MicroRNAs (miRNAs) are a class of non-coding RNAs
characterized by very short sequences, typically containing
approximately 20–25 bases (1–3).
Traditionally, miRNAs exert their biological functions by binding
to specific sites on mRNA molecules, thereby influencing the
transcription of downstream genes. This binding occurs through
essentially complementary pairing with gene sequences on the target
mRNAs, organizing the translation process of the bound mRNA and
thus regulating the expression levels of corresponding proteins. In
addition to the classical mechanism of gene expression
downregulation, miRNAs exhibit other non-classical regulatory
mechanisms that warrant further investigation (4–7).
Research into miRNAs has seen a significant increase, revealing
their involvement in the development of various systemic diseases
such as cardiovascular (8),
digestive (9) and respiratory
(10) diseases. Moreover, they
play a unique and biologically significant role in numerous
biological processes.
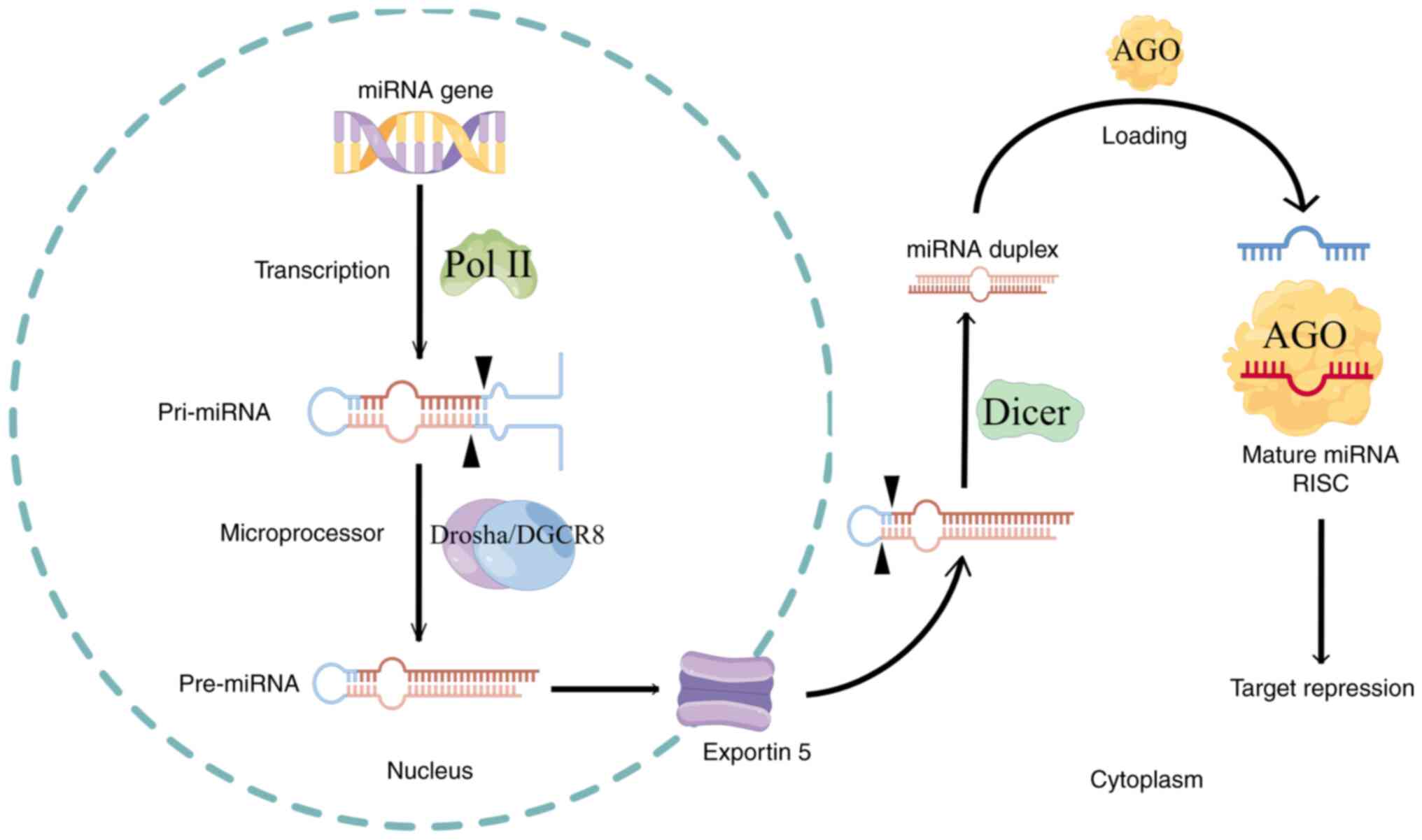
Previous research suggests that the canonical
pathways for miRNA production play critical roles in multiple
biological processes. Genes containing miRNAs undergo
transcription, translation and initial processing into pre-miRNAs
within the nucleus. These pre-miRNAs then traverse the nuclear
membrane via transmembrane proteins into the cytoplasm, where they
undergo further processing and modifications to become mature
miRNAs. Initially, genes harboring miRNAs are transcribed by RNA
polymerase II into primary miRNAs (pri-miRNAs), characterized by
one or more hairpin structures with 5′ caps and 3′ polyadenylate
(A) tails (11–13). Within the nucleus, a microprocessor
complex, consisting of a DROSHA endonuclease molecule and two
molecular chaperones, including DGCR8 with an RNase III structural
domain, cleaves and modifies pri-miRNAs into 50–70 nucleotide
pre-miRNAs (14). Subsequently,
pre-miRNAs are transported across the nuclear membrane to the
cytoplasm by the transmembrane protein exportin 5. In the
cytoplasm, Dicer, containing two RNase III endonuclease structures,
cleaves pre-miRNAs into mature miRNA duplexes ~22 nucleotides in
length (15). Finally, with the
assistance of molecular chaperones such as HSC70/HSP90 (16,17),
duplex miRNAs form a silencing complex known as the RNA-induced
silencing complex (RISC), which includes the Argonaute (AGO)
protein possessing shearing functionality (18,19).
This complex catalyzes the separation of duplex miRNAs into two
single-stranded miRNAs by disrupting the hydrogen bonds of the base
complementary sequences. The strand with a stronger binding
affinity to the 5′ end of the AGO protein within its inner pocket
is designated as the guide strand (20,21),
while the other strand, known as the passenger strand (22), undergoes degradation by cytoplasmic
RNA enzymes. Once a stable RISC structure forms, miRNAs bind to
specific sites on mRNA molecules, thereby silencing mRNA
transcription and exerting biological effects. This process
delineates the classical formation pathway of miRNAs.
It is generally accepted that miRNAs interact with
target genes by inhibiting translation. However, some miRNAs
interact with target genes with less complementarity, overlooking
the fact that certain miRNAs can lead to target gene degradation
(23). Numerous studies suggest
that, contrary to the common misconception, miRNAs typically do not
fully silence target gene mRNAs but rather decrease their
expression (24,25). However, over time, miRNAs exhibit
both temporal and tissue expression specificity, which explains why
mere inhibition or promotion does not fully reflect the biological
functions of miRNAs (26). A
number of studies have reported that miRNAs have seven
non-classical regulatory molecular mechanisms (27); however, this does not fully outline
the non-classical regulatory functions of miRNAs. Despite the
current limited understanding of the biological functions of
miRNAs, further investigation into cellular function regulation by
miRNAs is warranted (2,28,29).
Cardiovascular endothelial cells serve dual roles in
blood coagulation, with both anticoagulant and procoagulant
functions, thereby preserving the equilibrium necessary for proper
blood flow (30). Initially, the
intact endothelium acts as a physical barrier, segregating
platelets, clotting factors and the highly procoagulant endothelial
matrix from the bloodstream (31).
Furthermore, endothelial cells contribute to anti-platelet
aggregation mechanisms. They produce prostacyclin (32–34)
and nitric oxide (NO) (35–37),
which inhibit platelet aggregation, along with adenosine
diphosphatase (38–40), which degrades ADP and prevents
platelet agglutination. Additionally, the vascular endothelium
synthesizes antithrombin and coagulation factors. It generates
thrombomodulin, which binds to circulating thrombin, triggering the
activation of the anticoagulant factor protein C. This synergistic
action with protein S, also synthesized by the endothelium, leads
to the inactivation of coagulation factors V and VIII (41–44).
They synthesize membrane-associated heparin-like molecules that
bind to antithrombin III and inactivate thrombin, as well as
coagulation factors X and IX (45,46).
Additionally, VECs promote fibrinolysis. They internalize
tissue-type plasminogen activator, leading to the degradation of
fibrin deposited on the endothelial cell surface into fibrin
degradation products. The latter exhibits anticoagulant effects
(47–49). Damage to the cardiovascular
endothelium constitutes one of the three major pathological
processes of thrombosis. Following endothelial cell injury,
subendothelial collagen becomes exposed, activating platelets and
coagulation factor VII, thereby initiating the endogenous
coagulation pathway (50,51). Simultaneously, injured endothelial
cells release tissue factor (TF), which activates coagulation
factor VII and triggers the exogenous coagulation pathway (52,53).
Consequently, VECs play a multidimensional role in thrombus
formation and regulation, serving as key cells in maintaining the
dynamic balance between the coagulation and anticoagulation
systems. Recently, miRNAs have been recognized as common risk
factors for deep vein thrombosis (DVT) (54). Therefore, in the present review,
the diverse functions of different miRNAs in endothelial cells are
summarized and recent advances in their pathogenesis and clinical
application in DVT are discussed.
Several studies on DVT have highlighted the role of
miRNAs in directly regulating or indirectly influencing the
function of venous VECs, thus impacting thrombosis (30,54).
In recent years, researchers have employed genetic techniques to
initially screen for aberrantly expressed miRNAs in the blood of
patients with DVT. Subsequently, the target genes these miRNAs act
upon, their effects on the function of VECs and their involvement
in DVT formation have been investigated. These miRNAs play a
crucial role in regulating the function of VECs, inhibiting
apoptosis and ultimately preventing thrombus formation. For
instance, a previous study revealed that miR-9-5p (55) reduces TRPM7 expression by
activating the PI3K/Akt/autophagy pathway, thereby enhancing
endothelial progenitor cell migration, invasion and angiogenesis.
Conversely, the upregulation of histone deacetylase 3 can elevate
miR-19b levels, which in turn mediates peroxisome
proliferator-activated receptor γ to deactivate nuclear factor-κB
(NF-κB), thus mitigating inflammation (56). Decreased levels of miR-125a-5p
(57) have been shown to lead to
increased expression of myeloid cell leukemia sequence 1, promoting
VEC migration and angiogenesis, consequently inhibiting DVT. miRNAs
with similar biological functions include miR-9 (55,58),
miR-19b (56,59), miR-29c-3p (60), miR-125a-5p (57), miR-126 (61), miR-143-3p (62), miR-150 (63–65),
miR-205 (66), miR-411 (67) and miR-3120 (68), which primarily regulate the
autophagic pathway in PI3K/Akt cells, thus exerting influence on
this pathway. Li and Ni (69)
found that miR-26a regulates the NF-κB signaling pathway by binding
to protein kinase C-δ mRNA, thereby inhibiting the expression
levels of inflammatory factor mRNAs and reducing the risk of DVT.
Sun et al (70) revealed
that miR-103a-3p can inhibit the expression of inflammatory factors
such as TF, plasminogen activator inhibitors, interleukin (IL)-6
and IL-8, ultimately disrupting the inflammatory response.
miR-5189-3p can inhibit apoptosis of VECs through the Notch
signaling pathway (71);
miR-195/582 (72) maintains the
homeostasis of the intracellular environment by targeting and
inhibiting the 3′-UTR of post-transcriptional nitric oxide synthase
3 in VECs, which in turn inhibits NO release. The miR-125a-3p/IL-1
receptor type 1 axis and miR-136-5p inhibit the secretion of
inflammatory mediators in the blood of rat femoral veins, resulting
in morphological changes in thrombus length and weight, and
inhibition of thrombus formation (73,74).
A number of miRNAs inhibit thrombosis by promoting the
proliferation of VECs, increasing their viability and attenuating
cellular damage. These miRNAs are summarized as miR-21 (75,76),
miR-195 (77), miR-204-5p
(78), miR-296-5p (79), miR-342-3p (80) and miR-361-5p (81). The aforementioned studies
demonstrated that among various miRNAs, some could inhibit VEC
injury, improve endothelial cell inflammation and autophagy and
inhibit thrombus formation. Therefore, miRNA mimics or inhibitors
may serve as potential anticoagulants (Fig. 1; Table
I).
Functionally, miRNAs can enhance the secretion of
inflammatory factors, proliferation, migration and VEC
angiogenesis. They accelerate injury and apoptosis of VECs and
cause shape elongation, aggregation and cytoskeletal rearrangement
of VECs, thereby promoting DVT formation. Previous studies have
reported that miR-122 (82),
miR-181a-5p (83), miR-195-5p
(84), miR-206 (85), miR-338-5p (86), miR-383-5p (87), miR-448 (88), miR-483-3p (89), miR-525p-5p (90) and let-7e-5p (91) can act on their respective target
proteins to promote DVT formation. These aforementioned studies
suggest that miRNAs can promote the release of inflammatory factors
and accelerate oxidative stress-mediated cellular injury, leading
to the loss of barrier and material transport functions and
exacerbation of ischemic and hypoxic damage to the vascular
microenvironment. The various damaging effects aforementioned
disrupt the blood system balance and the procoagulant state
accelerates DVT formation.
Additionally, miRNAs have been found to promote
thrombus formation in patients with cancer. Venous thromboembolism
(VTE) poses a significant risk to individuals with cancer, leading
to increased morbidity and mortality, with miRNAs playing a role in
this process (92–94). For instance, miRNA-135a directly
affects forkhead box M1 and metastasis suppressor 1, thereby
promoting metastasis of hepatocellular carcinoma cells and
facilitating portal vein thrombosis (95). Oto et al (96) identified seven miRNAs, including
miR-423-5p, as biological markers capable of predicting venous
thrombosis in pancreatic ductal adenocarcinoma and distal
extrahepatic cholangiocarcinoma. Similarly, four miRNAs, including
miR-3652, were found to be significantly downregulated in patients
with colorectal cancer with VTE, suggesting their potential as
novel predictive biomarkers (97).
Morelli et al (98)
observed a positive correlation between plasma levels of miR-145
and the absence of VTE, proposing miR-145 as a potential target for
VTE prevention (98). These
studies collectively demonstrate the dysregulated expression of
miRNAs in the plasma of patients with cancer, their involvement in
cancer cell metastasis and their role in promoting
cancer-associated thrombosis formation. Consequently, numerous
investigations have centered on utilizing miRNAs as biological
markers for cancer-associated thrombosis (99) (Table
II).
Fibrinolytic enzyme activation within the newly
formed thrombus and the release of lysozyme from cell
disintegration allow for gradual thrombus dissolution. The lysis of
thrombi depends on numerous factors and previous studies suggest
that miRNAs are among them. miR-21 (75), miR-92a-3p (100), miR-126 (61,101), miRNA-136-5p (102) and miR-361-5p (81) increase the expression level of
fibrinolytic enzymes, inhibiting the production and activity of
fibrinogen and thrombin while promoting thrombus lysis.
Additionally, seven additional miRNAs have been reported to be
aberrantly expressed in patients with DVT after anticoagulation
therapy and stable thrombosis (103). These studies could provide new
targets and insights for thrombolysis. While miRNAs have the
function of promoting DVT, this effect did not result in complete
thrombus lysis and miRNAs were not identified as the most direct or
critical factors for thrombus dissolution. The aforementioned
studies have only demonstrated reduced thrombus size in animal
models and have not been applied in a clinical setting (Table III).
miRNAs play a crucial role as transcriptional
regulators within the human blood system, with considerable
research centered on human umbilical vein endothelial cells or
endothelial progenitor cells. Consequently, the present review
emphasized the role of miRNAs in the regulation of VEC injury,
which in turn affects DVT formation by modulating cellular injury.
Previous studies have highlighted the involvement of miRNAs in
various facets of thrombosis. miRNAs not only impact the function
of VECs but also influence platelet activation (104), coagulation factor secretion
(105,106) and fibrinolytic enzyme function
(107). However, of note, while
miRNAs are not primary determinants in thrombus formation and
dissolution, they play a significant regulatory role in these
processes. These investigations reveal that individual miRNAs can
modulate different proteins in VECs and conversely, the same
proteins may be subject to regulation by multiple miRNAs. This
complexity suggests that the mechanisms through which miRNAs
regulate thrombosis are highly intricate.
With the increasing focus on research regarding DVT,
a growing number of researchers have turned their attention to
utilizing miRNAs as biomarkers for DVT (108). Currently, miRNAs are primarily
extracted and detected from plasma (109). In patients with DVT, various
factors influence changes in miRNA expression in plasma (110), including: i) Active release from
VECs, ii) platelet and red blood cell lysis, iii) cellular exosome
secretion and iv) the influence of other unknown factors. miRNAs
that enter the circulatory system are primarily in the form of
nucleic acid-protein complexes (111). Conversely, miRNAs present as
monomers are small in molecular weight, convey limited genetic
information and are highly susceptible to degradation. Therefore,
the majority of miRNAs detected in the circulatory system are in
the form of nucleic acid-protein complexes rather than miRNA
monomers (110). Detecting
circulating miRNAs relies on two main methods: i) Microarrays/chips
and ii) reverse transcription-quantitative PCR (RT-qPCR) (112). While microarrays excel at
screening hundreds of miRNAs per sample and offer quantitative
analysis, their complexity, high cost and limitation to small
sample sizes hinder their clinical use, relegating them mainly to
research purposes (113). RT-qPCR
stands out due to its affordability, ease of use and ability to
perform both relative and absolute quantification, rendering it the
preferred choice for clinical applications (114). Therefore, the current clinical
approach for miRNA detection involves initially using miRNA
microarrays to screen out miRNAs with aberrant expression profiles,
followed by validation through RT-qPCR to identify the target
miRNAs (112).
As miRNAs are highly conserved genetically and
stable within the circulatory system, investigations have been
initiated into the utilization of aberrantly expressed miRNAs in
plasma of patients, as markers for early DVT diagnosis (108). miRNAs present in the circulatory
system offer diagnostic advantages over traditional biomarkers for
several reasons: i) Circulating miRNAs demonstrate greater
stability and reproducibility; ii) they impose less damage to
samples, are more convenient to detect and are easier to analyze
and iii) alterations in circulating miRNA expression occur earlier,
facilitating early detection and diagnosis (115). The high early diagnostic value of
miR-582, miR-195, miR-532 (116)
and miR-488 (88) in serum for the
development of DVT has been well-documented. Similarly,
upregulation of miR-424-5p and miR-125a-5p expression levels,
alongside downregulation of miR-136-5p and miR-223-3p expression
levels, has been observed in patients with DVT compared with
healthy participants (108,117). These alterations are associated
with a hypercoagulable state of the blood, indicating potential
diagnostic value for these miRNAs. By contrast, the diagnostic
potential of miRNAs has been investigated by examining their
relationship with D-dimers. For instance, the simultaneous
detection of D-dimer and miR-96 (118) or miRNA-320a/b (119) has been shown to enhance the
diagnostic accuracy of DVT. However, this does not imply that
miRNAs surpass D-dimers in diagnostic value; the latter remain
indispensable. In summary, the fluctuation in miRNA levels in blood
during DVT is influenced by various factors and their specificity
is not high. Consequently, studies utilizing miRNAs as diagnostic
markers for VTE are still in the experimental stage and have not
been extensively implemented in clinical settings. Nevertheless,
monitoring changes in miRNA expression levels may offer a more
precise assessment of risk levels in patients with DVT (54). Further research into miRNAs is
likely to uncover their diagnostic potential to a greater
extent.
Recent studies on miRNA therapy for patients with
DVT have revealed significant progress (120). Research into miRNA therapy for
various diseases has indicated that miRNA overexpression
effectively regulates the transcriptional process of various genes,
inhibits tissue inflammatory responses and reduces adverse effects
in patients with DVT (121,122). A previous study suggested that
when miRNAs enter the pulmonary veins following a DVT episode, they
regulate and control cardiomyocyte activity, thus protecting the
heart from DVT-induced damage (103). There is potential for treating
vascular endothelial cell injury through miRNA upregulation or
downregulation, which could pave the way for new therapeutic
approaches to DVT intervention. However, challenges persist for
miRNA-based therapies, including the need for specific miRNA
delivery within tissues, targeted receptor binding and dose
optimization (61). To date, the
direct miRNA use for DVT therapy has not been reported.
Nonetheless, in the future, integrating miRNAs into novel
therapeutic strategies may revolutionize DVT treatment.
miRNAs play a crucial role in regulating DVT
formation via VECs, rendering them a valuable addition to clinical
studies on DVT. These small RNA molecules exert biological effects
on DVT formation by modulating VECs. However, there have been no
reports of direct antagonistic effects between functionally
distinct miRNAs regulating DVT formation. Consequently, the
complexity of the miRNA regulatory network in DVT surpasses
previous understanding. The vascular endothelium, being in direct
contact with the vein wall, maintains a dynamic balance between
procoagulation and anticoagulation. miRNAs influence the function
of VECs through various pathways, including autophagy and
inflammation, disturbing this delicate balance between pro- and
anti-coagulation systems. Ultimately, this disruption indirectly
regulates thrombus formation. It is unclear whether miRNAs mainly
act on venous VECs, thereby regulating the biological function of
these cells and potentially exacerbating the coagulation process
following vascular endothelium damage. Thus, miRNAs do not directly
participate in the endogenous coagulation pathway but rather
modulate the process of thrombus formation. Furthermore, the
expression levels of miRNAs in plasma are influenced by numerous
factors, diminishing their sensitivity and specificity. There is no
evidence indicating that the diagnostic efficacy of miRNAs
surpasses that of traditional coagulation function tests.
Similarly, while the thrombolytic potential of miRNAs may equal or
exceed that of conventional thrombolytic drugs, no targeted
thrombolytic medications or therapeutic approaches utilizing miRNAs
have been developed. Consequently, there has been no significant
clinical breakthrough or application of miRNAs in the diagnosis and
treatment of DVT. Nevertheless, studying the biological
interactions between miRNAs and VECs is crucial for advancing the
comprehension of the intricate process of thrombosis. Continuous
research and development of extraction, detection and analytical
techniques for miRNAs will provide robust support and technical
assurances for DVT research. As exploration in this field advances,
the future holds promise for expanded applications of miRNA-based
disease prevention and treatment. Investigating the regulatory
mechanisms of miRNAs in thrombosis holds significant value,
offering new insights for early DVT diagnosis and identifying fresh
targets for thrombolysis and personalized therapy.
Not applicable.
The present study was supported by The National Natural Science
Foundation of China (grant no. 82160375), The Natural Science
Foundation of Jiangxi Province (grant no. 20202BABL206035) and The
Jiangxi Ganzhou Science and Technology Program Project (grant no.
2023LNS26841).
Not applicable.
JMo conceived the theme and reviewed the manuscript.
CF and FH wrote the first draft of the manuscript. ZW, MY and JMa
revised parts of the manuscript. DW, TG and FZ helped revise the
manuscript and reviewed the literature. All authors read and
approved the final version of the manuscript. Data authentication
is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Bartel DP: Metazoan MicroRNAs. Cell.
173:20–51. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matsuyama H and Suzuki HI: Systems and
synthetic microRNA biology: From biogenesis to disease
pathogenesis. Int J Mol Sci. 21:1322019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mohr AM and Mott JL: Overview of microRNA
biology. Semin Liver Dis. 35:3–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Thamotharan S, Chu A, Kempf K, Janzen C,
Grogan T, Elashoff DA and Devaskar SU: Differential microRNA
expression in human placentas of term intra-uterine growth
restriction that regulates target genes mediating angiogenesis and
amino acid transport. PLoS One. 12:e01764932017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lucas T, Schäfer F, Müller P, Eming SA,
Heckel A and Dimmeler S: Light-inducible antimiR-92a as a
therapeutic strategy to promote skin repair in healing-impaired
diabetic mice. Nat Commun. 8:151622017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Siomi H and Siomi MC: Posttranscriptional
regulation of microRNA biogenesis in animals. Mol Cell. 38:323–332.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karreth FA, Tay Y, Perna D, Ala U, Tan SM,
Rust AG, DeNicola G, Webster KA, Weiss D, Perez-Mancera PA, et al:
In vivo identification of tumor-suppressive PTEN ceRNAs in an
oncogenic BRAF-induced mouse model of melanoma. Cell. 147:382–395.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vegter EL, van der Meer P, de Windt LJ,
Pinto YM and Voors AA: MicroRNAs in heart failure: From biomarker
to target for therapy. Eur J Heart Fail. 18:457–468. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jung H, Kim JS, Lee KH, Tizaoui K,
Terrazzino S, Cargnin S, Smith L, Koyanagi A, Jacob L, Li H, et al:
Roles of microRNAs in inflammatory bowel disease. Int J Biol Sci.
17:2112–2123. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weidner J, Bartel S, Kılıç A, Zissler UM,
Renz H, Schwarze J, Schmidt-Weber CB, Maes T, Rebane A,
Krauss-Etschmann S and Rådinger M: Spotlight on microRNAs in
allergy and asthma. Allergy. 76:1661–1678. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee Y, Jeon K, Lee JT, Kim S and Kim VN:
MicroRNA maturation: Stepwise processing and subcellular
localization. EMBO J. 21:4663–4670. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nguyen TA, Jo MH, Choi YG, Park J, Kwon
SC, Hohng S, Kim VN and Woo JS: Functional anatomy of the human
microprocessor. Cell. 161:1374–1387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nature reviews. Nat Rev Mol Cell Biol. 15:509–524.
2014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Iwasaki S, Kobayashi M, Yoda M, Sakaguchi
Y, Katsuma S, Suzuki T and Tomari Y: Hsc70/Hsp90 chaperone
machinery mediates ATP-dependent RISC loading of small RNA
duplexes. Mol Cell. 39:292–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Frank F, Sonenberg N and Nagar B:
Structural basis for 5′-nucleotide base-specific recognition of
guide RNA by human AGO2. Nature. 465:818–822. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Suzuki HI, Katsura A, Yasuda T, Ueno T,
Mano H, Sugimoto K and Miyazono K: Small-RNA asymmetry is directly
driven by mammalian Argonautes. Nat Struct Mol. 22:512–521. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Khvorova A, Reynolds A and Jayasena SD:
Functional siRNAs and miRNAs exhibit strand bias. Cell.
115:209–216. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Schwarz DS, Hutvágner G, Du T, Xu Z,
Aronin N and Zamore PD: Asymmetry in the assembly of the RNAi
enzyme complex. Cell. 115:199–208. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chiang HR, Schoenfeld LW, Ruby JG, Auyeung
VC, Spies N, Baek D, Johnston WK, Russ C, Luo S, Babiarz JE, et al:
Mammalian microRNAs: Experimental evaluation of novel and
previously annotated genes. Genes Dev. 24:992–1009. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Giraldez AJ, Mishima Y, Rihel J, Grocock
RJ, Van Dongen S, Inoue K, Enright AJ and Schier AF: Zebrafish
MiR-430 promotes deadenylation and clearance of maternal mRNAs.
Science. 312:75–79. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Baek D, Villen J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Selbach M, Schwanhäusser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dragomir MP, Knutsen E and Calin GA:
SnapShot: Unconventional miRNA functions. Cell. 174:1038–1038.e1.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Neubauer K and Zieger B: Endothelial cells
and coagulation. Cell Tissue Res. 387:391–398. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Krüger-Genge A, Blocki A, Franke RP and
Jung F: Vascular endothelial cell biology: An update. Int J Mol
Sci. 20:44112019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Di Nisio M, van Es N and Büller HR: Deep
vein thrombosis and pulmonary embolism. Lancet. 388:3060–3073.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Juchem G, Weiss DR, Knott M, Senftl A,
Förch S, Fischlein T, Kreuzer E, Reichart B, Laufer S and Nees S:
Regulation of coronary venular barrier function by blood borne
inflammatory mediators and pharmacological tools: Insights from
novel microvascular wall models. Am J Physiol Heart Circ Physiol.
302:H567–H581. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Moncada S, Gryglewski R, Bunting S and
Vane JR: An enzyme isolated from arteries transforms prostaglandin
endoperoxides to an unstable substance that inhibits platelet
aggregation. Nature. 263:663–665. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Moncada S, Higgs EA and Vane JR: Human
arterial and venous tissues generate prostacyclin (prostaglandin
x), a potent inhibitor of platelet aggregation. Lancet. 1:18–20.
1977. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cines DB, Pollak ES, Buck CA, Loscalzo J,
Zimmerman GA, McEver RP, Pober JS, Wick TM, Konkle BA, Schwartz BS,
et al: Endothelial cells in physiology and in the pathophysiology
of vascular disorders. Blood. 91:3527–3561. 1998.PubMed/NCBI
|
|
35
|
Panza JA, Quyyumi AA, Brush JE Jr and
Epstein SE: Abnormal endothelium-dependent vascular relaxation in
patients with essential hypertension. N Engl J Med. 323:22–27.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marti CN, Gheorghiade M, Kalogeropoulos
AP, Georgiopoulou VV, Quyyumi AA and Butler J: Endothelial
dysfunction, arterial stiffness, and heart failure. J Am Coll
Cardiol. 60:1455–1469. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Radomski MW, Palmer RM and Moncada S:
Endogenous nitric oxide inhibits human platelet adhesion to
vascular endothelium. Lancet. 2:1057–1058. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Deaglio S and Robson SC: Ectonucleotidases
as regulators of purinergic signaling in thrombosis, inflammation,
and immunity. Adv Pharmacol. 61:301–332. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Marcus AJ, Safier LB, Hajjar KA, Ullman
HL, Islam N, Broekman MJ and Eiroa AM: Inhibition of platelet
function by an aspirin-insensitive endothelial cell ADPase.
Thromboregulation by endothelial cells. J Clin Invest.
88:1690–1696. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fuentes E and Palomo I: Extracellular ATP
metabolism on vascular endothelial cells: A pathway with
pro-thrombotic and anti-thrombotic molecules. Vascul Pharmacol.
75:1–6. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Esmon CT: Structure and functions of the
endothelial cell protein C receptor. Crit Care Med. 32 (Suppl
5):S298–S301. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Giri H, Panicker SR, Cai X, Biswas I,
Weiler H and Rezaie AR: Thrombomodulin is essential for maintaining
quiescence in vascular endothelial cells. Proc Natl Acad Sci USA.
118:e20222481182021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Iba T and Levy JH: Inflammation and
thrombosis: Roles of neutrophils, platelets and endothelial cells
and their interactions in thrombus formation during sepsis. J
Thromb Haemost. 16:231–241. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Griffin JH, Evatt B, Zimmerman TS, Kleiss
AJ and Wideman C: Deficiency of protein C in congenital thrombotic
disease. J Clin Invest. 68:1370–1373. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ofosu FA, Modi GJ, Smith LM, Cerskus AL,
Hirsh J and Blajchman MA: Heparan sulfate and dermatan sulfate
inhibit the generation of thrombin activity in plasma by
complementary pathways. Blood. 64:742–747. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mann KG, Butenas S and Brummel K: The
dynamics of thrombin formation. Arterioscler Thromb Vasc Biol.
23:17–25. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Loskutoff DJ and Edgington TE: Synthesis
of a fibrinolytic activator and inhibitor by endothelial cells.
Proc Natl Acad Sci USA. 74:3903–3907. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huber D, Cramer EM, Kaufmann JE, Meda P,
Massé JM, Kruithof EK and Vischer UM: Tissue-type plasminogen
activator (t-PA) is stored in Weibel-Palade bodies in human
endothelial cells both in vitro and in vivo. Blood. 99:3637–3645.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Henderson SJ, Weitz JI and Kim PY:
Fibrinolysis: Strategies to enhance the treatment of acute ischemic
stroke. J Thromb Haemost. 16:1932–1940. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Levi M and van der Poll T: Coagulation and
sepsis. Thromb Res. 149:38–44. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Levi M and van der Poll T: Inflammation
and coagulation. Crit Care Med. 38 (Suppl 2):S26–S34. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Mackman N, Tilley RE and Key NS: Role of
the extrinsic pathway of blood coagulation in hemostasis and
thrombosis. Arterioscler Thromb Vasc Biol. 27:1687–1693. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Zelaya H, Rothmeier AS and Ruf W: Tissue
factor at the crossroad of coagulation and cell signaling. J Thromb
Haemost. 16:1941–1952. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sun LL, Li WD, Lei FR and Li XQ: The
regulatory role of microRNAs in angiogenesis-related diseases. J
Cell Mol Med. 22:4568–4587. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhou DM, Sun LL, Zhu J, Chen B, Li XQ and
Li WD: MiR-9 promotes angiogenesis of endothelial progenitor cell
to facilitate thrombi recanalization via targeting TRPM7 through
PI3K/Akt/autophagy pathway. J Cell Mol Med. 24:4624–4632. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang J, Xu X, Li P, Zhang B and Zhang J:
HDAC3 protects against atherosclerosis through inhibition of
inflammation via the microRNA-19b/PPARγ/NF-κB axis.
Atherosclerosis. 323:1–12. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yu J, Jin Y, Xu C, Fang C, Zhang Z, Chen L
and Xu G: Downregulation of miR-125a-5p promotes endothelial
progenitor cell migration and angiogenesis and alleviates deep vein
thrombosis in mice via upregulation of MCL-1. Mol Biotechnol.
65:1664–1678. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhu J, Sun LL, Li WD and Li XQ:
Clarification of the role of miR-9 in the angiogenesis, migration,
and autophagy of endothelial progenitor cells through RNA sequence
analysis. Cell Transplant. 29:9636897209639362020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liang HZ, Li SF, Zhang F, Wu MY, Li CL,
Song JX, Lee C and Chen H: Effect of endothelial microparticles
induced by hypoxia on migration and angiogenesis of human umbilical
vein endothelial cells by delivering MicroRNA-19b. Chin Med J
(Engl). 131:2726–2733. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Lou Z, Ma H, Li X, Zhang F, Du K and Wang
B: Hsa_circ_0001020 accelerates the lower extremity deep vein
thrombosis via sponging miR-29c-3p to promote MDM2 expression.
Thromb Res. 211:38–48. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Meng Q, Wang W, Yu X, Li W, Kong L, Qian
A, Li C and Li X: Upregulation of MicroRNA-126 contributes to
endothelial progenitor cell function in deep vein thrombosis via
its target PIK3R2. J Cell Biochem. 116:1613–1623. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Zhang W, Chen P, Zong H, Ding Y and Yan R:
MiR-143-3p targets ATG2B to inhibit autophagy and promote
endothelial progenitor cells tube formation in deep vein
thrombosis. Tissue Cell. 67:1014532020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Wang W, Zhu X, Du X, Xu A, Yuan X, Zhan Y,
Liu M and Wang S: MiR-150 promotes angiogensis and proliferation of
endothelial progenitor cells in deep venous thrombosis by targeting
SRCIN1. Microvasc Res. 123:35–41. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Du X, Hu N, Yu H, Hong L, Ran F, Huang D,
Zhou M, Li C and Li X: miR-150 regulates endothelial progenitor
cell differentiation via Akt and promotes thrombus resolution. Stem
Cell Res Ther. 11:3542020. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wang W, Li C, Li W, Kong L, Qian A, Hu N,
Meng Q and Li X: MiR-150 enhances the motility of EPCs in vitro and
promotes EPCs homing and thrombus resolving in vivo. Thromb Res.
133:590–598. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Sun LL, Xiao L, Du XL, Hong L, Li CL, Jiao
J, Li WD and Li XQ: MiR-205 promotes endothelial progenitor cell
angiogenesis and deep vein thrombosis recanalization and resolution
by targeting PTEN to regulate Akt/autophagy pathway and MMP2
expression. J Cell Mol Med. 23:8493–8504. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ai P, Shen B, Pan H, Chen K, Zheng J and
Liu F: MiR-411 suppressed vein wall fibrosis by downregulating
MMP-2 via targeting HIF-1α. J Thromb Thrombolysis. 45:264–273.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Li WD, Zhou DM, Sun LL, Xiao L, Liu Z,
Zhou M, Wang WB and Li XQ: LncRNA WTAPP1 promotes migration and
angiogenesis of endothelial progenitor cells via MMP1 through
MicroRNA 3120 and Akt/PI3K/autophagy pathways. Stem Cells.
36:1863–1874. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Li Z and Ni J: Role of microRNA-26a in the
diagnosis of lower extremity deep vein thrombosis in patients with
bone trauma. Exp Ther Med. 14:5069–5074. 2017.PubMed/NCBI
|
|
70
|
Sun S, Chai S, Zhang F and Lu L:
Overexpressed microRNA-103a-3p inhibits acute lower-extremity deep
venous thrombosis via inhibition of CXCL12. IUBMB Life. 72:492–504.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lu J, Fang Q and Ge X: Role and mechanism
of mir-5189-3p in deep vein thrombosis of lower extremities. Ann
Vasc Surg. 77:288–295. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Qin JZ, Wang SJ and Xia C: microRNAs
regulate nitric oxide release from endothelial cells by targeting
NOS3. J Thromb Thrombolysis. 46:275–282. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yang B and Zhang Z: Suppression of long
intergenic non-protein coding RNA 1123 constrains lower extremity
deep vein thrombosis via microRNA-125a-3p to target interleukin 1
receptor type 1. Bioengineered. 13:13452–13461. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ou M, Hao S, Chen J, Zhao S, Cui S and Tu
J: Downregulation of interleukin-6 and C-reactive protein underlies
a novel inhibitory role of microRNA-136-5p in acute lower extremity
deep vein thrombosis. Aging (Albany NY). 12:21076–21090. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Du X, Hong L, Sun L, Sang H, Qian A, Li W,
Zhuang H, Liang H, Song D, Li C, et al: miR-21 induces endothelial
progenitor cells proliferation and angiogenesis via targeting FASLG
and is a potential prognostic marker in deep venous thrombosis. J
Transl Med. 17:2702019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang W, Yuan X, Xu A, Zhu X, Zhan Y, Wang
S and Liu M: Human cancer cells suppress behaviors of endothelial
progenitor cells through miR-21 targeting IL6R. Microvasc Res.
120:21–28. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Mo J, Zhang D and Yang R: MicroRNA-195
regulates proliferation, migration, angiogenesis and autophagy of
endothelial progenitor cells by targeting GABARAPL1. Biosci Rep.
36:e003962016. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Ding M, Chi G, Li F, Wang B, Shao C and
Song W: Up-regulated miR-204-5p promoted the migration, invasion,
and angiogenesis of endothelial progenitor cells to enhance the
thrombolysis of rats with deep venous thrombosis by targeting
SPRED1. Exp Cell Res. 411:1129852022. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Pan Z, Zhang Y, Li C, Yin Y, Liu R, Zheng
G, Fan W, Zhang Q, Song Z, Guo Z, et al: MiR-296-5p ameliorates
deep venous thrombosis by inactivating S100A4. Exp Biol Med
(Maywood). 246:2259–2268. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Pan Z, Chen Q, Ding H and Li H:
MicroRNA-342-3p loaded by human umbilical cord mesenchymal stem
cells-derived exosomes attenuates deep vein thrombosis by
downregulating EDNRA. J Thromb Thrombolysis. 54:411–419. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yang X, Song Y, Sun Y, Wang M and Xiang Y:
Down-regulation of miR-361-5p promotes the viability, migration and
tube formation of endothelial progenitor cells via targeting FGF1.
Biosci Rep. 40:BSR202005572020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Li HQ, Pan ZY, Yang Z, Zhang DB and Chen
Q: Overexpression of MicroRNA-122 resists oxidative stress-induced
human umbilical vascular endothelial cell injury by inhibition of
p53. Biomed Res Int. 2020:97916082020.PubMed/NCBI
|
|
83
|
He X, Liu Y, Li Y and Wu K: Long
non-coding RNA crnde promotes deep vein thrombosis by sequestering
miR-181a-5p away from thrombogenic Pcyox1l. Thromb J. 21:442023.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Jin J, Wang C, Ouyang Y and Zhang D:
Elevated miR-195-5p expression in deep vein thrombosis and
mechanism of action in the regulation of vascular endothelial cell
physiology. Exp Ther Med. 18:4617–4624. 2019.PubMed/NCBI
|
|
85
|
Li Y, Ge J, Yin Y, Yang R, Kong J and Gu
J: Upregulated miR-206 aggravates deep vein thrombosis by
regulating GJA1-mediated autophagy of endothelial progenitor cells.
Cardiovasc Ther. 2022:99663062022. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zhang Y, Zhang Z, Wei R, Miao X, Sun S,
Liang G, Chu C, Zhao L, Zhu X, Guo Q, et al: IL (interleukin)-6
contributes to deep vein thrombosis and is negatively regulated by
miR-338-5p. Arterioscler Thromb Vasc Biol. 40:323–334. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wang H, Lin S, Yang Y, Zhao M, Li X and
Zhang L: Significant role of long non-coding RNA MALAT1 in deep
vein thrombosis via the regulation of vascular endothelial cell
physiology through the microRNA-383-5p/BCL2L11 axis. Bioengineered.
13:13728–13738. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wang P, Dai J and Li D: Peripheral blood
levels of miR-448 and SIRT1 in patients with deep venous thrombosis
and their relationship. Clin Lab. 68:2022. View Article : Google Scholar
|
|
89
|
Kong L, Hu N, Du X, Wang W, Chen H, Li W,
Wei S, Zhuang H, Li X and Li C: Upregulation of miR-483-3p
contributes to endothelial progenitor cells dysfunction in deep
vein thrombosis patients via SRF. J Transl Med. 14:232016.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhu X, Chen B and Xu H: By modulating
miR-525-5p/Bax axis, LINC00659 promotes vascular endothelial cell
apoptosis. Immun Inflamm Dis. 11:e7642023. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Kong L, Du X, Hu N, Li W, Wang W, Wei S,
Zhuang H, Li X and Li C: Downregulation of let-7e-5p contributes to
endothelial progenitor cell dysfunction in deep vein thrombosis via
targeting FASLG. Thromb Res. 138:30–36. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Mulder FI, Horváth-Puhó E, van Es N, van
Laarhoven HWM, Pedersen L, Moik F, Ay C, Büller HR and Sørensen HT:
Venous thromboembolism in cancer patients: A population-based
cohort study. Blood. 137:1959–1969. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Tavares V, Neto BV, Marques IS, Assis J,
Pereira D and Medeiros R: Cancer-associated thrombosis: What about
microRNAs targeting the tissue factor coagulation pathway? Biochim
Biophys Acta Rev Cancer. 1879:1890532024. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Lazar S and Goldfinger LE: Platelet
microparticles and miRNA transfer in cancer progression: Many
targets, modes of action, and effects across cancer stages. Front
Cardiovasc Med. 5:132018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liu S, Guo W, Shi J, Li N, Yu X, Xue J, Fu
X, Chu K, Lu C, Zhao J, et al: MicroRNA-135a contributes to the
development of portal vein tumor thrombus by promoting metastasis
in hepatocellular carcinoma. J Hepatol. 56:389–396. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Oto J, Navarro S, Larsen AC, Solmoirago
MJ, Plana E, Hervás D, Fernández-Pardo Á, España F, Kristensen SR,
Thorlacius-Ussing O and Medina P: MicroRNAs and neutrophil
activation markers predict venous thrombosis in pancreatic ductal
adenocarcinoma and distal extrahepatic cholangiocarcinoma. Int J
Mol Sci. 21:8402020. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Anijs RJS, Laghmani EH, Ünlü B, Kiełbasa
SM, Mei H, Cannegieter SC, Klok FA, Kuppen PJK, Versteeg HH and
Buijs JT: Tumor-expressed microRNAs associated with venous
thromboembolism in colorectal cancer. Res Pract Thromb Haemost.
6:e127492022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Morelli VM, Snir O, Hindberg KD, Hveem K,
Brækkan SK and Hansen JB: High microRNA-145 plasma levels are
associated with decreased risk of future incident venous
thromboembolism-The HUNT study. Blood. blood.2023022285. 2024.(Epub
ahead of print). View Article : Google Scholar
|
|
99
|
Anijs RJS, Nguyen YN, Cannegieter SC,
Versteeg HH and Buijs JT: MicroRNAs as prognostic biomarkers for
(cancer-associated) venous thromboembolism. J Thromb Haemost.
21:7–17. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Feng Y, Lei B, Zhang H, Niu L, Li X, Luo X
and Zhang F: Long noncoding RNA TUG1 induces angiogenesis of
endothelial progenitor cells and dissolution of deep vein
thrombosis. Thromb J. 20:542022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Kuhnert F, Mancuso MR, Hampton J,
Stankunas K, Asano T, Chen CZ and Kuo CJ: Attribution of vascular
phenotypes of the murine Egfl7 locus to the microRNA miR-126.
Development. 135:3989–3993. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Feng Y, Lei B, Zhang H, Niu L, Li X, Luo X
and Zhang F: MicroRNA-136-5p from endothelial progenitor
cells-released extracellular vesicles mediates TXNIP to promote the
dissolution of deep venous thrombosis. Shock. 57:714–721. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Jin QQ, Sun JH, Du QX, Lu XJ, Zhu XY, Fan
HL, Hölscher C and Wang YY: Integrating microRNA and messenger RNA
expression profiles in a rat model of deep vein thrombosis. Int J
Mol Med. 40:1019–1028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Edelstein LC, McKenzie SE, Shaw C,
Holinstat MA, Kunapuli SP and Bray PF: MicroRNAs in platelet
production and activation. J Thromb Haemost. 11 (Suppl
1):S340–S350. 2013. View Article : Google Scholar
|
|
105
|
Jankowska KI, Sauna ZE and Atreya CD: Role
of microRNAs in hemophilia and thrombosis in humans. Int J Mol Sci.
21:35982020. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Hembrom AA, Srivastava S, Garg I and Kumar
B: MicroRNAs in venous thrombo-embolism. Clin Chim Acta. 504:66–72.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Marques-Rocha JL, Samblas M, Milagro FI,
Bressan J, Martínez JA and Marti A: Noncoding RNAs, cytokines, and
inflammation-related diseases. FASEB J. 29:3595–3611. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Wang X, Sundquist K, Elf JL, Strandberg K,
Svensson PJ, Hedelius A, Palmer K, Memon AA, Sundquist J and Zöller
B: Diagnostic potential of plasma microRNA signatures in patients
with deep-vein thrombosis. Thromb Haemost. 116:328–336. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Monguió-Tortajada M, Gálvez-Montón C,
Bayes-Genis A, Roura S and Borràs FE: Extracellular vesicle
isolation methods: Rising impact of size-exclusion chromatography.
Cell Mol Life Sci. 76:2369–2382. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Felekkis K and Papaneophytou C: Challenges
in using circulating Micro-RNAs as biomarkers for cardiovascular
diseases. Int J Mol Sci. 21:5612020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
He Y, Lin J, Kong D, Huang M, Xu C, Kim
TK, Etheridge A, Luo Y, Ding Y and Wang K: Current state of
circulating MicroRNAs as cancer biomarkers. Clin Chem.
61:1138–1155. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ban E and Song EJ: Considerations and
suggestions for the reliable analysis of miRNA in plasma using
qRT-PCR. Genes (Basel). 13:3282022. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Masubuchi T, Endo M, Iizuka R, Iguchi A,
Yoon DH, Sekiguchi T, Qi H, Iinuma R, Miyazono Y, Shoji S, et al:
Construction of integrated gene logic-chip. Nat Nanotechnol.
13:933–940. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Andrews WJ, Brown ED, Dellett M, Hogg RE
and Simpson DA: Rapid quantification of microRNAs in plasma using a
fast real-time PCR system. Biotechniques. 58:244–252. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Chandrasekaran AR, Punnoose JA, Zhou L,
Dey P, Dey BK and Halvorsen K: DNA nanotechnology approaches for
microRNA detection and diagnosis. Nucleic Acids Res.
47:10489–10505. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Qin J, Liang H, Shi D, Dai J, Xu Z, Chen
D, Chen X and Jiang Q: A panel of microRNAs as a new biomarkers for
the detection of deep vein thrombosis. J Thromb Thrombolysis.
39:215–221. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Xu L, Ji C, Miao X, Ge J, Li F and Xu C:
Combination of circulating miR-125a-5p, miR-223-3p and D-dimer as a
novel biomarker for deep vein thrombosis. Am J Med Sci.
364:601–611. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Xie X, Liu C, Lin W, Zhan B, Dong C, Song
Z, Wang S, Qi Y, Wang J and Gu Z: Deep vein thrombosis is
accurately predicted by comprehensive analysis of the levels of
microRNA-96 and plasma D-dimer. Exp Ther Med. 12:1896–1900. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Jiang Z, Ma J, Wang Q, Wu F, Ping J and
Ming L: Combination of circulating miRNA-320a/b and D-dimer
improves diagnostic accuracy in deep vein thrombosis patients. Med
Sci Monit. 24:2031–2037. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Sun LL, Liu Z, Ran F, Huang D, Zhang M, Li
XQ and Li WD: Non-coding RNAs regulating endothelial progenitor
cells for venous thrombosis: Promising therapy and innovation. Stem
Cell Res Ther. 15:72024. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Gareri C, De Rosa S and Indolfi C:
MicroRNAs for restenosis and thrombosis after vascular injury. Circ
Res. 118:1170–1184. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lu Y, Thavarajah T, Gu W, Cai J and Xu Q:
Impact of miRNA in atherosclerosis. Arterioscler Thromb Vasc Biol.
38:e159–e170. 2018. View Article : Google Scholar : PubMed/NCBI
|