Introduction
The mammalian signal transducer and activator of
transcription (STAT) family of proteins comprises seven members,
namely STAT1, STAT2, STAT3, STAT4, STAT5A, STAT5B and STAT6, which
function to initiate the transcription of genes, hence playing a
role in advance of all cytokine-driven signaling present in the
extracellular environment (1). The
coding genes for STAT family proteins are mapped in three
chromosomal clusters: Chromosomes 2, 12 and 17 (2); the molecular size ranges from 748
amino acids for STAT4 to 851 amino acids for STAT2 (https://www.uniprot.org/uniprotkb?query), as
summarized in Table I.
 | Table I.Chromosomal mapping and length in
amino acid residue numbers of STATs. |
Table I.
Chromosomal mapping and length in
amino acid residue numbers of STATs.
| Member of STAT
family | Chromosomal
localization of coding genes | Molecular length
(amino acids) |
|---|
| STAT1 | Chromosome 2 | 750 |
| STAT4 | Chromosome 2 | 748 |
| STAT2 | Chromosome 12 | 851 |
| STAT6 | Chromosome 12 | 847 |
| STAT3 | Chromosome 17 | 770 |
| STAT5A | Chromosome 17 | 794 |
| STAT5B | Chromosome 17 | 787 |
The molecules share the conserved domains, exerting
the activities of transducing intracellular signaling and
initiating transcription, as suggested by their names. From amino
(N)-terminus to carboxy (C)-terminus, the domains on the linear
structure include, the NH2-terminal domain of STAT provides
protein-protein interaction sites and is required for the
interactions of dimer-dimer to form tetramers or oligomers, a
coiled-coil domain, a DNA-binding domain, and the Src homology 2
(SH2) domain. The SH2 domain is required for the recruitment of
STATs to phosphorylated receptors. The tyrosine residues modified
by phosphorylation are Tyr 701 for STAT1, Tyr 690 for STAT2, Tyr
705 for STAT3, Tyr 693 for STAT4, Tyr 694 for STAT5 and Tyr 641 for
STAT6, required for SH-phosphotyrosine interaction. The
transactivation-domain on C-terminus is essential for co-factor
interactions (3). The scheme of
linear cluster of motifs in STAT proteins is depicted in Table II. The molecules of STATs are
recruited by activated receptors of different cytokines in forms of
homo- and heterodimers (4).
 | Table II.Motif composition of STAT family
members. |
Table II.
Motif composition of STAT family
members.
|
| Motif |
|---|
|
|
|
|---|
| STAT | Protein interact
domain | All-alpha
domain | Tetramerization
domain of TRPM | DNA binding
domain | Linker | SH2 domain | TAZ2 binding
domain |
|---|
| STAT1 | 2-119 | 144-308 | 266-286 | 322-458 | 481-558 | 578-622 | 715-738 |
| STAT2 | 2-122 | 146-308 | - | 321-456 | 482-557 | 578-648 | 783-838 |
| STAT3 | 2-119 | 145-313 | - | 326-464 | 488-565 | 584-651 | - |
| STAT4 | 2-119 | 144-308 | - | 321-454 | 478-554 | 573-627 | - |
| STAT5a | 3-123 | 146-324 | - | 336-469 | 492-574 | 595-669 | - |
| STAT5b | 2-123 | 146-324 | - | 336-469 | 492-574 | 595-668 | - |
| STAT6 | 2-113 | 182-242 | - | 277-413 | 436-518 | 538-616 |
655-847a |
STATs are transcription factors that latently exist
in the cytoplasm; they are activated upon phosphorylation by Janus
kinase (JAK). JAK is a family of intracellular tyrosine kinases,
comprising JAK1-3 and tyrosine kinase 2 (TYK2) that non-covalently
interact with the domain on membrane receptor or receptors of
growth factor extending to the intracellular portion; they are
triggered to activate when different cytokines bind their
receptors. JAKs catalyze phosphorylation of tyrosine residue to
transduce signals of a wide range of surface receptors on membrane.
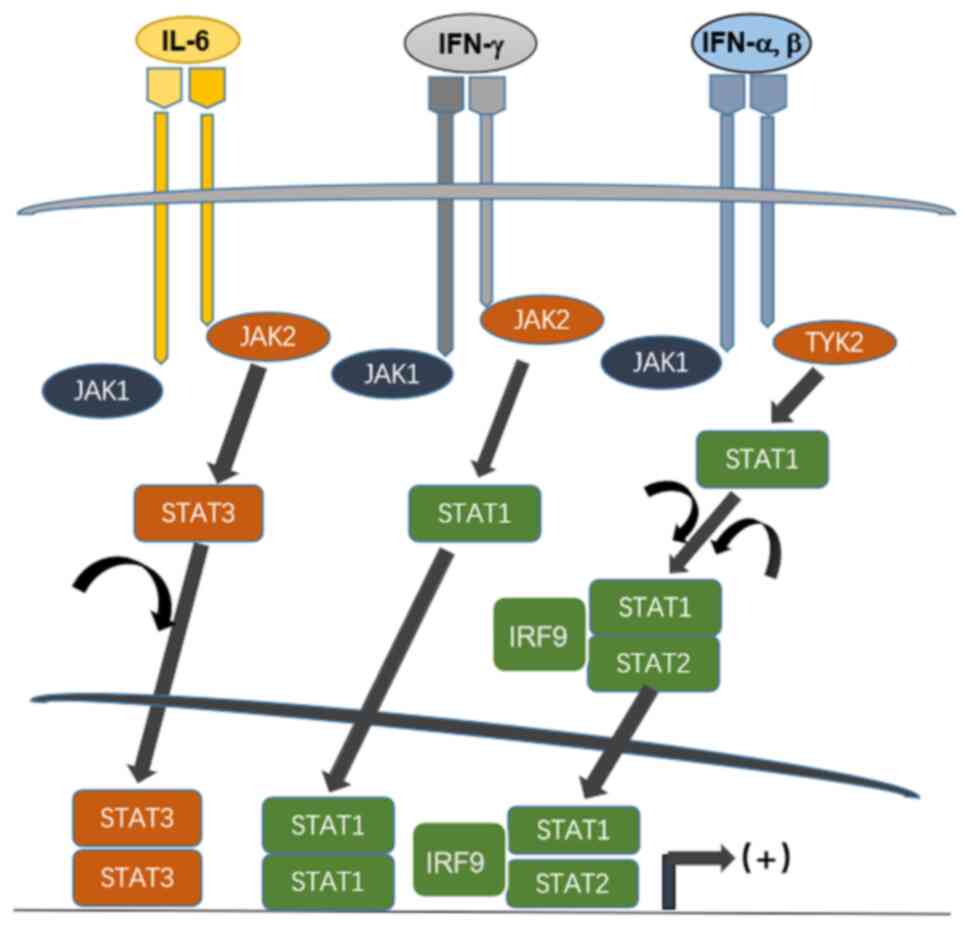
Upon phosphorylation, individual STAT molecules are dimerized and
translocate to the nucleus, initiating transcription (Fig. 1). The important role of STAT in the
regulation of immune response and other biological events has been
revealed in gene-targeting studies (5,6).
Three major mechanisms have been revealed to
negatively regulate STAT signaling: i) The JAK and STAT
dephosphorylation catalyzed by various tyrosine protein
phosphatases, such T-cell specific 45 (7–9); ii)
JAK inactivation by suppressor of cytokine signaling (SOCS) family
proteins; and iii) interaction with protein inhibitor of activated
STAT (PIAS), which has been identified as a negative regulator of
STAT signaling. Previous studies have shown that PIAS proteins act
on small ubiquitin, as modifier small ubiquitin-like modifier E3
ligase. In addition, biochemical studies have suggested that the
list of proteins either positively or negatively regulated by the
PIAS family through multiple mechanisms has rapidly expanded to
>60, with the majority being transcription factors (10,11).
Upon phosphorylation, STATs are homo- or
heterodimerized, translocate to the nucleus and coordinate with
other coactivators of transcription or transcription factors,
contributing to an increased initiation of transcription (Fig. 1). In cultured cells and
experimental animals, the ligand-dependent activation of STATs has
been observed as a transient process, lasting for minutes or hours.
Molecules of STATs, and STAT1, 3 and 5 in particular, are
persistently tyrosine-phosphorylated or tyrosine-activated. A
number of experiments have revealed the importance of STAT
activation for the control of growth. Data obtained strongly
suggest its role in controlling cell cycle progression and
initiation and occurrence of apoptosis. The JAK/STAT signaling
pathway controls processes at the cellular level that is of essence
in homeostasis. Alterations of this axis contribute to the
progression of cancer, inflammatory and autoimmune diseases
(11,12).
Evidence suggests the role of STAT family proteins
in inducing and maintaining a pro-carcinogenic inflammatory
microenvironment, for the initiation of malignant transformation
and during cancer progression (13). The importance of inflammation in
tumor initiation and malignant progression has been primarily
subject to investigation. Inflammatory conditions can initiate or
promote oncogenic transformation, genetic and epigenetic changes in
malignant cells can also generate an inflammatory microenvironment
that further supports tumor progression. In addition to
tumor-promoting role of inflammation, the importance of immune
responses and inflammatory mediators have been underscored in
suppressing tumorigenesis and tumor growth (14–16).
STAT3 and, to some extent, STAT5 and STAT6 are involved in
inhibiting antitumor immunity. In view of the effector molecules of
cell cycle and apoptosis regulated by JAK-STAT, it suggests that
the pathway also contributes to carcinogenesis through
proliferation promotion and programmed cell death potentiation
(13).
Modulation of the antiviral immunity mediated by
JAK/STAT pathway contributes to the signaling of interferons,
notably type I. The JAK-STAT pathway is also implicated in the
pathogenesis by two tumorigenic human herpesviruses, Epstein-Barr
virus (EBV) and Kaposi sarcoma-associated herpesvirus (KSHV).
The gamma group of human herpesviruses contains two
lymphotropic members, EBV and KSHV. In their genome, genes are
located next to the terminal repeat region code for membrane
proteins to transduce cellular signals. The coding products termed
terminal membrane proteins (TMPs) interact with the cellular
proteins involved in the activities of non-receptor protein
tyrosine kinases and tumor necrosis factor receptor-associated
factors. It has been demonstrated that persistent STAT activation
may be implicated in EBV-driven tumorigenesis in immunocompetent
individuals (17), as observed in
nasopharyngeal carcinoma, tightly associated with EBV infection and
endemic among certain regions in the world, including southern
China and Southeast Asia.
When the cytokine receptors are activated, JAKs
catalyze phosphorylation of tyrosine residue to transduce signals
of a wide range of surface receptors on membrane. As
aforementioned, the activity of STATs is modulated by a group of
inhibitor PIAS. PIAS1 and PIAS3 block the effects of STAT1 and
STAT3 respectively; it has been reported that the entry of lytic
cycle is prevented if STAT3 is downregulated by PIAS3. The high
expression of STAT3 has been revealed to facilitate the entry of
lytic cycle of EBV (Fig. 2A)
(18).
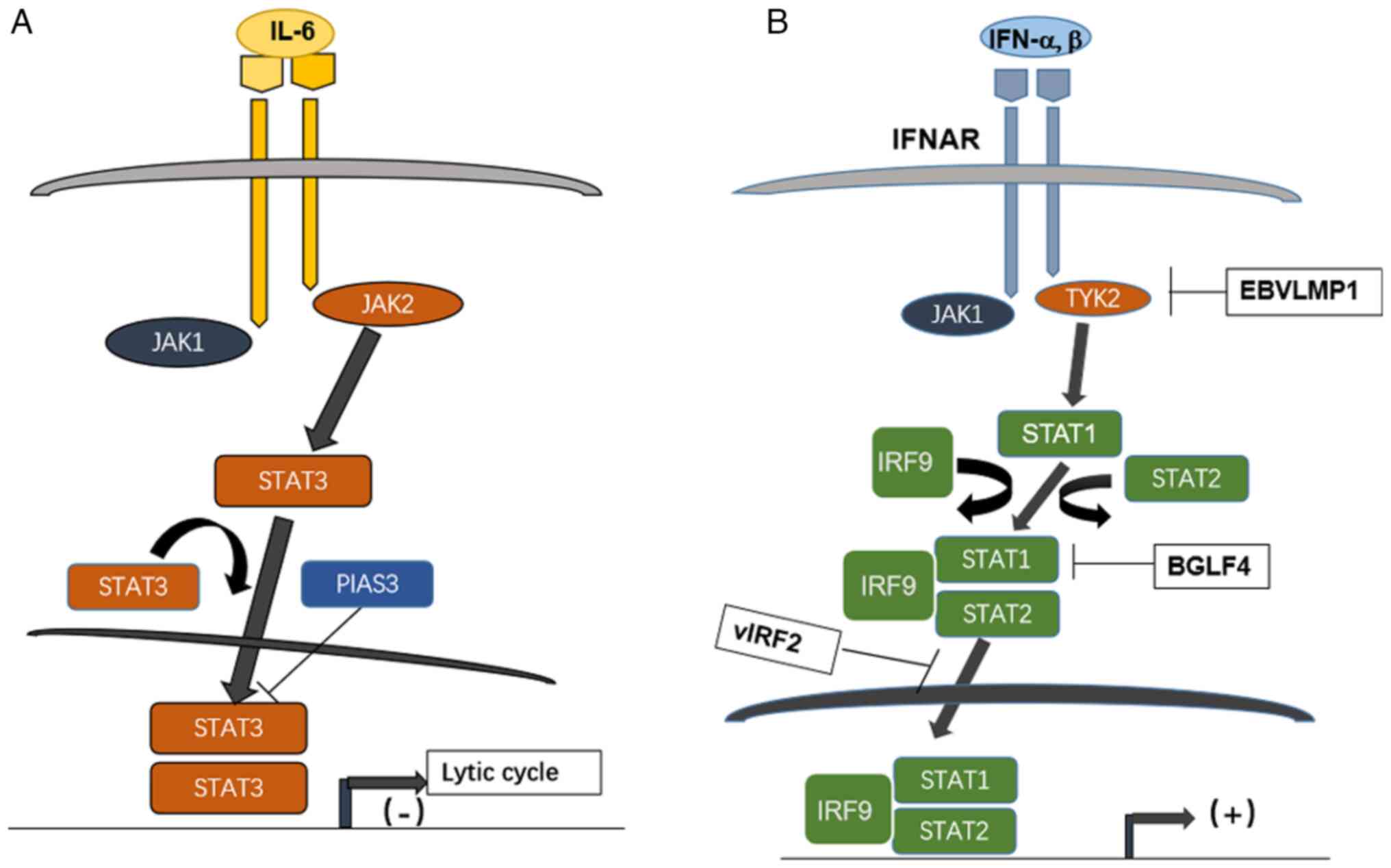 | Figure 2.Effect of STATs pathway interacting
with herpesviruses EBV and KSHV during lytic cycle and interferon
signaling. (A) High level of STAT3 activated by IL-6 on binding
with its cognate receptor regulates EBV entry to lytic cycle; it is
inhibited by the protein PIAS3. (B) The JAK/STAT pathway downstream
of type I interferon counteracted by herpesviruses EBV and KSHV
encoded products; IFNAR binds IFN-alpha and beta, then recruited
JAK1 and TYK2, the latter is inhibited by a EBV latent protein,
LMP1, while the heterotrimer STAT1-STAT2-IRF9 formed after
activation of JAK1/TYK2 is inhibited by EBV kinase BGLF4 and by
KSHV encoded vIRF2. STAT, signal transducer and activator of
transcription; EBV, Epstein Barr virus; KSHV, Kaposi
sarcoma-associated herpesvirus; PIAS3, protein inhibitor of
activated STAT3; JAK, Janus kinase; IFNAR, interferon alpha/beta
receptor; TYK, tyrosine kinase; vIRF, viral interferon regulatory
factor. |
An EBV encoded serine/threonine-protein kinase BGLF4
has been shown to enhance the production of extracellular viral
particles during EBV lytic replication (19,20).
BGLF4 effectively suppresses the activities of the
polyinosinic:polycytidylic acid poly (I:C)-stimulated IFN-beta
promoter and responsive element of interferon regulatory factor
(IRF) 3. Moreover, BGLF4 represses expression of endogenous
IFN-beta mRNA stimulated by the poly (I:C) and the phosphorylation
of STAT1 at Tyr701 (21). The
genetic coding product, viral IRF2 of KSHV/HHV-8 inhibits signaling
of interferon as reported (22).
The knowledge of Herpesviral interfering of JAK/STAT is summarized
in Fig. 2B with the employment of
the pathway mapping tool from the Kyoto Encyclopedia of Genes and
Genomes database (https://www.genome.jp/entry/K11220; map05167 KSHV
infection; map05169 EBV infection).
Several non-receptor tyrosine kinases, such as SH2
and the protooncogene homologous to retroviral ABL
transforming gene, forming a malignancy-related cytogenetic
aberration known as the Ph chromosome (19) have been found to phosphorylate
STATs. It has been revealed that sorafenib, a multikinase
inhibitor, inhibits cell proliferation and triggers apoptosis at
much lower concentrations in cells in which the fused protein
BCR/ABL is expressed (23). In
Ph+ leukemia cells, apoptosis is induced by sorafenib as
evidenced by that caspase-3 have been revealed and drops of
mitochondrial membrane potential have been specifically identified
in cells harboring BCR/ABL.
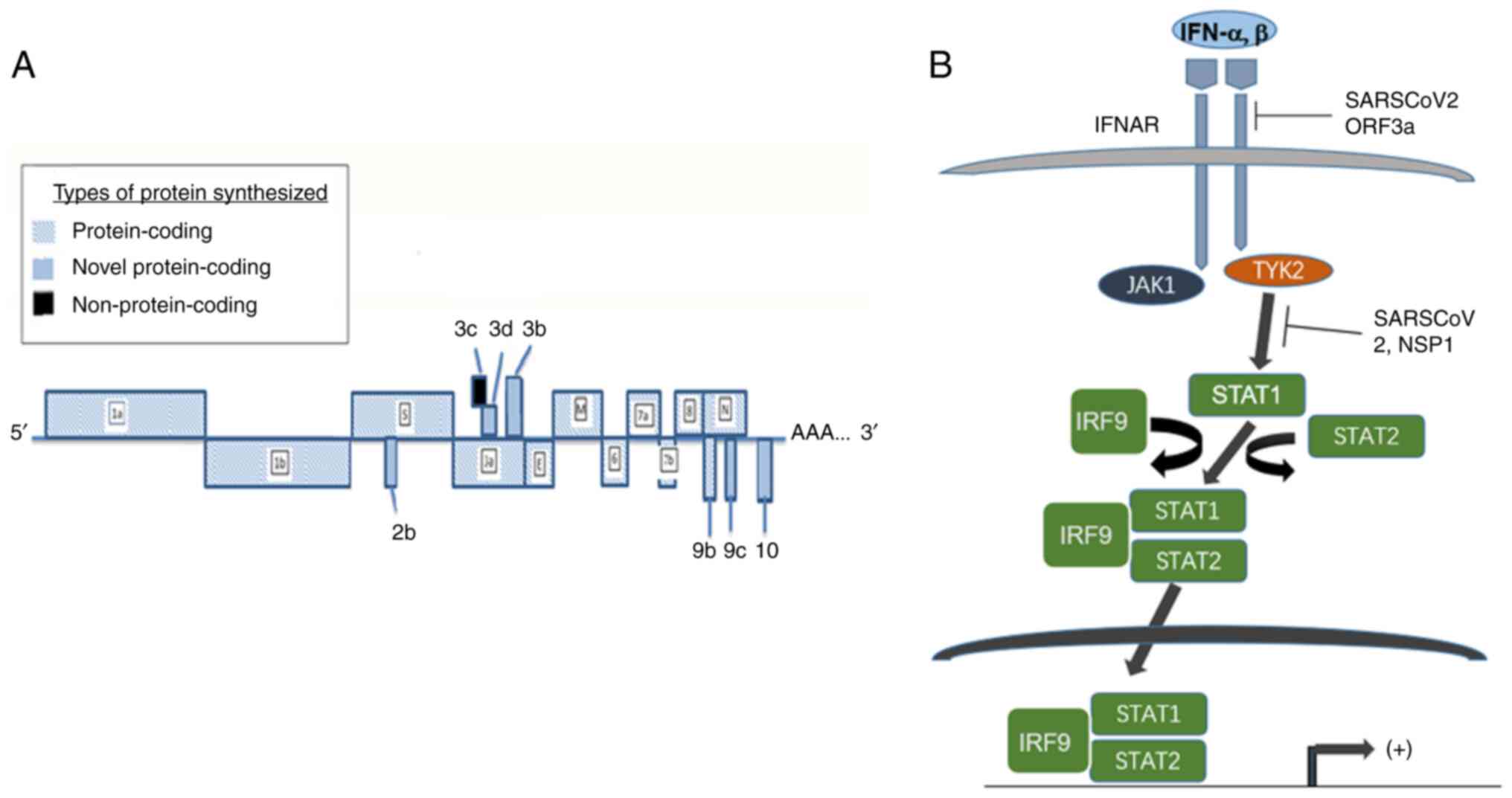
JAK/STAT pathway analysis has revealed that elements
of proximal signaling such as IFN receptor 1, JAK1 and TYK2, are
disrupted by SARS-CoV-2, leading to the inhibition of IFN-induced
STAT phosphorylation. The mechanisms underlying STAT inhibition
have been explained to uncover an immune evasion strategy against
SARS-CoV-2 and the pathway involved could be targeted by
anti-coronavirus therapy (24).
Coronavirus disease of 2019 (COVID-19) caused a
severe pandemic in the next three years. The clinical entity of
COVID-19 has been intensely studied (25), and it has been found that the
infection of lower respiratory tract and multiple internal organs
is characterized by cytokine release syndrome (CRS) with an
increased production of interleukins (ILs), such as IL-6, IL-2,
IL-7 and IL-10. Some of the cytokines involved in the pathogenesis
use a distinct intracellular JAK-mediated signaling pathway. The
inhibition of JAK, therefore, presents a marked therapeutic
potential for CRS, which is known as a common cause of adverse
clinical outcomes for COVID-19 (26,27).
A unique panel of cell culture models profiling
proteomic responses to SARS-CoV-2 infection has been established
and described using lung, liver, intestine, kidney, heart and brain
cells. The system enables the identification of proteins and
pathways in the cells that are widely targeted by SARS-CoV-2. Among
the pathways traced, the most notable was the JAK-STAT signaling
cascade, the key component in the interferon (IFN) response
pathway. The inhibition of STAT phosphorylation combined with its
nuclear translocation has been demonstrated in cells infected with
SARS-CoV-2 (28–31).
JAK/STAT pathway analysis has revealed that elements
of proximal signaling such as IFN receptor 1, JAK1 and TYK2, are
disrupted by SARS-CoV-2, leading to the inhibition of IFN-induced
STAT phosphorylation.
The JAK/STAT pathway has been implicated in
carcinogenesis and viral pathogenesis, notably in pulmonary
involvement from COVID-19. Progress in this field will be reviewed
in the present study.
Janus kinase/signal transducer and activator
of transcription (JAK/STAT) pathway is implicated in carcinogenesis
and malignancy progression
Malignancy is achieved by the functional alteration
of two types of cancer-related genes, that is, oncogenes and tumor
suppressor genes (TSGs). They acquire oncogenic potential in two
ways; gain-of-function for oncogenes and loss-of-function for
TSGs.
Clinically, patients with colorectal carcinoma or
other cancers get a prolonged and improved antitumor immunity from
immunotherapy when STAT1 is expressed in the nucleus (32). When JAK1 is dissociated from the
receptor of IFN-γ, the phosphorylation of STAT1 inhibition follows;
concomitantly, STAT1 is translocated to the nucleus to promote
tumor growth through the inhibition of apoptosis (33).
Small RNAs, such as microRNAs, which bind to the
untranslated region of mRNA, may directly or indirectly impact the
expression of STAT1. It has been revealed that STAT1 overexpression
significantly repressed the expression of miR-181a (34).
STAT1 is phosphorylated at the Tyr 701 residue by
activated JAK. Upon phosphorylation, STAT1 is then homodimerized
and translocates to the nucleus, where it binds to specific gamma
activated sequences (GAS), to stimulate the expression of
IFN-stimulated genes (ISGs) upon the initiation of transcription
(35–37). In some types of cells, STAT1 is
also phosphorylated at residue Ser727, resulting in an enhancement
of its transcriptional activity (38). During the induction of
transcription factor IFN regulatory factor 1 (IRF1) expression is
triggered by IFN-γ, IRF1 interacts with STAT1 and recruits the
STAT1-IRF1 complex to the elements of GAS (32,33,39,40).
Herpesvirus and coronavirus infections induce IFN-γ, which
regulates the activation of the JAK/STAT pathway. It has been
demonstrated that, in ovarian cancer (OC) cells, IFN-γ-induced
PD-L1 expression is dependent on JAK1, STAT1 and IRF1 signaling.
Furthermore, IFN-γ increases acetylation of the human PD-L1
promoter in OC and other cancer cells, and STAT1, Ser727-p-STAT1
and IRF1 are recruited to the promoter.
In OC, the elevated PD-L1 expression level is
associated with poor prognosis (31). It has been further demonstrated
that the levels of Tyr 701-p, STAT1 and Ser 727-p-STAT1 were
increased in OC tissues (41). In
OC cells, IFN-γ was found to increase STAT1, Tyr 701-p-STAT1 and
Ser727-STAT1 levels when the expression of PD-L1 was increased
(42).
STAT3 and other members of the STAT family are known
to be oncogenic, and the JAK-STAT3 pathway is known for its
potential to promote tumor cell proliferation, survival, invasion,
angiogenesis and immune suppression. Previously, it has been
revealed that the signaling of JAK/STAT3 contributes to
inflammation-mediated carcinogenesis, maintenance of cancer stem
cell (CSCs) phenotypes and of pre-metastatic niches (35,43–49).
CSCs are a cancer cell subpopulation that manifests a phenotype of
stem cells; they have the ability of sustaining self-renewal,
resembling normal stem cells.
Upon hetero- or homodimerization, the STAT molecules
bind fragments with regulatory function upstream to coding portion
in genomic sequence and initiate the transcription of target genes.
JAK2 activation by trans- or autophosphorylation induces the
cascade of activating downstream molecules, including STATs.
Experimental findings have shown that both extrinsic
(environmental) and intrinsic pathways link cancer. The intrinsic
pathways originate from genetic and epigenetic events in the tumor
cells. STATs, notably STAT3, are crucial for both extrinsic and
intrinsic pathways of inflammation (50,51).
Due to its oncogenic potential, which promotes tumor cell
proliferation and survival, STAT3 drives the transition from
chronic inflammation to malignancy (52). In a variety of hematological
malignancies, STAT5 and STAT6 are persistently activated (52–55).
In cases of chronic myelogenous leukemia, STAT5 has been
demonstrated to be persistently activated in malignant cells in the
presence of the classic chromosomal translocation, BCR/ABL
(56).
The phosphorylated form of STAT5 enhances the
transcription of regulatory factors in apoptosis, such as Bcl-X,
and cell cycle progression, such as cyclin D1, D2, 7E, p21Cip and
other inhibitors of cyclin dependent kinase (CDK) (57,58).
Mutations within the pseudokinase domain of JAK2, from valine to
phenylalanine at residue 617 (JAK2 V617F) is a major activating
mutation for JAK2 signaling (59,60).
The mutation pattern is illustrated in Fig. 2. JAK2 V617F is a frequent mutation
in myelo-proliferation disorder with absence of BCR/ABL
juxtaposition caused by the cytogenetic aberration of t (9:22)
observed in the Ph chromosome, in which some kinase activity is
affected (61).
JAK2 engages the pathway to activate multiple STAT
family members in addition to STAT3. Hyperproliferation, which is
induced by JAK2 V617F involves the activation of STAT3 together
with the downstream factors it targets (62). It has been reported that the
inhibition of cyclin D2 transcription and enhancement of
p27kip1 account for the growth arrest caused by the
inhibition of JAK2 V617F (63).
The p27kip1 downregulation caused by the expression of
JAK2 V617F has been found to be associated with the STAT5-induced
expression of Skp2, suggesting that p27kip1 degradation
could result from the overexpression of the ubiquitin ligase Skp2
directed at p27kip1 (62). p27kip1 has hence been
identified as a substrate of JAK2 (64).
Mutational activation frequently contributes to
malignant proliferation through events such as histone marker
modification. In fact, STAT3 is classified as oncogenic (14). Its engagement directing to
hallmarks of cancer, such as the proliferation, apoptosis evasion
and angiogenesis of tumor cells, has been reported (65). The activities control the
expression of pro-tumorigenic genes, such as cyclins D1 and D2,
c-Myc, MCL1 apoptosis regulator, survivin/baculoviral IAP repeat
containing 5 (BIRC5), B-cell lymphoma-extra large, hepatocyte
growth factor, hypoxia induced factor-α (HIF-1α and vascular
endothelial growth factor A (VEGF) (66–70).
Alterations in STAT3 expression affects the host
proliferation, as well as the drug resistance of cancer cells. When
treated with erlotinib, p-STAT3 levels are increased in epidermal
growth factor receptor (EGFR)-mutated non-small cell lung cancer
(NSCLC) cell lines and drug resistance is induced. Conversely, the
RNA interference-mediated knockdown of STAT3 enhances the
sensitivity of cells to erlotinib (71). Of note, the findings could be
extended to NSCLC, whose tumorigenesis is driven by KRAS mutation.
When treated with mitogen-activated protein kinase inhibitor, NSCLC
lines harboring a KRAS mutation exhibit an increased level of
p-STAT3, and drug resistance is induced (72).
NSCLC lines carrying EGFR and KRAS mutations secrete
cytokines, such as IL-6, which contribute to the underlying
mechanisms of STAT3 activation in NSCLC-related gp130/JAK
signaling. Autocrine JAK signaling-dependent STAT3 activation is
followed by the engagement of gp130, thereby promoting tumor cell
survival (71–73). In this context, in vivo and
in vitro tumor cell growth is decreased by knocking down
IL-6 or JAK1/2 (67,71,73).
Among the signaling pathways of inflammation
associated with tumor development, STAT3 phosphorylation plays a
key role (74). STAT3, an
oncogenic transcription factor is usually constitutively activated
in prostate and several other human cancers, such as breast cancer
(75–77). Due to the important role it plays
in cell survival and proliferation (33), STAT3 has been designated as a major
target of anticancer therapy. In hepatocellular carcinoma cells,
STAT3 also exerts anti-apoptotic and pro-proliferation effects
(78). Indeed, in obesity
(79) and fatty liver-associated
inflammation (80), the IL-6/STAT3
activation is regarded as a ‘bona fide’ tumor promoter.
The central role of STAT3 is to promote and maintain
the stem cell phenotype. In this population, STAT3 inhibits tumor
progression. In stem cells, it has been noted that histone (H)
methylation at lysine residues catalyzed by enhancer of zester
homolog 2 (EZH2) regulates STAT3 (47). As the catalytic subunit of polycomb
repressive complex 2, EZH2 has been known to exhibit
oncoprotein-like activity to bi- or trimethylate a repressive mark
on H3, lysine 27 of H3, to repress the transcription of the target
gene (81). Negative cell cycle
regulators, such as CDK inhibitor 1A or p21Waf1/Cip1 are
downregulated. Repressive effects on these molecules lead to the
promotion of malignancy through its activity of lysine transferase
on histones; the target genes that are implicated in cell
proliferation, apoptosis and regulation of cell cycle progression
include p15 INK4b-ARF, p16 INK4A, tumor
necrosis factor related apoptosis inducing ligand and Kruppel-like
factor 2 (82).
The overexpression of EZH2 disrupts cell
proliferation, apoptosis, migration and invasion. High EZH2 level
has also been found to be associated with an unfavorable clinical
outcome in cancers (83–86). It has been revealed that EZH2
promotes malignancy in NSCLC and EZH2 overexpression and is
associated with poor prognosis in patients with NSCLC. Data have
suggested that EZH2-siRNA increases the expression of
p15INK4B p21Waf1, and p27Kip1 in
NSCLC lines. Univariate analysis revealed that NSCLC patients with
a high EZH2 expression have a significantly inferior overall,
cancer-specific and disease-free survival. It has been demonstrated
that overexpressed EZH2 is essential for NSCLC progression, and the
levels of EZH2 may serve as a prognostic predicator of the same
cancer (87).
EZH2 also methylates the non-histone protein STAT3
and is involved in the acetylation of STAT3 which is catalyzed by
acetyltransferase p300, thus playing a role in the regulation of
the formation of a transcription complex bound to the target gene
promoters.
The authors have previously demonstrated that a zinc
finger motif containing protein ZMYND10 encoded by a frequently
lost TSG found in a variety of human tumors induces the
trimethylation of a histone repressive mark (H3K9) and
downregulates cyclins that promote cell cycle entry (88). Forkhead box A1 (FOXA1) is a
transcription factor which plays a role in the translation of the
epigenetic signatures into enhancer-driven transcriptional program
characterized by cell type specificity; FOXA1 is differentially
recruited to chromatin fragments and downregulated in several
cancers. The positive association between FOXA1 and CDKN2A/p16
INK4a, a negative regulator of the cell cycle that can
be observed in prostate and breast cancers weakly expressing EZH2,
epigenetically represses CDKN2A. It has been analyzed and revealed
in prostate and breast cancer cells that high expression of FOXA1
antagonizes the EZH2-mediated repression of CDKN2A, as further
depletion of FOXA1 reverts the effect of CDKN2A de-silencing caused
by the inhibition of EZH2. Concomitantly, the depletion of EZH2
suppresses the cancer cell cycle progression, while the presence of
FOXA1 and CDKN2A optimizes this regulation (89). The modulation of
proliferation-related molecules by tumor suppressors through
histone and non-histone protein modification remains to be
elucidated.
High level of STAT3 contributes to
refractory state to lytic cycle switch in Epstein-Barr virus (EBV)
and Kaposi sarcoma-associated herpesvirus (KSHV)
Upon viral infection, IFNs are secreted and the
JAK/STAT pathway is activated, which leads to the promotion of the
transcription of ISGs to defend against viral infection (18). Type I IFNs bind to the
IFN-activated receptor IFNAR1, leading to JAK activation. The
activated JAK in turn phosphorylates STAT1 and STAT2, enabling them
to form heterodimers to bind with IRF9, forming the complex of ISG
factor 3. The complex is further linked to IFN-stimulated response
elements and induces the transcription of ISGs (90).
Herpesviruses are characterized by the adoption of
two distinctive patterns of infection in their life cycle: Latent
and lytic infection. To date, two gamma-herpesviruses among eight
known human herpesviruses, the lymphotropic human herpesvirus 4
(HHV-4) or EBV and HHV-8 or KSHV are classified as tumorigenic
viruses. These viruses may harbor in the body of the host
throughout their life. During their latency phase, the viruses
express limited genome products, without rupture of the host cells.
The mechanisms underlying the switch from a latent to a lytic cycle
in EBV and KSHV have been studied in B cell lines cultured in
vitro.
In EBV, the activation of the lytic cycle is
controlled by two virally encoded transcription factors with Zip
motif; Z EB replication activator (ZEBRA) and replication and
transcription activator (Rta). The viral bZIP transcription factor
ZEBRA/Zta encoded by BamHI Z left frame 1 (BZLF1) regulates this
cycle by binding to two classes of ZEBRA response elements. ZEBRA
is a homodimeric protein related to the activating protein 1 (AP-1)
family of bZIP transcription factors (91). It regulates the EBV infection cycle
by contributing both in establishing viral latency and triggering
lytic replication. During pre-latency, ZEBRA is transiently
expressed at this time course, the EBV genome is hypomethylated and
it is critical for promoting the proliferation of quiescent naive
and memory B cells. During latency, when the EBV genome is
methylated, the expression of ZEBRA activates a second viral
transcription factor, which functions along with ZEBRA to trigger
lytic replication (92,93).
The lytic cycle activator of KSHV is the open
reading frame 50 encoded Rta homolog (93). Following the switch to a lytic
cycle, they express all of the genomic products, ending with the
lysis of the host cells and establishing infection when entering
new cells (94). The lytic cycle
has been linked to the oncogenic potential of the viruses.
The molecular mechanisms underlying the
latent-to-lytic cycle switch with the ultimate aim of utilizing
anti-viral therapy by increasing the number of cancer cells
latently infected with the virus being converted to the lytic phase
and thereby becoming sensitive to the antiviral agents. In fact,
oncolytic therapy faces a major challenge due to the high cells
fractionation rates in populations of latently infected cells,
which are resistant to agents inducing the lytic cycle (95).
The lytic cycle of EBV is characterized by the
expression of a cascade of regulated genes; these include
intermediate-early, early and late genes. Strategies for inducing
lytic infection of EBV in tumor cells are investigated as a
potential therapy against EBV-related tumors; the latent-to-lytic
cycle switch of EBV in infected B and epithelial cells is regulated
by a panel of cellular and viral proteins and the manner in which
lytic viral reactivation is harnessed may be utilized in the
therapeutic approach. When the viral lytic cycle is triggered, a
number of EBV-encoded proteins are expressed, including ZTA/ZEBRA,
EB1, BZLF or Zta and Rta, encoded by immediate-early genes that
stimulate the expression of early and late genes of EBV. The early
genes code for proteins required for the replication of EBV DNA,
whereas the late genes code for viral structural proteins to
package infectious viral particles. In addition to BZLF1-encoded
transcription factors, BZLF1 transcription activator (Zta) and Rta,
the expression of lytic EBV genes is modulated by cellular proteins
(96,97). RNA transcribed from BZLF1 is highly
upregulated in lytic cell populations compared with refractory or
untreated cells. This result has been confirmed as an effective
indicator for separating refractory and lytic EBV-harboring
cells.
It has been observed that a high level of STATs
maintains the latent infection state of EBV. An inhibitor of
activated STAT1, PIAS1, is a factor capable of restricting EBV by
inhibiting transcription factors of viral and cellular origins. It
has been demonstrated that PIAS1 inhibits the IRF-8 mediated
activation of lytic genes through their molecular interaction
(98). In the cell populations
that are refractory to the induction of the lytic cycle, the
preferential upregulation of STAT3 and Fos proto-oncogene,
AP-1 transcription factor subunit has been observed and the
expression of both factors is increased in folds in comparison with
untreated cells (98).
In EBV harboring HH516-16 cells, the regulation of
the lytic cycle has been investigated. When latently infected with
EBV, cells highly express STAT3 protein, predominantly in its
unphosphorylated form. When exposed to sodium butyrate (NaB), an
agent that induces the lytic cycle and a prototype of inhibitor of
histone deacetylase, entry to the lytic cycle is triggered.
HH516-16 cells are also treated with IFN-γ to determine the
possible phosphorylation at residue Y705 induced by the treatment,
or if the pathway is detectable (88). Since the STAT3 transcript is
increased primarily in refractory cells, it was examined whether
the increased STAT3 protein levels correspondingly existed in the
population of refractory cells. An increase in STAT3 protein in
these populations compared with the untreated cells in a
time-dependent manner was observed following NaB treatment. In the
subpopulation of lytic cells, STAT3 protein was not significantly
upregulated.
It has been observed in KSHV/HHV-8 that periodic
switching from the latent to the lytic cycle contributes to an
orderly expression of a large panel of viral genes to produce
infectious virions (18).
Clinico-epidemiological studies have revealed that the activation
of the KSHV lytic cycle critically contributed to the pathogenesis
of KS, pleural effusion lymphoma and multicentric Castleman disease
(99–104). The lytic activation of KSHV/HHV-8
is also correlated with the progression and prognosis of the
diseases.
Severe atypical respiratory
syndrome-coronavirus-2 (SARS-CoV-2) viral genomic coding products
stimulate transcription factors to synthesize pro-inflammatory
mediators
COVID-19, the disease caused by SARS-CoV-2, affects
the lining epithelia, leading to severe respiratory disease in
humans. It also infects other viscera, including the liver and
kidneys, by engaging with multiple intracellular signaling
pathways, leading to the production of mediators of inflammatory
response, and hence tissue damage. It was a remarkable feature of
pathogenesis when widespread thrombosis with microangiopathy in the
blood vessels of the lungs was identified during the clinical
course of COVID-19 (105).
SARS-CoV-2 has a genome containing a single stranded
RNA measuring 30 kb. Two open reading frames (ORFs), ORF 1a and 1b
synthesize 27 non-structural proteins upon translation. ORFs 3, 6,
7a, 7b, 8, 9a, 9c, 10 and 14 code for spike (S), envelope (E) and
nucleocapsid (N) proteins, as well as several accessory proteins.
The alignment of the coding region is demonstrated in Fig. 3. The accessory proteins encoded by
the aforementioned ORFs play an important role in the pathobiology
attributed by the virus (106).
These proteins assist the virus to establish infection to the
susceptible host cells and hijack the cellular machinery of the
host for particle assembly, amplification and pathogenesis of the
virus. During the infection, the S protein binds to the receptor of
angiotensin-converting enzyme 2 on the host cells (107); subsequently the cellular
transmembrane protease serine 2 (TMPRSS2) cleaves the viral S
protein, resulting in two subunits, S1 and S2. The two fragments
fuse with viral and cellular membranes and initiate virus
internalization (107) (Fig. 4).
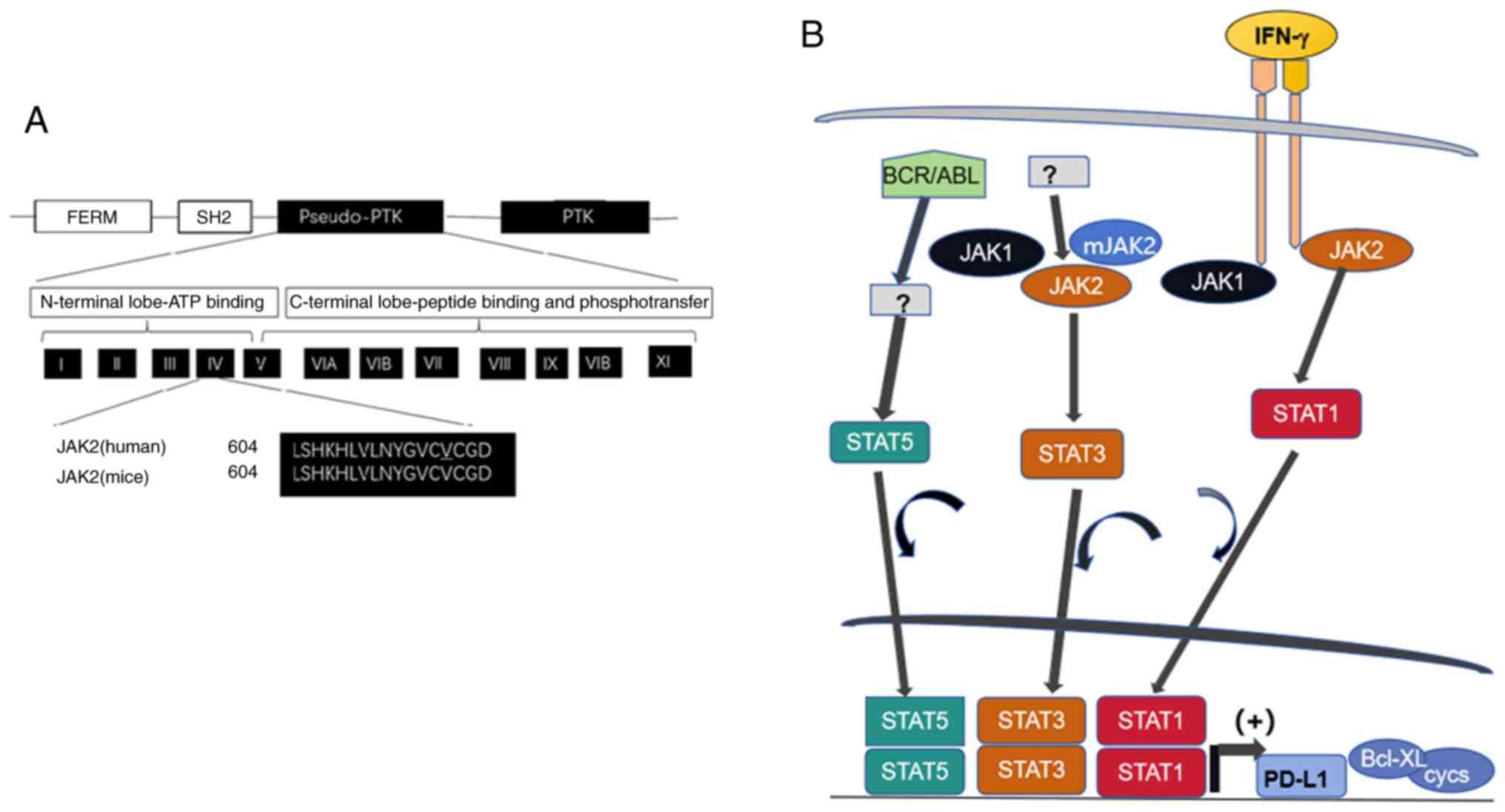 | Figure 3.Linear clustering of JAK2 motifs and
role of abnormal JAK/STAT signaling in carcinogenesis. (A) The JAK2
molecule contains motifs of FERM required for lower Michaelis
constant (Km; the concentration of saturated substrate in case of
the half of maximum catalytic speed of an enzyme being reached) of
JAK2 (V617F) towards substrates, SH2, pseudo-PTK and the functional
PTK. The pseudo-PTK domain starting with amino acid residue 604 is
identical in human and mouse. The mutation on 617 position of amino
acid sequence is responsible for hemopoietic disorders described.
(B) The impact of aberrant JAK/STAT pathway in carcinogenesis. High
expression of STAT1 downstream of IFN-g signaling induces PD-L1
expression, enabling the tumor cells to evade host antitumor
immunity. Mutant JAK2 with the lesion of V617F alters STAT3
signaling and fused protein BCR/ABL expressed from the translocated
gene on Ph1 chromosome induces abnormal STAT5 signaling to enhance
transcription of cyclins D1, D2, E and anti-apoptotic regulator
Bcl_xL, to promote cancer initiation. JAK, Janus kinase; STAT,
signal transducer and activator of transcription; FERM,
4.1/ezrin/radixin/moesin; SH2, Src homolog 2; PTK, protein tyrosine
kinase; PD-L1, programmed cell death ligand 1; Ph, Philadelphia;
Bcl xL, B-cell lymphoma-extra large. |
Previously, the viral peptides binding with major
histocompatibility complex-1 (MHC-1) molecules to downregulate
antiviral immune response have been described in human
immunodeficiency virus type 1 and KSHV (108,109). A similar interaction between
SARS-CoV-2 through ORF8 protein and human proteins has been
identified, considered to be essential for the SARS-CoV-2-mediated
immune evasion by MHC-1 suppression (110).
The coding products of the SARS-CoV-2 genome have
been investigated in relation to their pathogenesis of host
internal organs through engagement of intracellular pathways to
trigger production of pro-inflammatory mediators. NF-κB proteins
are a set of transcription factors that regulate inflammation and
cell death. The transcription factors comprise several subunits,
such as NF-κB 3/p65, together with negative regulator I-κB, which
dimerize with the subunits to repress their activities.
Modification on specific amino residues, such as phosphorylation,
leads to the NF-κB activation. The inhibition of the activity of
these transcription factors is removed on their degradation through
phosphorylation and then ubiquitination on their amino (N)
terminus. HK-2 cells derived from the proximal tubular epithelium
are susceptible to SARS-CoV-2 infection. The effect of ORF3A
expression on the effective activation of the NF-κB signaling
pathway by increasing the phosphorylation of subunit p65 has been
studied; it has been revealed that the SARS-CoV-2-encoded protein
stimulates phosphorylation on positions Ser 536 and Ser 276 of the
p65 protein. ORF3A also increases the amount of cleavage of
caspase-3 (111), suggesting its
role in triggering apoptosis, as identified in other cell lines. It
has been observed that the mRNA levels of the downstream targets of
TNF-α and IL-6 are increased in HK-2 cells by ORF3A. TNF-α is a
cytokine that activates the NF-κB pathway and causes tubular cell
injury, confirming the modulation of NF-κB by ORF3A. STAT3
phosphorylation on residue 705 leads to functional activation of
STAT3 induced by the cytokines TNF-α and IL-6, the expression of
STAT3 is increased by ORF3A expression. It has also been suggested
that TRIM59 expression is increased by SARS-CoV-2 infection in HK-2
cells. It has been noted that SARS-CoV-2 has a similar antagonistic
activity to that of IFN (112).
It has therefore been hypothesized that the pathophysiology of
SARS-CoV-2 infection is related to the effects of the virus on IFN
and JAK/STAT signaling. It has been proposed that COVID-19 is a
disease stemming from the dysregulation of STATs induced by
SARS-CoV-2, leading to a catastrophic cascade of internal organ
failure.
After the entry of susceptible cells, SARS-CoV-2
expresses the proteins non-structural protein 1 and ORF6 to inhibit
the activity of STAT1 (113).
While the activity of STAT1 is restricted, STAT3 becomes the
predominant form in signaling and triggers downstream pathways.
SOCS1 and SOCS3, are factors involved in the negative feedback of
type I IFN signaling. They are induced by both STAT1 and STAT3 and
inhibit the activity of JAKs (114). The binding of STAT1 and STAT3 to
DNA is prevented by PIAS1 and PIAS3, respectively (115). When STAT3 is aberrantly activated
and uncoupled from SOCS regulation, the role of PIAS3 becomes
crucial in regulating the activity of STAT3. Protein tyrosine
phosphorylases exert regulatory activities on activated JAKs and
STATs (116), but their role in
the control of viral infection requires further elucidation. In
lungs infected with SARS-CoV-2, EGFR levels are induced by acute
lung injury or reduced activity of STAT1; STAT3 is activated by
ligation or viral protein binding of EGFR (117). PIAS3 normally inhibits STAT3
activity; however, during the viral infection, PAI-1 is produced
(118) and an escalating cascade
in the PAI-1/STAT3 axis is in turn established.
Immunomodulation and corticosteroids for
intervention against cytokine production have been suggested as a
means to limiting or minimizing the hyperactive response of
inflammation (119).
Immunoregulatory antagonists of IL-1 or IL-6 and of JAK inhibitors
have so far been examined (120).
Clinical trials on antagonists and antibodies targeting several
cytokines at initial stages have produced promising results for the
treatment of COVID-19, which is characterized by abnormalities in
the expression of cytokines. The inhibition of JAK has been proven
to have the potential to suppress inflammatory response, and
endocytosis has been shown to mediate the surface viral receptor
during the pathogenesis of COVID-19 (121). Ruxolitinib, an inhibitor of JAK1
and JAK2 is an orally administered drug approved for the treatment
of myelofibrosis and polycythemia vera in Europe (122). Several newly developed agents
have been tested to determine their efficacy in treating COVID-19,
including Baricitinib (123),
Clazakizumab, Siluximab and Anakinra (124).
A recent study described a COVID-19 patient
suffering from systemic sclerosis (SSc) with pulmonary fibrosis
(125). It has been observed that
the respiratory function had rapidly improved 10 days after she was
administered Ruxolitinib. The reduction in pulmonary fibrosis was
compared before and after the diagnosis of COVID-19. JAK/STAT
signaling has been revealed to be involved in pathogenesis and
fibrosis modulation in SSc patients (125). The administration of ruxolitinib
is therefore recommended as a new therapeutic strategy for patients
with COVID-19 and lung fibrosis (122). To date, numerous clinical trials
are ongoing to evaluate the effectiveness of biopharmaceutical
drugs such as blockers of IL-1, inhibitors of IL-6 and JAKs in
anti-COVID-19 therapy. The rationale behind pathogenesis and
state-of-the-art therapeutic approach for blocking
hyperinflammation have been described (126).
The therapeutic efficacy of Vitamin (Vit) D in
COVID-19 has been tested. Ongoing studies indicate the antiviral
effect of Vit D supplements in the protection against SARS-CoV-2
infection, as well as its ability to reduce the risk of other viral
and bacterial infections, including influenza (127–129).
Vit D is a lipid soluble vitamin that maintains
calcium levels in the body. Vit D binds to the cognate receptor,
Vit D receptor (VDR) to form a complex and in turn binds to the
regions of a promoter on a genomic DNA sequence to modulate target
gene expression (130). In
addition, Vit D binds to VDR through a non-genomic activity,
activates several intracellular signaling pathways and directly
regulates the transcription of multiple genes, including immune
response-related genes. The ability of Vit D in vitro, in
vivo and in patients with severe COVID-19 to enhance host
IFN-a/b signaling have been reported (131). Higher levels of JAK/STAT
signaling pathway activity have been observed with significantly
higher antiviral ISGs at both the gene and protein levels; the
revealed regulatory role of Vit D on IFN type I suggests the
importance of maintaining a normal level of Vit D to prevent and
possibly treat SARS-CoV-2 infection. Additional mechanistic studies
are needed to fully elucidate the antiviral activity of Vit D
against COVID-19. The recent laboratory findings support the
promising use of Vit D as a therapeutic agent for COVID-19.
Conclusions
As a kinase molecule non-covalently associated with
the intracellular portion of membrane integral surface receptor of
ILs and other cytokines, JAK is activated upon ligation of the
receptors to activate a downstream cascade, leading with STATs. The
pathways involved are implicated in carcinogenesis, viral infection
and host antiviral defense against the infection of human
herpesviruses and SARS-CoV-2. The elucidation of the JAK/STAT
pathway would shed light into viral pathogenesis and precise
targeting therapy against cancers, herpesviruses and
coronaviruses.
Acknowledgements
The authors would like to thank Mr. Jiaqing Luo and
Ms. Yuan Zhang, undergraduates of biotechnology, Guangdong Medical
University, China for helpful suggestions on the content of the
manuscript.
Funding
Funding: No funding was received.
Availability of data and materials
Not applicable.
Authors' contributions
WL, YZ and SJS wrote draft of the manuscript. SJS
and PT assisted with figures preparation, searched for references
and revised the manuscript. XZ, GLH, BZ and ZH conceived ideas of
the study and corrected the drafts. Data authentication is not
applicable. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
JAK
|
Janus kinase
|
|
STAT
|
signal transducer and activator of
transcription
|
|
EBV
|
Epstein-Barr virus
|
|
KSHV
|
Kaposi sarcoma-associated
herpesvirus
|
|
ISG
|
IFN-stimulated gene
|
|
COVID-19
|
coronavirus infectious disease
2019
|
|
SARS-CoV-2
|
severe atypical respiratory
syndrome-coronavirus-2
|
References
|
1
|
Malemud CJ: The role of the JAK/STAT
signal pathway in rheumatoid arthritis. Ther Adv Musculoskelet Dis.
10:117–127. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ihle JN: The STAT family in cytokine
signaling. Curr Opin Cell Biol. 13:211–217. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Levy DE and Darnell JE Jr: Stats:
Transcriptional control and biological impact. Nat Rev Mol Cell
Biol. 3:651–662. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Darnell JE Jr: STATs and gene regulation.
Science. 277:1630–1635. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Simoncic PD, Lee-Loy A, Barber DL,
Tremblay ML and McGlade CJ: The T cell protein tyrosine phosphatase
is a negative regulator of Janus family kinases 1 and 3. Curr Biol.
12:446–453. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Gao Z, Jiang F, Yan H, Yang B, He
Q, Luo P, Xu Z and Yang X: JAK-STAT signaling as an ARDS
therapeutic target: Status and future trends. Biochem Pharmacol.
208:1153822023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen CW, Chang YH, Tsi CJ and Lin WW:
Inhibition of IFN-gamma-mediated inducible nitric oxide synthase
induction by the peroxisome proliferator-activated receptor gamma
agonist, 15-deoxy-delta 12,14-prostaglandin J2, involves inhibition
of the upstream Janus kinase/STAT1 signaling pathway. J Immunol.
171:979–988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wiede F, Shields BJ, Chew SH,
Kyparissoudis K, van Vliet C, Galic S, Tremblay ML, Russell SM,
Godfrey DI and Tiganis T: T cell protein tyrosine phosphatase
attenuates T cell signaling to maintain tolerance in mice. J Clin
Invest. 121:4758–4774. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
ten Hoeve J, de Jesus Ibarra-Sanchez M, Fu
Y, Zhu W, Tremblay M, David M and Shuai K: Identification of a
nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol.
22:5662–5668. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shuai K and Liu B: Regulation of JAK-STAT
signalling in the immune system. Nat Rev Immunol. 3:900–911. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shuai K: Modulation of STAT signaling by
STAT-interacting proteins. Oncogene. 19:2638–2644. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yan Z, Gibson SA, Buckley JA, Qin H and
Benveniste EN: Role of the JAK/STAT signaling pathway in regulation
of innate immunity in neuroinflammatory diseases. Clin Immunol.
189:4–13. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Koebel CM, Vermi W, Swann JB, Zerafa N,
Rodig SJ, Old LJ, Smyth MJ and Schreiber RD: Adaptive immunity
maintains occult cancer in an equilibrium state. Nature.
450:903–907. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Galon J, Costes A, Sanchez-Cabo F,
Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M,
Berger A, Wind P, et al: Type, density, and location of immune
cells within human colorectal tumors predict clinical outcome.
Science. 313:1960–1964. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dunn GP, Koebel CM and Schreiber RD:
Interferons, immunity and cancer immunoediting. Nat Rev Immunol.
6:836–848. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen H, Lee JM, Zong Y, Borowitz M, Ng MH,
Ambinder RF and Hayward SD: Linkage between STAT regulation and
Epstein-Barr virus gene expression in tumors. J Virol.
75:2929–2937. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang K, Lv DW and Li R: Cell receptor
activation and chemical induction trigger caspase-mediated cleavage
of PIAS1 to facilitate epstein-barr virus reactivation. Cell Rep.
21:3445–3457. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang JT, Doong SL, Teng SC, Lee CP, Tsai
CH and Chen MR: Epstein-Barr virus BGLF4 kinase suppresses the
interferon regulatory factor 3 signaling pathway. J Virol.
83:1856–1869. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang J, Guo W, Long C, Zhou H, Wang H and
Sun X: The split Renilla luciferase complementation assay is useful
for identifying the interaction of Epstein-Barr virus protein
kinase BGLF4 and a heat shock protein Hsp90. Acta Virol. 60:62–70.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li R, Wang L, Liao G, Guzzo CM, Matunis
MJ, Zhu H and Hayward SD: SUMO binding by the Epstein-Barr virus
protein kinase BGLF4 is crucial for BGLF4 function. J Virol.
86:5412–5421. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fuld S, Cunningham C, Klucher K, Davison
AJ and Blackbourn DJ: Inhibition of interferon signaling by the
Kaposi's sarcoma-associated herpesvirus full-length viral
interferon regulatory factor 2 protein. J Virol. 80:3092–3097.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Aurer I, Butturini A and Gale RP: BCR-ABL
rearrangements in children with Philadelphia chromosome-positive
chronic myelogenous leukemia. Blood. 78:2407–2410. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dan S, Naito M and Tsuruo T: Selective
induction of apoptosis in Philadelphia chromosome-positive chronic
myelogenous leukemia cells by an inhibitor of BCR-ABL tyrosine
kinase, CGP 57148. Cell Death Differ. 5:710–715. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Miller G, El-Guindy A, Countryman J, Ye J
and Gradoville L: Lytic cycle switches of oncogenic human
gammaherpesviruses. Adv Cancer Res. 97:81–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen N, Zhou M, Dong X, Qu J, Gong F, Han
Y, Qiu Y, Wang J, Liu Y, Wei Y, et al: Epidemiological and clinical
characteristics of 99 cases of 2019 novel coronavirus pneumonia in
Wuhan, China: A descriptive study. Lancet. 395:507–513. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luo W, Li YX, Jiang LJ, Chen Q, Wang T and
Ye DW: Targeting JAK-STAT signaling to control cytokine release
Syndrome in COVID-19. Trends Pharmacol Sci. 41:531–543. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xia H, Cao Z, Xie X, Zhang X, Chen JY,
Wang H, Menachery VD, Rajsbaum R and Shi PY: Evasion of type I
interferon by SARS-CoV-2. Cell Rep. 33:1082342020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuen CK, Lam JY, Wong WM, Mak LF, Wan X,
Chu H, Cai JP, Jin DY, To KK, Chan JF, et al: SARS-CoV-2 nsp13,
nsp14, nsp15 and orf6 function as potent interferon antagonists.
Emerg Microbes Infect. 9:1418–1428. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miorin L, Kehrer T, Sanchez-Aparicio MT,
Zhang K, Cohen P, Patel RS, Cupic A, Makio T, Mei M, Moreno E, et
al: SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and
antagonize interferon signaling. Proc Natl Acad Sci USA.
117:28344–28354. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen DY, Khan N, Close BJ, Goel RK, Blum
B, Tavares AH, Kenney D, Conway HL, Ewoldt JK, Chitalia VC, et al:
SARS-CoV-2 disrupts proximal elements in the JAK-STAT pathway. J
Virol. 95:e00862212021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Montero P, Milara J, Roger I and Cortijo
J: Role of JAK/STAT in interstitial lung diseases; molecular and
cellular mechanisms. Int J Mol Sci. 22:62112021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Simpson JA, Al-Attar A, Watson NF,
Scholefield JH, Ilyas M and Durrant LG: Intratumoral T cell
infiltration, MHC class I and STAT1 as biomarkers of good prognosis
in colorectal cancer. Gut. 59:926–933. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jia H, Song L, Cong Q, Wang J, Xu H, Chu
Y, Li Q, Zhang Y, Zou X, Zhang C, et al: The LIM protein AJUBA
promotes colorectal cancer cell survival through suppression of
JAK1/STAT1/IFIT2 network. Oncogene. 36:2655–2666. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang X, Li X, Tan F, Yu N and Pei H:
STAT1 Inhibits MiR-181a expression to suppress colorectal cancer
cell proliferation through PTEN/Akt. J Cell Biochem. 118:3435–3443.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Schroder K, Hertzog PJ, Ravasi T and Hume
DA: Interferon-gamma: An overview of signals, mechanisms and
functions. J Leukoc Biol. 75:163–189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stark GR and Darnell JE Jr: The JAK-STAT
pathway at twenty. Immunity. 36:503–514. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Varinou L, Ramsauer K, Karaghiosoff M,
Kolbe T, Pfeffer K, Müller M and Decker T: Phosphorylation of the
STAT1 transactivation domain is required for full-fledged
IFN-gamma-dependent innate immunity. Immunity. 19:793–802. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Garda-Diaz A, Shin DS, Moreno BH, Saco J,
Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X,
et al: Interferon receptor signaling pathways regulating PD-L1 and
PD-L2 expression. Cell Rep. 19:1189–1201. 2017. View Article : Google Scholar
|
|
40
|
Ivashkiv LB: IFNγ: Signalling, epigenetics
and roles in immunity, metabolism, disease and cancer
immunotherapy. Nat Rev Immunol. 18:545–558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Abiko K, Mandai M, Hamanishi J, Yoshioka
Y, Matsumura N, Baba T, Yamaguchi K, Murakami R, Yamamoto A, Kharma
B, et al: PD-L1 on tumor cells is induced in asdtes and promotes
peritoneal dissemination of ovarian cancer through CTL dysfunction,
din. Cancer Res. 19:1363–1374. 2013.PubMed/NCBI
|
|
42
|
Tian X, Guan W, Zhang L, Sun W, Zhou D,
Lin Q, Ren W, Nadeem L and Xu G: Physical interaction of STAT1
isoforms with TGF-β receptors leads to functional crosstalk between
two signaling pathways in epithelial ovarian cancer. J Exp Clin
Cancer Res. 37:1032018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Padmanabhan S, Gaire B, Zou Y, Uddin MM
and Vancurova I: IFNγ-induced PD-L1 expression in ovarian cancer
cells is regulated by JAK1, STAT1 and IRF1 signaling. Cell Signal.
97:1104002022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yu H, Kortylewski M and Pardoll D:
Crosstalk between cancer and immune cells: Role of STAT3 in the
tumour microenvironment. Nat Rev Immunol. 7:41–51. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Priceman SJ, Kujawski M, Shen S,
Cherryholmes GA, Lee H, Zhang C, Kruper L, Mortimer J, Jove R,
Riggs AD and Yu H: Regulation of adipose tissue T cell subsets by
Stat3 is crucial for diet-induced obesity and insulin resistance.
Proc Natl Acad Sci USA. 110:13079–13084. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Deng J, Liu Y, Lee H, Herrmann A, Zhang W,
Zhang C, Shen S, Priceman SJ, Kujawski M, Pal SK, et al:
S1PR1-STAT3 signaling is crucial for myeloid cell colonization at
future metastatic sites. Cancer Cell. 21:642–654. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Park EJ, Lee JH, Yu GY, He G, Ali SR,
Holzer RG, Osterreicher CH, Takahashi H and Karin M: Dietary and
genetic obesity promote liver inflammation and tumorigenesis by
enhancing IL-6 and TNF expression. Cell. 140:197–208. 2012.
View Article : Google Scholar
|
|
48
|
Carro MS, Lim WK, Alvarez MJ, Bollo RJ,
Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, et al:
The transcriptional network for mesenchymal transformation of brain
tumours. Nature. 463:318–325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Marotta LL, Almendro V, Marusyk A,
Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ,
Choudhury SA, Maruyama R, et al: The JAK2/STAT3 signaling pathway
is required for growth of CD44+CD24-stem cell-like breast cancer
cells in human tumors. J Clin Invest. 121:2723–2735. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Schroeder A, Herrmann A, Cherryholmes G,
Kowolik C, Buettner R, Pal S, Yu H, Müller-Newen G and Jove R: Loss
of androgen receptor expression promotes a stem-like cell phenotype
in prostate cancer through STAT3 signaling. Cancer Res.
74:1227–1237. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bollrath J, Phesse TJ, von Burstin VA,
Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T,
Canli O, Schwitalla S, et al: gp130-mediated Stat3 activation in
enterocytes regulates cell survival and cell-cycle progression
during colitis-associated tumorigenesis. Cancer Cell. 15:91–102.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Grivennikov S, Karin E, Terzic J, Mucida
D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H,
Eckmann L and Karin M: IL-6 and Stat3 are required for survival of
intestinal epithelial cells and development of colitis-associated
cancer. Cancer Cell. 15:103–113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu H and Jove R: The STATs of cancer-new
molecular targets come of age. Nat Rev Cancer. 4:97–105. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lin TS, Mahajan S and Frank DA: STAT
signaling in the pathogenesis and treatment of leukemias. Oncogene.
19:2496–2504. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Battle TE and Frank DA: The role of STATs
in apoptosis. Curr Mol Med. 2:381–392. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bruns HA and Kaplan MH: The role of
constitutively active Stat6 in leukemia and lymphoma. Crit Rev
Oncol Hematol. 57:245–253. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sorger H, Dey S, Vieyra-Garcia PA, Pölöske
D, Teufelberger AR, de Araujo ED, Sedighi A, Graf R, Spiegl B,
Lazzeri I, et al: Blocking STAT3/5 through direct or upstream
kinase targeting in leukemic cutaneous T-cell lymphoma. EMBO Mol
Med. 14:e152002022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhao R, Xing S, Li Z, Fu X, Li Q, Krantz
SB and Zhao ZJ: Identification of an acquired JAK2 mutation in
polycythemia vera. J Biol Chem. 280:22788–22792. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhao L, Ma Y, Seemann J and Huang LJ: A
regulating role of the JAK2 FERM domain in hyperactivation of
JAK2(V617F). Biochem J. 426:91–98. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kralovics R, Passamonti F, Buser AS, Teo
SS, Tiedt R, Passweg JR, Tichelli A, Cazzola M and Skoda RC: A
gain-of-function mutation of JAK2 in myeloproliferative disorders.
N Engl J Med. 352:1779–1790. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Levine RL, Wadleigh M, Cools J, Ebert BL,
Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, et
al: Activating mutation in the tyrosine kinase JAK2 in polycythemia
vera, essential thrombocythemia, and myeloid metaplasia with
myelofibrosis. Cancer Cell. 7:387–397. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Walz C, Crowley BJ, Hudon HE, Gramlich JL,
Neuberg DS, Podar K, Griffin JD and Sattler M: Activated Jak2 with
the V617F point mutation promotes G1/S phase transition. J Biol
Chem. 281:18177–18183. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wernig G, Gonneville JR, Crowley BJ,
Rodrigues MS, Reddy MM, Hudon HE, Walz C, Reiter A, Podar K, Royer
Y, et al: The Jak2V617F oncogene associated with myeloproliferative
diseases requires a functional FERM domain for transformation and
for expression of the Myc and Pim protooncogenes. Blood.
111:3751–3759. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Furuhata A, Kimura A, Shide K, Shimoda K,
Murakami M, Ito H, Gao S, Yoshida K, Tagawa Y, Hagiwara K, et al:
p27 deregulation by Skp2 overexpression induced by the JAK2V617
mutation. Biochem Biophys Res Commun. 383:411–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Jäkel H, Weinl C and Hengst L:
Phosphorylation of p27Kip1 by JAK2 directly links cytokine receptor
signaling to cell cycle control. Oncogene. 30:3502–3512. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Mohrherr J, Uras IZ, Moll HP and Casanova
E: STAT3: Versatile functions in non-small cell lung cancer.
Cancers (Basel). 12:11072020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bromberg J: Stat proteins and oncogenesis.
J Clin Investig. 109:1139–1142. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Huynh J, Etemadi N, Hollande F, Ernst M
and Buchert M: The JAK/STAT3 axis: A comprehensive drug target for
solid malignancies. Semin Cancer Biol. 45:13–22. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Lee HJ, Zhuang G, Cao Y, Du P, Kim HJ and
Settleman J: Drug resistance via feedback activation of Stat3
oncogene-addicted cancer cells. Cancer Cell. 26:207–221. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gao SP, Mark KG, Leslie K, Pao W, Motoi N,
Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B and Bromberg
JF: Mutations in the EGFR kinase domain mediate STAT3 activation
via IL-6 production in human lung adenocarcinomas. J Clin Investig.
117:3846–3856. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhu Z, Aref AR, Cohoon TJ, Barbie TU,
Imamura Y, Yang S, Moody SE, Shen RR, Schinzel AC, Thai TC, et al:
Inhibition of KRAS-driven tumorigenicity by interruption of an
autocrine cytokine circuit. Cancer Discov. 4:452–465. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Liu D, Huang Y, Zeng J, Chen B, Huang N,
Guo N, Liu L, Xu H, Mo X and Li W: Down-regulation of JAK1 by RNA
interference inhibits growth of the lung cancer cell line A549 and
interferes with the PI3K/mTOR pathway. J Cancer Res Clin Oncol.
137:1629–1640. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Xu Y, Jin J, Xu J, Shao YW and Fan Y: JAK2
variations and functions in lung adenocarcinoma. Tumour Biol.
39:10104283177111402017. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lee JH, Kim C, Baek SH, Ko JH, Lee SG,
Yang WM, Um JY, Sethi G and Ahn KS: Capsazepine inhibits JAK/STAT3
signaling, tumor growth, and cell survival in prostate cancer.
Oncotarget. 8:17700–17711. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lee JH, Kim JE, Kim BG, Han HH, Kang S and
Cho NH: STAT3-induced WDR1 overexpression promotes breast cancer
cell migration. Cell Signal. 28:1753–1760. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Subramaniam A, Shanmugam MK, Ong TH, Li F,
Perumal E, Chen L, Vali S, Abbasi T, Kapoor S, Ahn KS, et al:
Emodin inhibits growth and induces apoptosis in an orthotopic
hepatocellular carcinoma model by blocking activation of STAT3. Br
J Pharmacol. 170:807–821. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Paul A, Das S, Das J, Samadder A, Bishayee
K, Sadhukhan R and Khuda-Bukhsh AR: Diarylheptanoid-myricanone
isolated from ethanolic extract of Myrica cerifera shows anticancer
effects on HeLa and PC3 cell lines: Signalling pathway and drug-DNA
interaction. J Integr Med. 11:405–415. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
He G and Karin M: NF-kappaB and STAT3-key
players in liver inflammation and cancer. Cell Res. 21:159–168.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu Z, Chen T, Lu X, Xie H, Zhou L and
Zheng S: Overexpression of variant PNPLA3 gene at I148M position
causes malignant transformation of hepatocytes via IL-6-JAK2/STAT3
pathway in low dose free fatty acid exposure: A laboratory
investigation in vitro and in vivo. Am J Transl Res. 8:1319–1338.
2016.PubMed/NCBI
|
|
81
|
Miller AM, Wang H, Bertola A, Park O,
Horiguchi N, Ki SH, Yin S, Lafdil F and Gao B:
Inflammation-associated interleukin-6/signal transducer and
activator of transcription 3 activation ameliorates alcoholic and
nonalcoholic fatty liver diseases in interleukin-10-deficient mice.
Hepatol. 54:846–856. 2011. View Article : Google Scholar
|
|
82
|
Kim E, Kim M, Woo DH, Shin Y, Shin J,
Chang N, Oh YT, Kim H, Rheey J, Nakano I, et al: Phosphorylation of
EZH2 activates STAT3 signaling via STAT3 methylation and promotes
tumorigenicity of glioblastoma stem-like cells. Cancer Cell.
23:839–852. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Cao R, Wang L, Wang H, Xia L,
Erdjument-Bromage H, Tempst P, Jones RS and Zhang Y: Role of
histone H3 lysine 27 methylation in Polycomb-group silencing.
Science. 298:1039–1043. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Kim KH and Roberts CW: Targeting EZH2 in
cancer. Nat Med. 22:128–134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Cebria F, Kobayashi C, Umesono Y, Nakazawa
M, Mineta K, Ikeo K, Gojobori T, Itoh M, Taira M, Sánchez Alvarado
A and Agata K: The polycomb group protein EZH2 is involved in
progression of prostate cancer. Nature. 419:620–624.
2002.PubMed/NCBI
|
|
87
|
Cao W, Ribeiro Rde O, Liu D, Saintigny P,
Xia R, Xue Y, Lin R, Mao L and Ren H: EZH2 promotes malignant
behaviors via cell cycle dysregulation and its mRNA level
associates with prognosis of patient with non-small cell lung
cancer. PLoS One. 7:e529842012. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wu LJ, Zhang X, Wang J, Kong X, Zheng BY
and Yu H: HeZ: ZMYND10 downregulates cyclins B1 and D1 to arrest
cell cycle by trimethylating lysine 9 on histone 3. Life Res.
4:17–24. 2021. View Article : Google Scholar
|
|
89
|
Zhang Y and Tong T: FOXA1 antagonizes
EZH2-mediated CDKN2A repression in carcinogenesis. Biochem Biophys
Res Commun. 453:172–178. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Ganem D: KSHV infection and the
pathogenesis of Kaposi's sarcoma. Annu Rev Pathol. 1:273–296. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Farrell PJ, Rowe DT, Rooney CM and
Kouzarides T: Epstein-Barr virus BZLF1 trans-activator specifically
binds to a consensus AP-1 site and is related to c-fos. EMBO J.
8:127–132. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Feederle R, Kost M, Baumann M, Janz A,
Drouet E, Hammerschmidt W and Delecluse HJ: The Epstein-Barr virus
lytic program is controlled by the co-operative functions of two
transactivators. EMBO J. 19:3080–3089. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kenney SC and Mertz JE: Regulation of the
latent-lytic switch in Epstein-Barr virus. Semin Cancer Biol.
26:60–68. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Zhang Y, Ma R, Wang Y, Sun W, Yang Z, Han
M, Han T, Wu XA and Liu R: Viruses run: the evasion mechanisms of
the antiviral innate immunity by Hantavirus. Front Microbiol.
12:7591982021. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Mesev EV, LeDesma RA and Ploss A: Decoding
type I and III interferon signaling during viral infection. Nat.
Microbiol. 4:914–924. 2019.
|
|
96
|
Boneschi V, Brambilla L, Berti E, Ferrucci
S, Corbellino M, Parravicini C and Fossati S: Human herpesvirus 8
DNA in the skin and blood of patients with Mediterranean Kaposi's
sarcoma: Clinical correlations. Dermatology. 203:19–23. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Campbell TB, Borok M, Gwanzura L,
MaWhinney S, White IE, Ndemera B, Gudza I, Fitzpatrick L and
Schooley RT: Relationship of human herpesvirus 8 peripheral blood
virus load and Kaposi's sarcoma clinical stage. AIDS. 14:2109–2116.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Murray PG and Young LS: The Role of the
Epstein-Barr virus in human disease. Front Biosci. 7:d519–d540.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
99
|
Chen J, Ueda K, Sakakibara S, Okuno T,
Parravicini C, Corbellino M and Yamanishi K: Activation of latent
Kaposi's sarcoma-associated herpesvirus by demethylation of the
promoter of the lytic transactivator. Proc Natl Acad Sci USA.
98:4119–4124. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Fardet L, Blum L, Kerob D, Agbalika F,
Galicier L, Dupuy A, Lafaurie M, Meignin V, Morel P and Lebbé C:
Human herpesvirus 8-associated hemophagocytic lymphohistiocytosis
in human immunodeficiency virus-infected patients. Clin Infect Dis.
37:285–291. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
101
|
Grandadam M, Dupin N, Calvez V, Gorin I,
Blum L, Kernbaum S, Sicard D, Buisson Y, Agut H, Escande JP and
Huraux JM: Exacerbations of clinical symptoms in human
immunodeficiency virus type 1-infected patients with multicentric
Castleman's disease are associated with a high increase in Kaposi's
sarcoma herpesvirus DNA load in peripheral blood mononuclear cells.
J Infect Dis. 175:1198–1201. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
102
|
Oksenhendler E, Carcelain G, Aoki Y,
Boulanger E, Maillard A, Clauvel JP and Agbalika F: High levels of
human herpesvirus 8 viral load, human interleukin-6,
interleukin-10, and C reactive protein correlate with exacerbation
of multicentric Castleman disease in HIV-infected patients. Blood.
96:2069–2073. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Robles R, Lugo D, Gee L and Jacobson MA:
Effect of antiviral drugs used to treat cytomegalovirus end-organ
disease on subsequent course of previously diagnosed Kaposi's
sarcoma in patients with AIDS. J Acquir Immune Defic Syndr Hum
Retrovirol. 20:34–38. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
King CA, Li X, Barbachano-Guerrero A and
Bhaduri-McIntosh S: STAT3 regulates lytic activation of Kaposi's
sarcoma-associated herpesvirus. J Virol. 89:11347–11355. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Mousavizadeh L and Ghasemi S: Genotype and
phenotype of COVID-19: Their roles in pathogenesis. J Microbiol
Immunol Infect. 54:159–163. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Robson F, Khan KS, Le TK, Paris C,
Demirbag S, Barfuss P, Rocchi P and Ng WL: Coronavirus RNA
proofreading: molecular basis and therapeutic targeting. Mol Cell.
79:710–727. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Chou JM, Tsai JL, Hung JN, Chen IH, Chen
ST and Tsai MH: The ORF8 protein of SARS-CoV-2 modulates the spike
protein and its implications in viral transmission. Front
Microbiol. 13:8835972022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Kim D, Lee JY, Yang JS, Kim JW, Kim VN and
Chang H: The architecture of SARS-CoV-2 transcriptome. Cell.
181:914–921. e102020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Liu DX, Fung TS, Chong KK, Shukla A and
Hilgenfeld R: Accessory proteins of SARS-CoV and other
coronaviruses. Antiviral Res. 109:97–109. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Hoffmann M, Kleine-Weber H, Schroeder S,
Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH,
Nitsche A, et al: SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2
and is blocked by a clinically proven protease inhibitor. Cell.
181:271–280. e82020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Valcarcel A, Bensussen A, Álvarez-Buylla
ER and Díaz J: Structural analysis of SARS-CoV-2 ORF8 protein:
Pathogenic and therapeutic implications. Front Genet.
12:6932272021. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Leifert JA, Holler PD, Harkins S, Kranz DM
and Whitton JL: The cationic region from HIV tat enhances the
cell-surface expression of epitope/MHC class I complexes. Gene
Ther. 10:2067–2073. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Haque M, Ueda K, Nakano K, Hirata Y,
Parravicini C, Corbellino M and Yamanishi K: Major
histocompatibility complex class I molecules are down-regulated at
the cell surface by the K5 protein encoded by Kaposi's
sarcoma-associated herpesvirus/human herpesvirus-8. J Gen Virol.
82:1175–1180. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Selvaraj C, Dinesh DC, Pedone EM, Alothaim
AS, Vijayakumar R, Rudhra O and Singh SK: SARS-CoV-2 ORF8
dimerization and binding mode analysis with class I MHC:
computational approaches to identify COVID-19 inhibitors. Brief
Funct Genomics. 22:227–240. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Cai H, Chen Y, Feng Y, Asadi M, Kaufman L,
Lee K, Kehrer T, Miorin L, Garcia-Sastre A, Gusella GL, et al:
SARS-CoV-2 viral protein ORF3A injures renal tubules by interacting
with TRIM59 to induce STAT3 activation. Mol Ther. 31:774–787. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Ackermann M, Verleden SE, Kuehnel M,
Haverich A, Welte T, Laenger F, Vanstapel A, Werlein C, Stark H,
Tzankov A, et al: Pulmonary vascular endothelialitis, thrombosis,
and angiogenesis in Covid-19. N Engl J Med. 383:120–128. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Klok FA, Kruip MJHA, van der Meer NJM,
Arbous MS, Gommers DAMPJ, Kant KM, Kaptein FHJ, van Paassen J,
Stals MAM, Huisman MV and Endeman H: Incidence of thrombotic
complications in critically ill ICU patients with COVID-19. Thromb
Res. 191:145–147. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Matsuyama T, Kubli SP, Yoshinaga SK,
Pfeffer K and Mak TW: An aberrant STAT pathway is central to
COVID-19. Cell Death Differ. 27:3209–3225. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Martineau AR, Jolliffe DA, Hooper RL,
Greenberg L, Aloia JF, Bergman P, Dubnov-Raz G, Esposito S, Ganmaa
D, Ginde AA, et al: Vitamin D supplementation to prevent acute
respiratory tract infections: Systematic review and meta-analysis
of individual participant data. BMJ. 356:i65832017. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Mehta P, McAuley DF, Brown M, Sanchez E,
Tattersall RS and Manson JJ; HLH Across Speciality Collaboration, :
UK: COVID-19: Consider cytokine storm syndromes and
immunosuppression. Lancet. 395:1033–1034. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Jamilloux Y, Henry T, Belot A, Viel S,
Fauter M, El Jammal T, Walzer T, Francois B and Seve P: Should we
stimulate or suppress immune responses in COVID-19? Cytokine and
anti-cytokine interventions. Autoimmun Rev. 19:1025672020.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Richardson P, Griffin I, Tucker C, Smith
D, Oechsle O, Phelan A, Rawling M, Savory E and Stebbing J:
Baricitinib as potential treatment for 2019-nCoV acute respiratory
disease. Lancet. 395:e30–e31. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Vannucchi AM, Mortara A, D'Alessio A,
Morelli M, Tedeschi A, Festuccia MB, Monforte AD, Capochiani E,
Selleri C, Simonetti F, et al: JAK Inhibition with Ruxolitinib in
Patients with COVID-19 and severe pneumonia: multicenter clinical
experience from a compassionate use program in Italy. J Clin Med.
10:37522021. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Dobosh B, Zandi K, Giraldo DM, Goh SL,
Musall K, Aldeco M, LeCher J, Giacalone VD, Yang J, Eddins DJ, et
al: Baricitinib attenuates the proinflammatory phase of COVID-19
driven by lung-infiltrating monocytes. Cell Rep. 39:1109452022.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Ucciferri C, Auricchio A, Marinari S,
Vecchiet J and Falasca K: COVID-19 in a patient with SISTEMIC
sclerosis: The role of ruxolitinib. Eur J Inflammation. 19:1–4.
2021. View Article : Google Scholar
|
|
126
|
Ucciferri C, Vecchiet J and Falasca K:
Role of monoclonal antibody drugs in the treatment of COVID-19.
World J Clin Cases. 8:4280–4285. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Hashemi R, Morshedi M, Asghari Jafarabadi
M, Altafi D, Saeed Hosseini-Asl S and Rafie-Arefhosseini S:
Anti-inflammatory effects of dietary vitamin D. Neurol Genet.
4:e2782018. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hashemi R, Hosseini-Asl SS, Arefhosseini
SR and Morshedi M: The impact of vitamin D3 intake on inflammatory
markers in multiple sclerosis patients and their first-degree
relatives. PLoS One. 15:e02311452020. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Grant WB, Lahore H, McDonnell SL, Baggerly
CA, French CB, Aliano JL and Bhattoa HP: Evidence that Vitamin D
Supplementation Could Reduce Risk of Influenza and COVID-19
Infections and Deaths. Nutrients. 12:9882020. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Hii CS and Ferrante A: The Non-Genomic
Actions of Vitamin D. Nutrients. 8:1352016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Hafezi S, Saheb Sharif-Askari F, Saheb
Sharif-Askari N, Ali Hussain Alsayed H, Alsafar H, Al Anouti F,
Hamid Q and Halwani R: Vitamin D enhances type I IFN signaling in
COVID-19 patients. Sci Rep. 12:177782022. View Article : Google Scholar : PubMed/NCBI
|