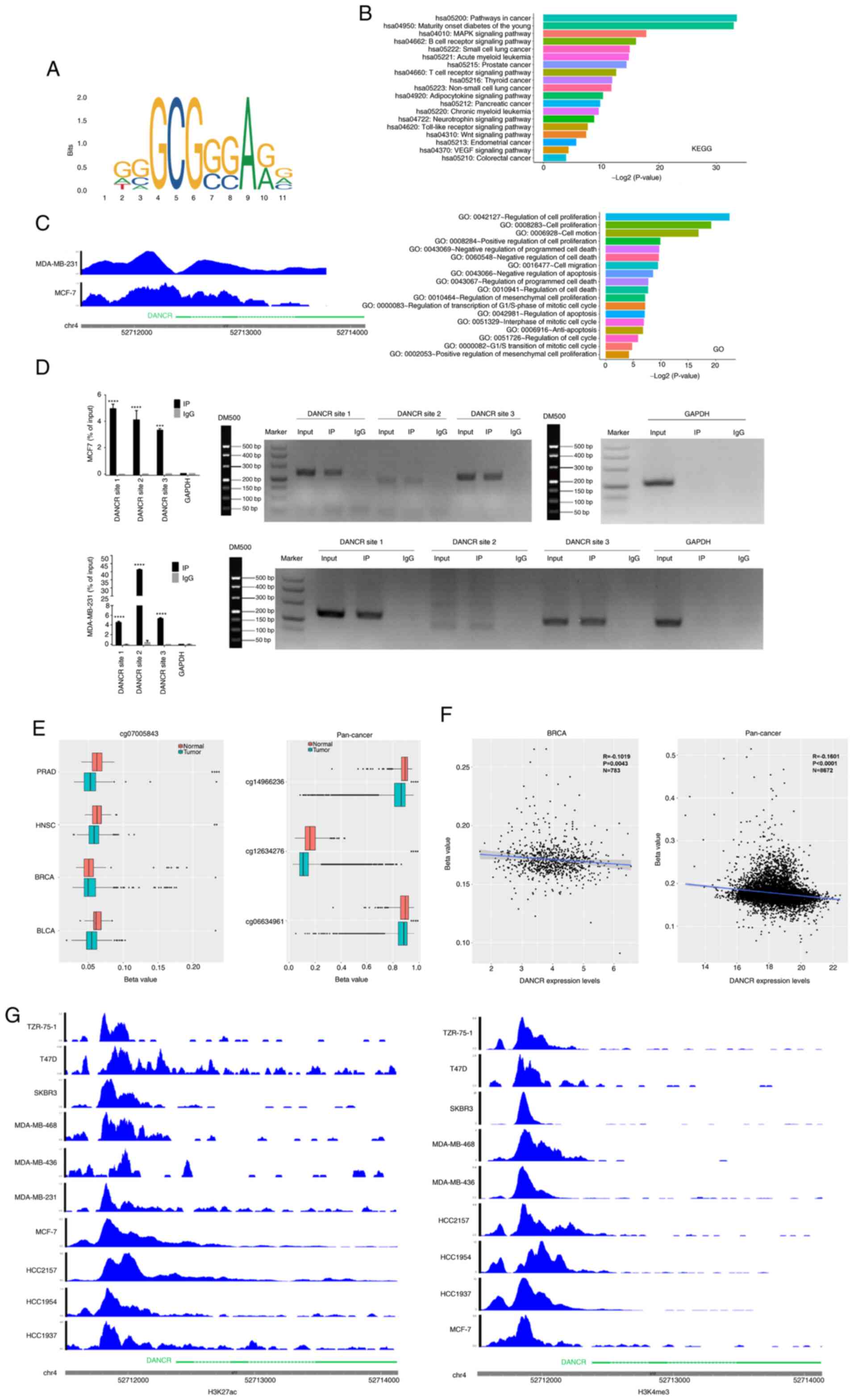
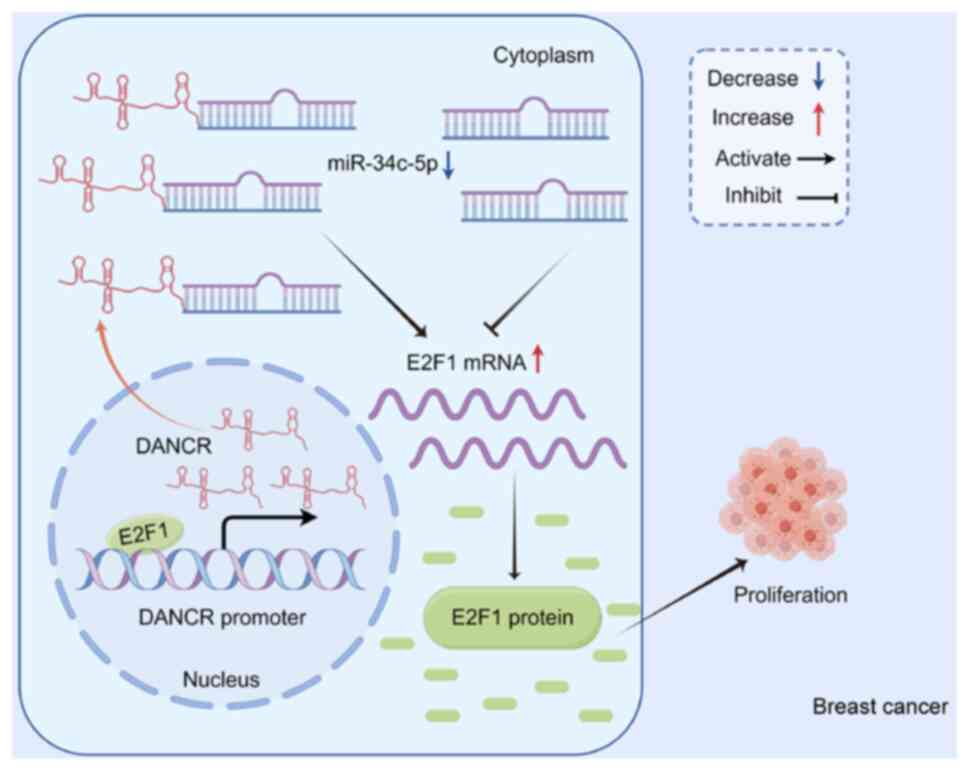
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ramamoorthi G, Kodumudi K, Gallen C,
Zachariah NN, Basu A, Albert G, Beyer A, Snyder C, Wiener D, Costa
RLB and Czerniecki BJ: Disseminated cancer cells in breast cancer:
Mechanism of dissemination and dormancy and emerging insights on
therapeutic opportunities. Semin Cancer Biol. 78:78–89. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu L, Jiang S, Yu S, Liu X, Pu S, Xie P,
Chen H, Liao X, Wang K and Wang B: Increased SIX-1 expression
promotes breast cancer metastasis by regulating
lncATB-miR-200s-ZEB1 axis. J Cell Mol Med. 24:5290–5303. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu L, Tian Q, Gao H, Wu K, Wang B, Ge G,
Jiang S, Wang K, Zhou C, He J, et al: PROX1 promotes breast cancer
invasion and metastasis through WNT/β-catenin pathway via
interacting with hnRNPK. Int J Biol Sci. 18:2032–2046. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang W, Xia J, Xie S, Zou R, Pan S, Wang
ZW, Assaraf YG and Zhu X: Long non-coding RNAs as a determinant of
cancer drug resistance: Towards the overcoming of chemoresistance
via modulation of lncRNAs. Drug Resist Updat. 50:1006832020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chu Z, Huo N, Zhu X, Liu H, Cong R, Ma L,
Kang X, Xue C, Li J, Li Q, et al: FOXO3A-induced LINC00926
suppresses breast tumor growth and metastasis through inhibition of
PGK1-mediated Warburg effect. Mol Ther. 29:2737–2753. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang Y, Mo W, Ding X and Ding Y: Long
non-coding RNAs in breast cancer stem cells. Med Oncol. 40:1772023.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karthikeyan SK, Xu N, Ferguson Rd JE,
Rais-Bahrami S, Qin ZS, Manne U, Netto GJ, S Chandrashekar D and
Varambally S: Identification of androgen response-related lncRNAs
in prostate cancer. Prostate. 83:590–601. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen X, Luo R, Zhang Y, Ye S, Zeng X, Liu
J, Huang D, Liu Y, Liu Q, Luo ML, et al: Long noncoding RNA DIO3OS
induces glycolytic-dominant metabolic reprogramming to promote
aromatase inhibitor resistance in breast cancer. Nat Commun.
13:71602022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou L, Jiang J, Huang Z, Jin P, Peng L,
Luo M, Zhang Z, Chen Y, Xie N, Gao W, et al: Hypoxia-induced lncRNA
STEAP3-AS1 activates Wnt/β-catenin signaling to promote colorectal
cancer progression by preventing m6A-mediated
degradation of STEAP3 mRNA. Mol Cancer. 21:1682022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tong X, Gu PC, Xu SZ and Lin XJ: Long
non-coding RNA-DANCR in human circulating monocytes: A potential
biomarker associated with postmenopausal osteoporosis. Biosci
Biotechnol Biochem. 79:732–737. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gan X, Ding D, Wang M, Yang Y, Sun D, Li
W, Ding W, Yang F, Zhou W and Yuan S: DANCR deletion retards the
initiation and progression of hepatocellular carcinoma based on
gene knockout and patient-derived xenograft in situ hepatoma mice
model. Cancer Lett. 550:2159302022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lamere AT and Li J: Inference of gene
co-expression networks from single-cell RNA-sequencing data.
Methods Mol Biol. 1935:141–153. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Luo ZH, Walid AA, Xie Y, Long H, Xiao W,
Xu L, Fu Y, Feng L and Xiao B: Construction and analysis of a
dysregulated lncRNA-associated ceRNA network in a rat model of
temporal lobe epilepsy. Seizure. 69:105–114. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ruan Y, Li Y, Liu Y, Zhou J, Wang X and
Zhang W: Investigation of optimal pathways for preeclampsia using
network-based guilt by association algorithm. Exp Ther Med.
17:4139–4143. 2019.PubMed/NCBI
|
|
16
|
Thiel D, Conrad ND, Ntini E, Peschutter
RX, Siebert H and Marsico A: Identifying lncRNA-mediated regulatory
modules via ChIA-PET network analysis. BMC Bioinformatics.
20:2922019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhong G, Su S, Li J, Zhao H, Hu D, Chen J,
Li S, Lin Y, Wen L, Lin X, et al: Activation of Piezo1 promotes
osteogenic differentiation of aortic valve interstitial cell
through YAP-dependent glutaminolysis. Sci Adv. 9:eadg04782023.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiao YF, Li BS, Liu JJ, Wang SM, Liu J,
Yang H, Hu YY, Gong CL, Li JL and Yang SM: Role of lncSLCO1C1 in
gastric cancer progression and resistance to oxaliplatin therapy.
Clin Transl Med. 12:e6912022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tao W, Wang C, Zhu B, Zhang G and Pang D:
LncRNA DANCR contributes to tumor progression via targetting
miR-216a-5p in breast cancer: lncRNA DANCR contributes to tumor
progression. Biosci Rep. 39:BSR201816182019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bi Y, Guo S, Xu X, Kong P, Cui H, Yan T,
Ma Y, Cheng Y, Chen Y, Liu X, et al: Decreased ZNF750 promotes
angiogenesis in a paracrine manner via activating
DANCR/miR-4707-3p/FOXC2 axis in esophageal squamous cell carcinoma.
Cell Death Dis. 11:2962020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pan Z, Wu C, Li Y, Li H, An Y, Wang G, Dai
J and Wang Q: LncRNA DANCR silence inhibits SOX5-medicated
progression and autophagy in osteosarcoma via regulating
miR-216a-5p. Biomed Pharmacother. 122:1097072020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu X, Peng WX, Zhou H, Jiang J, Zhou X,
Huang D, Mo YY and Yang L: IGF2BP2 regulates DANCR by serving as an
N6-methyladenosine reader. Cell Death Differ. 27:1782–1794. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xiong M, Wu M, Peng D, Huang W, Chen Z, Ke
H, Chen Z, Song W, Zhao Y, Xiang AP, et al: LncRNA DANCR represses
Doxorubicin-induced apoptosis through stabilizing MALAT1 expression
in colorectal cancer cells. Cell Death Dis. 12:242021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang X, Xie K, Zhou H, Wu Y, Li C, Liu Y,
Liu Z, Xu Q, Liu S, Xiao D and Tao Y: Role of non-coding RNAs and
RNA modifiers in cancer therapy resistance. Mol Cancer. 19:472020.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang ZJ, Liu R, Han XJ, Qiu CL, Dong GL,
Liu ZQ, Liu LH, Luo Y and Jiang LP: Knockdown of the long
non-coding RNA MALAT1 ameliorates TNF-α-mediated endothelial cell
pyroptosis via the miR-30c-5p/Cx43 axis. Mol Med Rep. 27:902023.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shi SJ, Wang LJ, Yu B, Li YH, Jin Y and
Bai XZ: LncRNA-ATB promotes trastuzumab resistance and
invasion-metastasis cascade in breast cancer. Oncotarget.
6:11652–11663. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xue X, Yang YA, Zhang A, Fong KW, Kim J,
Song B, Li S, Zhao JC and Yu J: LncRNA HOTAIR enhances ER signaling
and confers tamoxifen resistance in breast cancer. Oncogene.
35:2746–2755. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ghafouri-Fard S, Khoshbakht T, Hussen BM,
Baniahmad A, Taheri M and Samadian M: A review on the role of DANCR
in the carcinogenesis. Cancer Cell Int. 22:1942022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xu Y, Bao Y, Qiu G, Ye H, He M and Wei X:
METTL3 promotes proliferation and migration of colorectal cancer
cells by increasing SNHG1 stability. Mol Med Rep. 28:2172023.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xue ST, Zheng B, Cao SQ, Ding JC, Hu GS,
Liu W and Chen C: Long non-coding RNA LINC00680 functions as a
ceRNA to promote esophageal squamous cell carcinoma progression
through the miR-423-5p/PAK6 axis. Mol Cancer. 21:692022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ghaemi Z, Mowla SJ and Soltani BM: Novel
splice variants of LINC00963 suppress colorectal cancer cell
proliferation via miR-10a/miR-143/miR-217/miR-512-mediated
regulation of PI3K/AKT and Wnt/β-catenin signaling pathways.
Biochim Biophys Acta Gene Regul Mech. 1866:1949212023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang S, Wang X, Zhou X, Hou L, Wu J, Zhang
W, Li H, Gao C and Sun C: ncRNA-mediated ceRNA regulatory network:
Transcriptomic insights into breast cancer progression and
treatment strategies. Biomed Pharmacother. 162:1146982023.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou Y, Meng X, Chen S, Li W, Li D, Singer
R and Gu W: IMP1 regulates UCA1-mediated cell invasion through
facilitating UCA1 decay and decreasing the sponge effect of UCA1
for miR-122-5p. Breast Cancer Res. 20:322018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu
J, Miao N, Shen J and Peng T: lncRNA DANCR promotes tumor
progression and cancer stemness features in osteosarcoma by
upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 405:46–55.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lu G, Li Y, Ma Y, Lu J, Chen Y, Jiang Q,
Qin Q, Zhao L, Huang Q, Luo Z, et al: Long noncoding RNA LINC00511
contributes to breast cancer tumourigenesis and stemness by
inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res.
37:2892018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tao WY, Wang CY, Sun YH, Su YH, Pang D and
Zhang GQ: MicroRNA-34c suppresses breast cancer migration and
invasion by targeting GIT1. J Cancer. 7:1653–1662. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang S, Miyakawa A, Wickström M, Dyberg
C, Louhivuori L, Varas-Godoy M, Kemppainen K, Kanatani S, Kaczynska
D, Ellström ID, et al: GIT1 protects against breast cancer growth
through negative regulation of Notch. Nat Commun. 13:15372022.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang J, Zhang J, Liu W, Ge R, Gao T, Tian
Q, Mu X, Zhao L and Li X: UBTF facilitates melanoma progression via
modulating MEK1/2-ERK1/2 signalling pathways by promoting GIT1
transcription. Cancer Cell Int. 21:5432021. View Article : Google Scholar : PubMed/NCBI
|