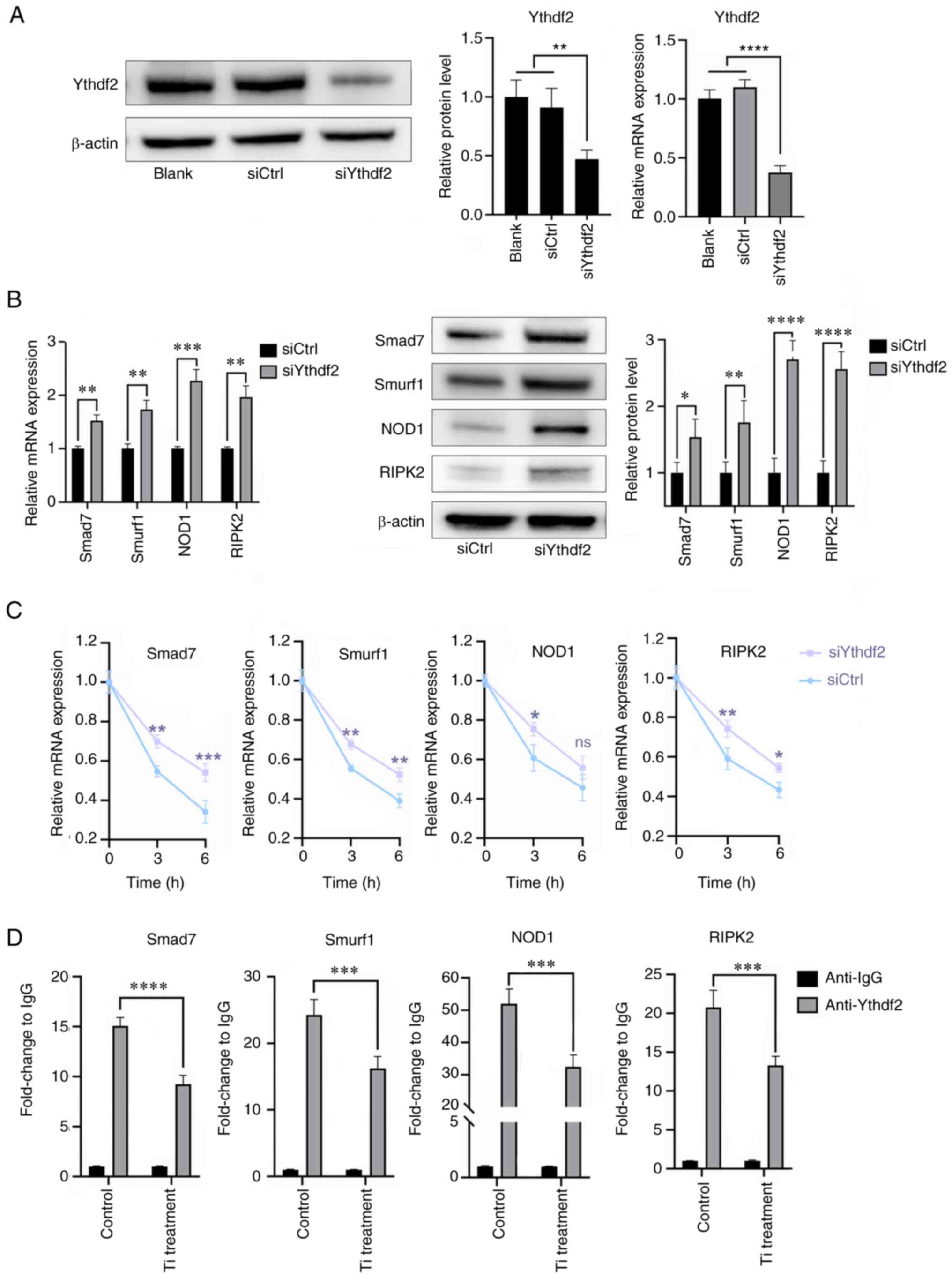 | Figure 8.Ythdf2 participates in the
methyltransferase-like 3-mediated bioactivities in titanium
particle treatment. (A) Knockdown of Ythdf2 and verification of
transfection efficiency using reverse transcription-quantitative
PCR and western blotting. (B) Ythdf2 knockdown promoted the
expression of Smad7, Smurf1, NOD1 and RIPK2. (C) Ythdf2 knockdown
enhanced the mRNA stabilities of Smad7, Smurf1, NOD1 and RIPK2 at 3
h. (D) Methylated RNA immunoprecipitation-quantitative PCR results
demonstrated that titanium particle treatment decreased the
relative expression levels of Smad7, Smurf1, NOD1 and RIPK2
precipitated by Ythdf2. Data are representative of three
independent experiments and are presented as the mean ± standard
deviation. *P<0.05, **P<0.01, ***P<0.001 and
****P<0.0001. Ctrl, control; NOD, nucleotide binding
oligomerization domain; RIPK2, receptor interacting
serine/threonine kinase 2; si, small interfering RNA; Smurf1, SMAD
specific E3 ubiquitin protein ligase 1; Ythdf2, YTH domain family
2; Ti, titanium particle. |
Discussion
Bone homeostasis is a dynamic balance regulated by
osteoblasts and osteoclasts. However, dysregulation of the balance
between osteoblasts and osteoclasts may result in pathological bone
loss, in which osteogenic inhibition and inflammatory responses
serve a vital role (57,58). The activation of inflammatory
responses promotes the secretion of several cytokines and the
regulation of specific cell types, including osteoclasts,
osteoblasts, macrophages and immune cells, thus leading to
pathological osteolysis (57,59).
The results of the present study revealed that titanium particles
inhibited osteogenesis and proinflammatory responses, and these
results are consistent with previous reports (10,22,60).
Furthermore, the results of the present study demonstrated that
titanium particles mediated the osteoblast-osteoclast
communication, leading to enhanced osteoclast activity, and these
underlying mechanisms were primarily explored. Notably, osteoblasts
secrete several soluble factors, including macrophage
colony-stimulating factor, RANKL, OPG and WNT5A, to act on
osteoclasts (57). The elevated
RANKL/OPG ratio may contribute to the enhanced osteoclast activity.
However, additional mechanisms, such as secreted exosomes and
membrane-bound mediators, were not further explored in the present
study.
Previous studies have reported the critical role of
m6A methylation in bone remodeling and inflammation
(48,61). To explore the potential role of
m6A methylation in titanium particle-induced
bioactivities, total m6A levels were determined.
Notably, the differentially expressed enzyme Mettl3 is the most
critical methyltransferase and previous studies have indicated that
Mettl3 exerts regulatory effects in several physiological and
pathological processes (42–47).
The results of the present study demonstrated that Mettl3 knockdown
induced the inhibition of osteogenesis and proinflammatory
responses, and these effects were comparable to those induced by
titanium particles. In addition, Mettl3 overexpression reversed the
bioactivities induced by titanium particles. Previous studies have
demonstrated that Mettl3-mediated m6A modification
regulated bone metabolism, including cell differentiation, and
abnormal Mettl3 expression levels may induce bone metabolic
diseases (44,46,48).
Collectively, the results of the present study highlighted that
Mettl3 may function as an upstream regulator in titanium
particle-induced bioactivities.
Notably, the canonical Smad-dependent pathway serves
a key role in osteogenesis and the activation and translocation of
the Smad1/5/9 complex triggers the subsequent expression of
osteogenesis-related genes (62,63).
Inhibitory Smads negatively regulate Smad signaling by preventing
Smad1/5/9 activation and degrading Smad1/5/9 with the assistance of
the E3 ubiquitin ligase, Smurf1 (62,64).
Thus, RT-qPCR and western blotting revealed the decreased levels of
Smad1/5/9 phosphorylation, along with the increased expression of
Smad7 and Smurf1, following treatment with titanium particles or
Mettl3 knockdown. Notably, enhanced mRNA stability may account for
the increased expression of Smad7 and Smurf1. To further confirm
the regulation of Mettl3, MeRIP-qPCR was performed. The results of
the present study indicated that the m6A enrichment of
these two transcripts was significantly reduced following titanium
particle treatment or Mettl3 knockdown. Thus, titanium particle
treatment may suppress the mRNA decay of Smad7 and Smurf1 via
Mettl3 downregulation, leading to inhibition of Smad-dependent
signaling.
Previous studies have demonstrated that titanium
particles may concurrently activate two classical proinflammatory
pathways, including the MAPK and NF-κB pathways, and promote the
expression of downstream proinflammatory cytokines (26,27,65).
The results of the present study demonstrated that titanium
particle treatment induced the activation of these two
inflammation-associated pathways, while Mettl3 overexpression
reversed the activation, suggesting that Mettl3 was involved in the
activation of the MAPK and NF-κB signaling pathways. To further
explore the target of titanium particles in these pathways, the
mRNA stability of numerous molecules was determined. A previous
study identified the activation of the NLR signaling pathway in
Mettl3 knockdown macrophages via RNA sequencing analysis (66). Activation of key members of the NLR
pathway, including NOD1, NOD2 and RIPK2, may trigger the subsequent
activation of the MAPK and NF-κB signaling pathways (67–69).
Thus, it was hypothesized that titanium particles may impact the
NLR pathway, leading to the subsequent activation of inflammatory
responses. Notably, titanium particle treatment increased the mRNA
expression and stability of NOD1 and RIPK2. In addition, the
m6A enrichment of these two transcripts was
significantly reduced following titanium particle treatment.
Inhibition of the NLR pathway inhibited the activation of MAPK and
NF-κB pathways and the expression of proinflammatory cytokines,
suggesting that titanium particles exert their effect via
downregulation of Mettl3, which activates the NLR pathway and leads
to the subsequent activation of inflammatory responses.
The m6A reader proteins are required to
recognize and bind to the m6A modified transcript to
regulate gene expression (70).
Several m6A readers have been identified, including YTH
domain-containing family proteins, heterogeneous nuclear
ribonucleoproteins, insulin like growth factor 2 mRNA binding
protein families and eukaryotic initiation factor (71). Ythdf2, a main member of the YTH
domain-containing family proteins, is the most extensively studied
m6A reader. Ythdf2 promotes targeted mRNA decay by
recruiting the C-C motif chemokine receptor 4 negative on TATA-less
deadenylase complex (72).
Increasing evidence has indicated that Ythdf2 serves a critical
role in pathological processes, including stress, viral infection
and the inflammatory response (66,73–76).
Considering that titanium particle treatment regulates RNA decay of
specific mRNAs, it was hypothesized that Ythdf2may recognize the
methylated modification of these mRNAs and promote the targeted
mRNA decay. The results of the present study demonstrated that the
knockdown of Ythdf2 led to significantly increased expression and
enhanced stabilities of Smad7, Smurf1, NOD1 and RIPK2 transcripts
at 3 h, and Smad7, Smurf1 and RIPK2 transcripts at 6 h. MeRIP-qPCR
results (Figs. 5D and 6D) showed that titanium particle
treatment could reduce the m6A sites of these four
mRNAs, meanwhile, binding sites between Ythdf2 and targeted mRNAs
was significantly decreased following titanium particle treatment
(Fig. 8D). Taken together, it
might be concluded that the reduced m6A sites of mRNAs
led to the reduced recognition from Ythdf2, therefore reduced the
mRNAs decay and increased the expression of these four molecules.
Furthermore, it is possible that there are other reader proteins
that participate in the titanium particle treatment. Previous
studies have reported several m6A methylation-regulated
bioactivities involving more than one reader protein (77–79),
including YTH N6-methyladenosine RNA binding protein F1, which
promotes mRNA translation, and YTH N6-methyladenosine RNA binding
protein F3, which mediates the translation or degradation of
targeted RNA, which could be potential readers that also
participate in the titanium particle-induced bone loss (71). Further studies are needed to
uncover the whole mechanism.
In addition, it is important to point out that the
real-life concentration range of titanium particles within the body
is wide (21). The in vivo
concentration of particles is affected by various factors,
including prosthesis type, implant time, sample location and
detection method (80). It should
be noted that the particle concentrations from different studies
are difficult to be integrated as a specific range due to different
measurement systems (81).
Measured concentrations from several representative studies include
720–820 ppm (82), 66–1,734 µg/g
tissue (83) and 103–5,759 µg/g
tissue (84). It is possible to
observe local high particle concentrations at specific timepoints
for various reasons such as implant cracks (85). On the other hand, the cell density
used in in vitro experiments can not simulate the real-life
condition, and thus, it is difficult to directly employ the
real-life concentration for in vitro experiments. The
concentration used in the present study was drawn from previous
studies (10,22,60);
however, this specific concentration may not be physiologically
relevant, which constitutes a potential limitation of the present
study.
In conclusion, the results of the present study
demonstrated that titanium particles reduced the expression of
Mettl3, a key methyltransferase, and reduced the m6A
modification of specific mRNAs. The reduced methylation of these
transcripts decreased the specific sites recognized by reader
protein Ythdf2, therefore decreased the mRNA degradation mediated
by Ythdf2. And that might account for the increased expression
levels of Smad7, Smurf1, NOD1 and RIPK2 transcripts, leading to the
inhibition of osteogenesis and proinflammatory responses. These
findings highlighted that Mettl3 may act as an upstream regulatory
molecule in titanium particle-induced osteolysis. Based on the
critical functions of Mettl3 in various types of cancer, Mettl3 has
been considered a potential target in cancer treatment (86). Therefore, future studies might
focus on the feasibility of employing Mettl3 as a therapeutic
target of PPO, so that multiple pathways can be regulated
simultaneously and to reach the targeted effect. The in vivo
application and ideal drug concentration are also topics for
further research. Thus, the present study may provide novel
insights into potential therapeutic targets for the aseptic
loosening of titanium prostheses.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by The Science and Technology
Bureau of Nansha, Guangzhou (grant no. 2021ZD004).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XL, YY, XY and FD contributed to the study design.
XL, YY, YH and XZ performed the experiments. EL and ZZ assisted in
the data collection. ZZ and RX assisted in the data analysis. XL
and YY wrote the manuscript and confirm the authenticity of all the
raw data. All authors have read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ollivere B, Wimhurst JA, Clark IM and
Donell ST: Current concepts in osteolysis. J Bone Joint Surg Br.
94:10–15. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hodges NA, Sussman EM and Stegemann JP:
Aseptic and septic prosthetic joint loosening: Impact of
biomaterial wear on immune cell function, inflammation, and
infection. Biomaterials. 278:1211272021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tay ML, Matthews BG, Monk AP and Young SW:
Disease progression, aseptic loosening and bearing dislocations are
the main revision indications after lateral unicompartmental knee
arthroplasty: A systematic review. J ISAKOS. 7:132–141. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eger M, Sterer N, Liron T, Kohavi D and
Gabet Y: Scaling of titanium implants entrains inflammation-induced
osteolysis. Sci Rep. 7:396122017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
McArthur BA, Scully R, Patrick Ross F,
Bostrom MPG and Falghren A: Mechanically induced periprosthetic
osteolysis: A systematic review. HSS J. 15:286–296. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eliaz N: Corrosion of metallic
biomaterials: A review. Materials (Basel). 12:4072019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Prestat M and Thierry D: Corrosion of
titanium under simulated inflammation conditions: Clinical context
and in vitro investigations. Acta Biomater. 136:72–87. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Delanois RE, Mistry JB, Gwam CU, Mohamed
NS, Choksi US and Mont MA: Current epidemiology of revision total
knee arthroplasty in the United States. J Arthroplasty.
32:2663–2668. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Goodman SB: Wear particles, periprosthetic
osteolysis and the immune system. Biomaterials. 28:5044–5048. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng K, Bai J, Li N, Li M, Sun H, Zhang
W, Ge G, Liang X, Tao H, Xue Y, et al: Protective effects of
sirtuin 3 on titanium particle-induced osteogenic inhibition by
regulating the NLRP3 inflammasome via the GSK-3β/β-catenin
signalling pathway. Bioact Mater. 6:3343–3357. 2021.PubMed/NCBI
|
|
11
|
Agarwal S: Osteolysis-basic science,
incidence and diagnosis. Curr Orthop. 18:220–231. 2004. View Article : Google Scholar
|
|
12
|
Dattani R: Femoral osteolysis following
total hip replacement. Postgrad Med J. 83:312–316. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mouhyi J, Dohan Ehrenfest DM and
Albrektsson T: The peri-implantitis: Implant surfaces,
microstructure, and physicochemical aspects. Clin Implant Dent
Relat Res. 14:170–183. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Derks J and Tomasi C: Peri-implant health
and disease. A systematic review of current epidemiology. J Clin
Periodontol. 42 (Suppl 16):S158–S171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bauer TW: Particles and periimplant bone
resorption. Clin Orthop Relat Res. 138–143. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Voggenreiter G, Leiting S, Brauer H,
Leiting P, Majetschak M, Bardenheuer M and Obertacke U:
Immuno-inflammatory tissue reaction to stainless-steel and titanium
plates used for internal fixation of long bones. Biomaterials.
24:247–254. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kotsakis GA and Olmedo DG:
Peri-implantitis is not periodontitis: Scientific discoveries shed
light on microbiome-biomaterial interactions that may determine
disease phenotype. Periodontol. 2000.86:231–240. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Magone K, Luckenbill D and Goswami T:
Metal ions as inflammatory initiators of osteolysis. Arch Orthop
Trauma Surg. 135:683–695. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Man K, Jiang LH, Foster R and Yang XB:
Immunological responses to total hip arthroplasty. J Funct
Biomater. 8:332017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guglielmotti MB, Olmedo DG and Cabrini RL:
Research on implants and osseointegration. Periodontol.
2000.79:178–189. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mombelli A, Hashim D and Cionca N: What is
the impact of titanium particles and biocorrosion on implant
survival and complications? A critical review. Clin Oral Implants
Res. 29 (Suppl 18):S37–S53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xiong L, Liu Y, Zhu F, Lin J, Wen D, Wang
Z, Bai J, Ge G, Xu C, Gu Y, et al: Acetyl-11-keto-β-boswellic acid
attenuates titanium particle-induced osteogenic inhibition via
activation of the GSK-3β/β-catenin signaling pathway. Theranostics.
9:7140–7155. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shah R, Penmetsa DSL, Thomas R and Mehta
DS: Titanium corrosion: Implications for dental implants. Eur J
Prosthodont Restor Dent. 24:171–180. 2016.PubMed/NCBI
|
|
24
|
Urban RM, Jacobs JJ, Tomlinson MJ,
Gavrilovic J, Black J and Peoc'h M: Dissemination of wear particles
to the liver, spleen, and abdominal lymph nodes of patients with
hip or knee replacement. J Bone Joint Surg Am. 82:457–476. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Choi MG, Koh HS, Kluess D, O'Connor D,
Mathur A, Truskey GA, Rubin J, Zhou DX and Sung KL: Effects of
titanium particle size on osteoblast functions in vitro and in
vivo. Proc Natl Acad Sci USA. 102:4578–4583. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fritz EA, Jacobs JJ, Glant TT and Roebuck
KA: Chemokine IL-8 induction by particulate wear debris in
osteoblasts is mediated by NF-kappaB. J Orthop Res. 23:1249–1257.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen D, Li Y, Guo F, Lu Z, Hei C, Li P and
Jin Q: Protective effect of p38 MAPK inhibitor on wear
debris-induced inflammatory osteolysis through downregulating
RANK/RANKL in a mouse model. Genet Mol Res. 14:40–52. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Geng D, Wu J, Shao H, Zhu S, Wang Y, Zhang
W, Ping Z, Hu X, Zhu X, Xu Y and Yang H: Pharmaceutical inhibition
of glycogen synthetase kinase 3 beta suppresses wear debris-induced
osteolysis. Biomaterials. 69:12–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gu Y, Wang Z, Shi J, Wang L, Hou Z, Guo X,
Tao Y, Wu X, Zhou W, Liu Y, et al: Titanium particle-induced
osteogenic inhibition and bone destruction are mediated by the
GSK-3β/β-catenin signal pathway. Cell Death Dis. 8:e28782017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang L, Bai J, Wang Q, Ge G, Lin J, Xu N,
Xu C, Xu Y, Wang Y and Geng D: Inhibition of protein phosphatase 2A
attenuates titanium-particle induced suppression of bone formation.
Int J Biol Macromol. 142:142–151. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu Z, Xie Q, Huang Y, Zhang S and Chen Y:
Aucubin suppresses titanium particles-mediated apoptosis of
MC3T3-E1 cells and facilitates osteogenesis by affecting the
BMP2/Smads/RunX2 signaling pathway. Mol Med Rep. 18:2561–2570.
2018.PubMed/NCBI
|
|
32
|
Wang J, Tao Y, Ping Z, Zhang W, Hu X, Wang
Y, Wang L, Shi J, Wu X, Yang H, et al: Icariin attenuates
titanium-particle inhibition of bone formation by activating the
Wnt/β-catenin signaling pathway in vivo and in vitro. Sci Rep.
6:238272016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Geng T, Sun S, Chen X, Wang B, Guo H,
Zhang S and Jin Q: Strontium ranelate reduces the progression of
titanium particle-induced osteolysis by increasing the ratio of
osteoprotegerin to receptor activator of nuclear factor-κB ligand
in vivo. Mol Med Rep. 17:3829–3836. 2018.PubMed/NCBI
|
|
34
|
Batista PJ: The RNA modification
N6-methyladenosine and its implications in human
disease. Genomics Proteomics Bioinformatics. 15:154–163. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schwartz S, Mumbach MR, Jovanovic M, Wang
T, Maciag K, Bushkin GG, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5′ sites. Cell
Rep. 8:284–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang P, Doxtader KA and Nam Y: Structural
basis for cooperative function of Mettl3 and Mettl14
methyltransferases. Mol Cell. 63:306–317. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zheng G, Dahl JA, Niu Y, Fedorcsak P,
Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al: ALKBH5
is a mammalian RNA demethylase that impacts RNA metabolism and
mouse fertility. Mol Cell. 49:18–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shi H, Wei J and He C: Where, when, and
how: Context-dependent functions of RNA methylation writers,
readers, and erasers. Mol Cell. 74:640–650. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xu K, Yang Y, Feng GH, Sun BF, Chen JQ, Li
YF, Chen YS, Zhang XX, Wang CX, Jiang LY, et al: Mettl3-mediated
m6A regulates spermatogonial differentiation and meiosis
initiation. Cell Res. 27:1100–1114. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Lin Z, Hsu PJ, Xing X, Fang J, Lu Z, Zou
Q, Zhang KJ, Zhang X, Zhou Y, Zhang T, et al:
Mettl3-/Mettl14-mediated mRNA N6-methyladenosine
modulates murine spermatogenesis. Cell Res. 27:1216–1230. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu Y, Xie L, Wang M, Xiong Q, Guo Y, Liang
Y, Li J, Sheng R, Deng P, Wang Y, et al: Mettl3-mediated
m6A RNA methylation regulates the fate of bone marrow
mesenchymal stem cells and osteoporosis. Nat Commun. 9:47722018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tian C, Huang Y, Li Q, Feng Z and Xu Q:
Mettl3 regulates osteogenic differentiation and alternative
splicing of vegfa in bone marrow mesenchymal stem cells. Int J Mol
Sci. 20:5512019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang Y, Gu X, Li D, Cai L and Xu Q:
METTL3 regulates osteoblast differentiation and inflammatory
response via smad signaling and MAPK signaling. Int J Mol Sci.
21:1992019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Song H, Song J, Cheng M, Zheng M, Wang T,
Tian S, Flavell RA, Zhu S, Li HB, Ding C, et al: METTL3-mediated
m6A RNA methylation promotes the anti-tumour immunity of
natural killer cells. Nat Commun. 12:55222021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang M, Xu S, Liu L, Zhang M, Guo J, Yuan
Y, Xu J, Chen X and Zou J: m6A methylation regulates osteoblastic
differentiation and bone remodeling. Front Cell Dev Biol.
9:7833222021. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nachbur U, Stafford CA, Bankovacki A, Zhan
Y, Lindqvist LM, Fiil BK, Khakham Y, Ko HJ, Sandow JJ, Falk H, et
al: A RIPK2 inhibitor delays NOD signalling events yet prevents
inflammatory cytokine production. Nat Commun. 6:64422015.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Tan X, Wei LJ, Fan GJ, Jiang YN and Yu XP:
Effector responses of bovine blood neutrophils against Escherichia
coli: Role of NOD1/NF-κB signalling pathway. Vet Immunol
Immunopathol. 168:68–76. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nguyen TTT, Shang E, Shu C, Kim S, Mela A,
Humala N, Mahajan A, Yang HW, Akman HO, Quinzii CM, et al: Aurora
kinase A inhibition reverses the Warburg effect and elicits unique
metabolic vulnerabilities in glioblastoma. Nat Commun. 12:52032021.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li D, Yang J, Malik V, Huang Y, Huang X,
Zhou H and Wang J: An RNAi screen of RNA helicases identifies
eIF4A3 as a regulator of embryonic stem cell identity. Nucleic
Acids Res. 50:12462–12479. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ratnadiwakara M and Änkö ML: mRNA
Stability assay using transcription inhibition by actinomycin D in
mouse pluripotent stem cells. Bio Protoc. 8:e30722018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou Z, Cao Y, Yang Y, Wang S and Chen F:
METTL3-mediated m6A modification of lnc KCNQ1OT1
promotes doxorubicin resistance in breast cancer by regulating
miR-103a-3p/MDR1 axis. Epigenetics. 18:22170332023. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Luo S, Liao C, Zhang L, Ling C, Zhang X,
Xie P, Su G, Chen Z, Zhang L, Lai T and Tang J: METTL3-mediated m6A
mRNA methylation regulates neutrophil activation through targeting
TLR4 signaling. Cell Rep. 42:1122592023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kim JM, Lin C, Stavre Z, Greenblatt MB and
Shim JH: Osteoblast-osteoclast communication and bone homeostasis.
Cells. 9:20732020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Redlich K and Smolen JS: Inflammatory bone
loss: Pathogenesis and therapeutic intervention. Nat Rev Drug
Discov. 11:234–250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Li Y, Ling J and Jiang Q: Inflammasomes in
alveolar bone loss. Front Immunol. 12:6910132021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiang Y, Jia T, Gong W, Wooley PH and Yang
SY: Titanium particle-challenged osteoblasts promote
osteoclastogenesis and osteolysis in a murine model of
periprosthestic osteolysis. Acta Biomater. 9:7564–7572. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Luo J, Xu T and Sun K: N6-methyladenosine
RNA modification in inflammation: Roles, mechanisms, and
applications. Front Cell Dev Biol. 9:6707112021. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Wu M, Chen G and Li YP: TGF-β and BMP
signaling in osteoblast, skeletal development, and bone formation,
homeostasis and disease. Bone Res. 4:160092016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Afzal F, Pratap J, Ito K, Ito Y, Stein JL,
van Wijnen AJ, Stein GS, Lian JB and Javed A: Smad function and
intranuclear targeting share a Runx2 motif required for osteogenic
lineage induction and BMP2 responsive transcription. J Cell
Physiol. 204:63–72. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yan X, Liu Z and Chen Y: Regulation of
TGF-beta signaling by Smad7. Acta Biochim Biophys Sin (Shanghai).
41:263–272. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Deng Z, Zhang R, Li M, Wang S, Fu G, Jin
J, Wang Z, Ma Y and Zheng Q: STAT3/IL-6 dependent induction of
inflammatory response in osteoblast and osteoclast formation in
nanoscale wear particle-induced aseptic prosthesis loosening.
Biomater Sci. 9:1291–1300. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Cai Y, Yu R, Kong Y, Feng Z and Xu Q:
METTL3 regulates LPS-induced inflammatory response via the NOD1
signaling pathway. Cell Signal. 93:1102832022. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Caruso R, Warner N, Inohara N and Núñez G:
NOD1 and NOD2: Signaling, host defense, and inflammatory disease.
Immunity. 41:898–908. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Pei G and Dorhoi A: NOD-like receptors:
Guards of cellular homeostasis perturbation during infection. Int J
Mol Sci. 22:67142021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kersse K, Bertrand MJ, Lamkanfi M and
Vandenabeele P: NOD-like receptors and the innate immune system:
Coping with danger, damage and death. Cytokine Growth Factor Rev.
22:257–276. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yang Y, Hsu PJ, Chen YS and Yang YG:
Dynamic transcriptomic m6A decoration: Writers, erasers,
readers and functions in RNA metabolism. Cell Res. 28:616–624.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Jiang X, Liu B, Nie Z, Duan L, Xiong Q,
Jin Z, Yang C and Chen Y: The role of m6A modification in the
biological functions and diseases. Signal Transduct Target Ther.
6:742021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M,
Ma J and Wu L: YTHDF2 destabilizes m(6)A-containing RNA through
direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun.
7:126262016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR
and Qian SB: Dynamic m(6)A mRNA methylation directs translational
control of heat shock response. Nature. 526:591–594. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Winkler R, Gillis E, Lasman L, Safra M,
Geula S, Soyris C, Nachshon A, Tai-Schmiedel J, Friedman N,
Le-Trilling VTK, et al: m6A modification controls the
innate immune response to infection by targeting type I
interferons. Nat Immunol. 20:173–182. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Mapperley C, van de Lagemaat LN, Lawson H,
Tavosanis A, Paris J, Campos J, Wotherspoon D, Durko J, Sarapuu A,
Choe J, et al: The mRNA m6A reader YTHDF2 suppresses
proinflammatory pathways and sustains hematopoietic stem cell
function. J Exp Med. 218:e202008292021. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Fang C, He M, Li D and Xu Q: YTHDF2
mediates LPS-induced osteoclastogenesis and inflammatory response
via the NF-κB and MAPK signaling pathways. Cell Signal.
85:1100602021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Tsuchiya K, Yoshimura K, Inoue Y, Iwashita
Y, Yamada H, Kawase A, Watanabe T, Tanahashi M, Ogawa H, Funai K,
et al: YTHDF1 and YTHDF2 are associated with better patient
survival and an inflamed tumor-immune microenvironment in
non-small-cell lung cancer. Oncoimmunology. 10:19626562021.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wu R, Liu Y, Zhao Y, Bi Z, Yao Y, Liu Q,
Wang F, Wang Y and Wang X: m6A methylation controls
pluripotency of porcine induced pluripotent stem cells by targeting
SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner.
Cell Death Dis. 10:1712019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y,
Qi M, Lu Z, Shi H, Wang J, et al: Ythdc2 is an
N6-methyladenosine binding protein that regulates
mammalian spermatogenesis. Cell Res. 27:1115–1127. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Keegan GM, Learmonth ID and Case CP:
Orthopaedic metals and their potential toxicity in the arthroplasty
patient: A review of current knowledge and future strategies. J
Bone Joint Surg Br. 89:567–573. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Gornet MF, Singh V, Schranck FW, Skipor AK
and Jacobs JJ: Serum metal concentrations in patients with titanium
ceramic composite cervical disc replacements. Spine (Phila Pa
1976). 42:366–371. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Day JS, Baxter RM, Ramsey ML, Morrey BF,
Connor PM, Kurtz SM and Steinbeck MJ: Characterization of wear
debris in total elbow arthroplasty. J Shoulder Elbow Surg.
22:924–931. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Chassot E, Irigaray JL, Terver S and
Vanneuville G: Contamination by metallic elements released from
joint prostheses. Med Eng Phys. 26:193–199. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Lukina E, Laka A, Kollerov M, Sampiev M,
Mason P, Wagstaff P, Noordeen H, Yoon WW and Blunn G: Metal
concentrations in the blood and tissues after implantation of
titanium growth guidance sliding instrumentation. Spine J.
16:380–388. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Safioti LM, Kotsakis GA, Pozhitkov AE,
Chung WO and Daubert DM: Increased levels of dissolved titanium are
associated with peri-implantitis-a cross-sectional study. J
Periodontol. 88:436–442. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol
Oncol. 13:1172020. View Article : Google Scholar : PubMed/NCBI
|