A characteristic feature of psoriasis is the
hyper-proliferation of keratinocytes, fibroblasts, and endothelial
cells, triggering immune system activation, inflammatory response
and new blood vessel formation (neovascularization or angiogenesis)
(10,13). Genetic predispositions and
environmental influences are significant factors that trigger these
effects (3,11). Genetic studies reveal that >10%
of patients with psoriasis have a familial connection (14–16).
Numerous genome-wide association studies (GWAS) conducted across
various nations have identified >100 susceptibility loci
associated with psoriasis, including human leukocyte antigen
(HLA)-Cw6, tyrosine kinase 2 (TK2), interleukin-12B
(IL-12B), interleukin-23 receptor (IL-23R) and late
Cornified Envelope 3B/C (LCE3B/3C) (17–20).
However, in Taiwan, no definitive genetic susceptibility loci or
associated processes have been identified.
In clinical research, the polygenic risk score (PRS)
is used as an analytical tool (21). Furthermore, the increase in sample
size for GWAS has enhanced the power and effectiveness of the PRS.
Therefore, a PRS based on GWAS is essential for personalized
medicine (22). The PRS provides
an accurate and efficient means of predicting the health status and
susceptibility of an individual to disease (23). In addition to evaluating the
genetic composition of an individual, PRS can provide valuable
information regarding the efficacy of therapy and the likelihood of
positive or negative outcomes, even before symptoms are evident
(21,24). Furthermore, PRS provides an
opportunity for early detection of abnormal test results before
they occur (25,26). Through this approach, healthcare
practitioners can prescribe preventive measures that are customized
to the genetic profile of each patient (23,27,28).
In the present study, GWAS, PRS and phenome-wide
association study (PheWAS) were conducted on gene variations in
patients with psoriasis using the Taiwanese gene database of China
Medical University Hospital (CMUH). A meta-analysis of single
nucleotide polymorphism (SNP) sites associated with psoriasis in
datasets from CMUH and BioBank Japan (BBJ) was also conducted
(29). Notably, the genetic
diversity among individuals from Taiwan is significantly different
from that in other racial groups.
The electronic medical records of the CMUH were
analyzed for clinical data of patients with psoriasis between 2003
and 2020. All the included patients were validated by physicians
with specific expertise in the field of psoriasis. The present
study used the following International Classification of Diseases
(ICD) diagnostic clinical modification (CM) codes: L40.0, L40.1,
L40.4, L40.50, L40.8, L40.9, L41.3, L41.4, L41.5, L41.8 and L41.9;
and the following ICD-9-CM codes: 696.1, 696.10, 696.2 and 696.8
(30). The number between ICD and
CM indicates the version of ICD. Currently, the clinical practice
of Taiwan includes two versions of ICD CM, versions 9 and 10. These
two versions (ICD-9-CM and ICD-10-CM) have different codes, and
therefore, their specifications are required. In Taiwan, clinicians
mostly continue to record diagnoses using ICD-9 codes, which are
then converted to ICD-10 codes through backend programs. This
practice aligns with government regulations that mandated a
complete transition to ICD-10 use for reporting purposes after 2015
(31). Regarding psoriasis
diagnosis, there are no significant differences between ICD-9 and
ICD-10. However, there is a primary difference in the coding for
psoriasis between the two versions in terms of the level of detail
and specificity. Under ICD-9, psoriasis was primarily coded as
696.1 for psoriatic arthropathy and 696.0 for other types of
psoriasis without specifying the type of psoriasis. On the other
hand, the ICD-10 offers a more detailed classification of
psoriasis, allowing healthcare professionals to accurately specify
the type and site of psoriasis. This increased specificity in
ICD-10 enables improved tracking of the prevalence and treatment
outcomes of various types of psoriasis, which in turn facilitates
more accurate public health surveillance and research on psoriasis
treatment and its effectiveness (32,33).
A schematic representation of the study design used
to investigate psoriasis is shown in Fig. 1. The Precision Medicine Project was
approved by the ethics committee of the institutional review board
(approval nos. CMUH110-REC3-005 and CMUH111-REC1-176) and was
approved by the Institutional Review Board of China Medical
University Hospital (Taichung, Taiwan). Patient data, including
genetic information (genetic variations detected by the TPMv1 SNP
array), laboratory tests [C-reactive protein and erythrocyte
sedimentation rate (ESR)], diagnoses, age and sex, were compiled
for subsequent statistical analysis. The inclusion criteria were as
follows: i) Patients with the ICD-9 and ICD-10 diagnostic codes for
psoriasis; and ii) patients diagnosed with psoriasis at least three
times at China Medical University Hospital. Examples of excluded
autoimmune skin diseases include lupus erythematosus,
dermatomyositis, systemic sclerosis (scleroderma), vitiligo and
alopecia areata.
The association between the SNP array and psoriasis
was investigated using PLINK (V.1.90). A Manhattan plot and
quantile-quantile plot (QQ) (https://cran.r-project.org/web/packages/qqman/index.html;
version 0.1.9) was generated using the R programming language
(https://cran.r-project.org/; version: R
4.1.0) within the R Studio (https://posit.co/downloads/; version: R 1.4.1717)
integrated development environment (38,39).
Table SI presents the GWAS
analysis results. The Gene Symbol and Entrez Gene Name in the last
two columns are the information obtained after inputting into the
Ingenuity Pathway Analysis (IPA) software and then merged into
Table SI.
In Taiwan, under the regulations of the National
Health Insurance, billing can be conducted solely with ICD codes,
which do not categorize the severity of conditions (40). This applies to both the ICD-9 and
ICD-10. In the present study, only ICD diagnosis codes were used
for categorization, with diagnostic data spanning 19 years and
being recorded by numerous physicians. Therefore, distinguishing
severity levels among participants is challenging. Upon
consultation with professional physicians, it was determined that
only a minority would document severity levels in an unstructured
manner for research purposes, further complicating the analysis
owing to the unstructured nature of the data. Therefore, this
limitation was added to the manuscript.
GWAS was employed to analyze biological networks
across the genome, with a significance threshold of
P<5×10−8 (PLINK; V.1.90). This analysis led to the
identification of a comprehensive set of 8,585 SNPs associated with
psoriasis. A molecular network was constructed using core analysis
in the IPA software (https://digitalinsights.qiagen.com/product-login/;
version: 107193442; Qiagen Sciences, Inc.). The statistical
significance of the available networks was assessed using Fisher's
exact test with a significance level of P<0.05 (39,41,42).
The HLA genotypes for each participant in the study
were computed using HLA genotype imputation via attribute tagging
(HIBAG-R) (https://bioconductor.org/packages/release/bioc/html/HIBAG.html;
V1.5). HIBAG allowed for the determination of haplotypes and
diplotypes for individuals, while maintaining P>0.90. Chi-square
analysis was conducted to examine the HLA haplotypes and diplotypes
to investigate the association between these genetic factors and
the incidence of psoriasis. Bonferroni correction was applied to
address multiple testing (39,43).
To calculate the PRS, the CMUH cohort was divided
into three distinct datasets including base, target and validation
(8:1:1 ratio). PRS analysis was performed on 1,799 psoriasis
patients and 53,952 controls in the base group, 225 psoriasis
patients and 6,744 controls in the target group, and 224 psoriasis
patients and 6,744 controls in validation group. A primary dataset
was used to investigate the association between the variables under
study and psoriasis using PLINK1.9. The PRS was calculated based on
the target dataset and the PRSice2 (https://choishingwan.github.io/PRSice/; v2.3.3) tool
while excluding variants with a missing rate of >1% and those
deviating from the Hardy-Weinberg equilibrium with P<0.001. The
present study utilized reference data from the 1,000 Genomes Phase
3 for the East Asian population (https://www.internationalgenome.org/data;
v5b.20130502). The PRS was calculated by normalizing the z-scores.
The PRS, clinical data, or a combination of both was used to
construct logistic regression models. The models were subsequently
adjusted for sex, age, HLA-A*02:07 and HLA-C*06:02
(22).
A genetic association between SNPs from PRS and
diseases was constructed through logistic regression models, using
the ‘PheWAS’ package (https://github.com/PheWAS/PheWAS; v0.12) in R
programming language within the R Studio integrated development
environment (https://cran.r-project.org/; R 4.1.0). PheWAS results
were defined using the phecode schema with ICD diagnostic codes
(10,750 unique ICD-10 codes and 3113 ICD-9 codes). Statistical
significance of the available networks was assessed using Fisher's
exact test with a significance level of P<5E-05 (44).
Baseline continuous and categorical variables were
examined using statistical tests such as the unpaired Student's
t-test, χ2 test and Fisher's exact test. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analyses were performed using SPSS (https://www.ibm.com/spss; version 22) and R (version R
4.1.0) software (46).
The present study presents the results of a GWAS
involving 67,440 controls and 2,248 cases. The control group
included 39,930 men (59.2%) and 27,510 women (40.8%), with a mean
age of 45.75±16.798 years. The psoriasis cohort included 1,331 men
(59.2%) and 917 women (40.8%), with a mean age of 45.75±16.801
years. The distribution of age and sex did not differ significantly
between the groups (Table I).
Habit-related clinical features such as smoking, alcohol
consumption and betel consumption were not statistically
significant. However, the comparison of ESR values between female
patients with psoriasis and those in the control group revealed a
statistically significant difference (P<5.34×10−11).
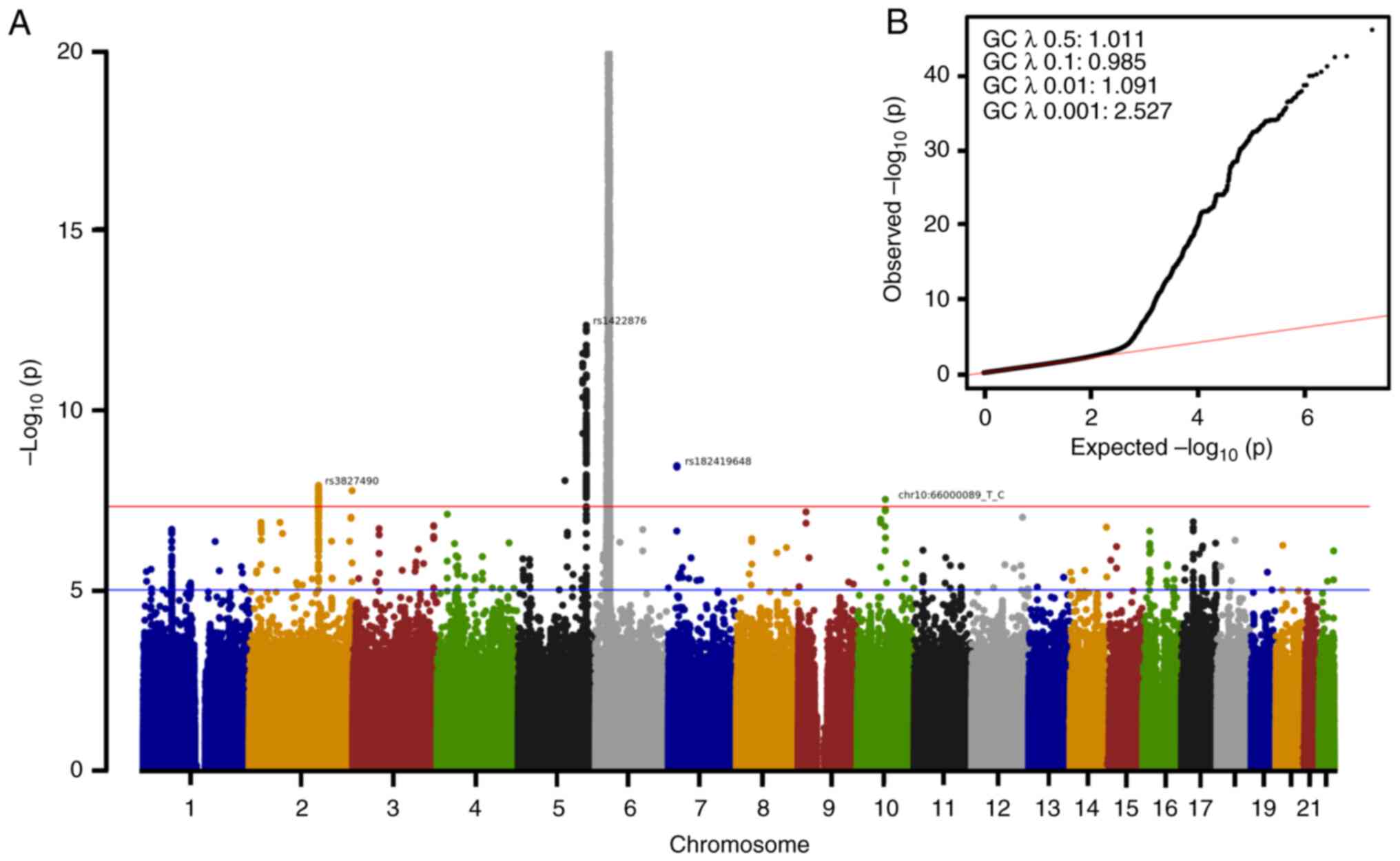
The Manhattan (Fig. 2A) and QQ
(Fig. 2B) plots are useful tools
for visualizing GWAS results. The Manhattan plots (Fig. 2A) demonstrated the strongest
associations between all the SNPs on a genomic scale. Notably,
psoriasis was associated with 8,585 SNPs at a specific position:
14,064,987 (Table SI;
P<5×10−8). As shown in Table II, SNP-induced amino acid
mutations were significantly associated with psoriasis
(P<5×10−8). Missense mutations were found in a
variety of genes; furthermore, the rs34182778 variant was
characterized by a DNA insertion within the corneodesmosin
(CDSN) gene, whereas the rs200838925 variant was
characterized by a DNA deletion within the mucin 22 (MUC22).
The aforementioned findings suggested that these loci and gene
mutations may share a common function, pathway or relevance to
psoriasis. In the present study, the following 92 novel genomic
markers for psoriasis were identified through GWAS and PRS
(Table SI). Additionally, 61
previously reported genomic markers associated with psoriasis in
Taiwanese populations were identified (Table SI): ATP binding cassette subfamily
F member 1 (47), advanced
glycosylation end-product specific receptor (AGER) (48), allograft inflammatory factor 1
(49), Annexin A6 (50), butyrophilin-like 2 (BTNL2)
(51), complement C2 (52), chromosome 6 open reading frame 15
(53,54), complement factor B (52,55),
fibroblast activation protein α (56), HLA complex group 26 (57), HLA complex group 27 (57), HLA complex group 9 (58), HLA complex P5 (59,60),
HLA complex P5B (59,60), major histocompatibility complex,
class I, A (HLA-A) (61),
major histocompatibility complex, class I, B (HLA-B)
(62,63), major histocompatibility complex,
class I, C (HLA-C) (53,54),
major histocompatibility complex, class II, DM β (HLA-DMB)
(64), major histocompatibility
complex, class II, DQ α1 (HLA-DQA1) (65), major histocompatibility complex,
class II, DQ α2 (HLA-DQA2) (66), major histocompatibility complex,
class II, DQ β1 (HLA-DQB1) (67), major histocompatibility complex,
class II, DR β1 (HLA-DRB1) (67), major histocompatibility complex,
class I, E (HLA-E) (68),
major histocompatibility complex, class I, F (HLA-F)
(69), major histocompatibility
complex, class I, G (HLA-G) (70), heat shock protein family A
(Hsp70) member 1A (HSPA1A/HSPA1B) (71), Hsp70 member 1-like (71), interleukin 12B (IL12B)
(72,73), leukocyte specific transcript 1
(74), MHC class I
polypeptide-related sequence A (MICA) (75), MICA antisense RNA 1 (75), MHC class I polypeptide-related
sequence B (MICB) (76),
MICB divergent transcript (76),
myelin oligodendrocyte glycoprotein (77), mucin (MUC)21 (78), MUC22 (78), natural cytotoxicity triggering
receptor 3 (NCR3) (79),
negative elongation factor complex member E (80), notch receptor 4 (81–83),
nurim (84), psoriasis
susceptibility 1 candidate 1 (PSORS1C1) (85), PSORS1C2 (53,54,86),
PSORS1C3 (57), ring finger
protein 39 (87), transporter 1,
ATP binding cassette subfamily B member (TAP1) (64), TAP2 (64), transcription factor 19 (57), tumor necrosis factor (TNF)
(88,89), TNFAIP3 interacting protein 1
(TNIP1) (60), tenascin XB
(TNXB) (88,89), tripartite motif containing 10
(TRIM10) (90),
TRIM15 (91), TRIM26
(92), TRIM27 (92), TRIM40 (92), TSBP1 and BTNL2 antisense RNA1
(93), ubiquitin D (UBD)
(94) and ubiquitin-like domain
containing CTD phosphatase 1 (95).
It is well established that certain HLA alleles and
diplotypes are associated with an increased risk of psoriasis.
High-resolution imputation was used to analyze the HLA genotypes
and their corresponding allele frequencies associated with
psoriasis. Table III shows the
diplotypes identified in the sample and those that exhibited a
significant association with psoriasis are listed in Table IV. Table SII presents raw data regarding HLA
genotypes and allele frequencies that were significantly associated
with psoriasis. The findings of the present study indicated that
the HLA-A*02:07 (adjusted P=3.69×10−34) and
HLA-C*06:02 (adjusted P=2.96×10−29) alleles
exhibited the strongest association with psoriasis in the Taiwanese
population.
A comprehensive analysis was carried out using
Bioinformatics IPA software to analyze 8,585 SNPs associated with
psoriasis. These SNPs were assessed for significance in the context
of the entire genome, using a threshold of P<5×10−8.
The present study findings revealed that psoriasis is characterized
by multiple key pathways, including immune responses (antigen
presentation, PD-1/PD-L1 cancer immunotherapy, IL-10 signaling,
interplay between dendritic cells and natural killer cells and
Th1/Th2 activation) and inflammatory signaling (multiple sclerosis
and neuroinflammatory signaling pathways), which are shown in
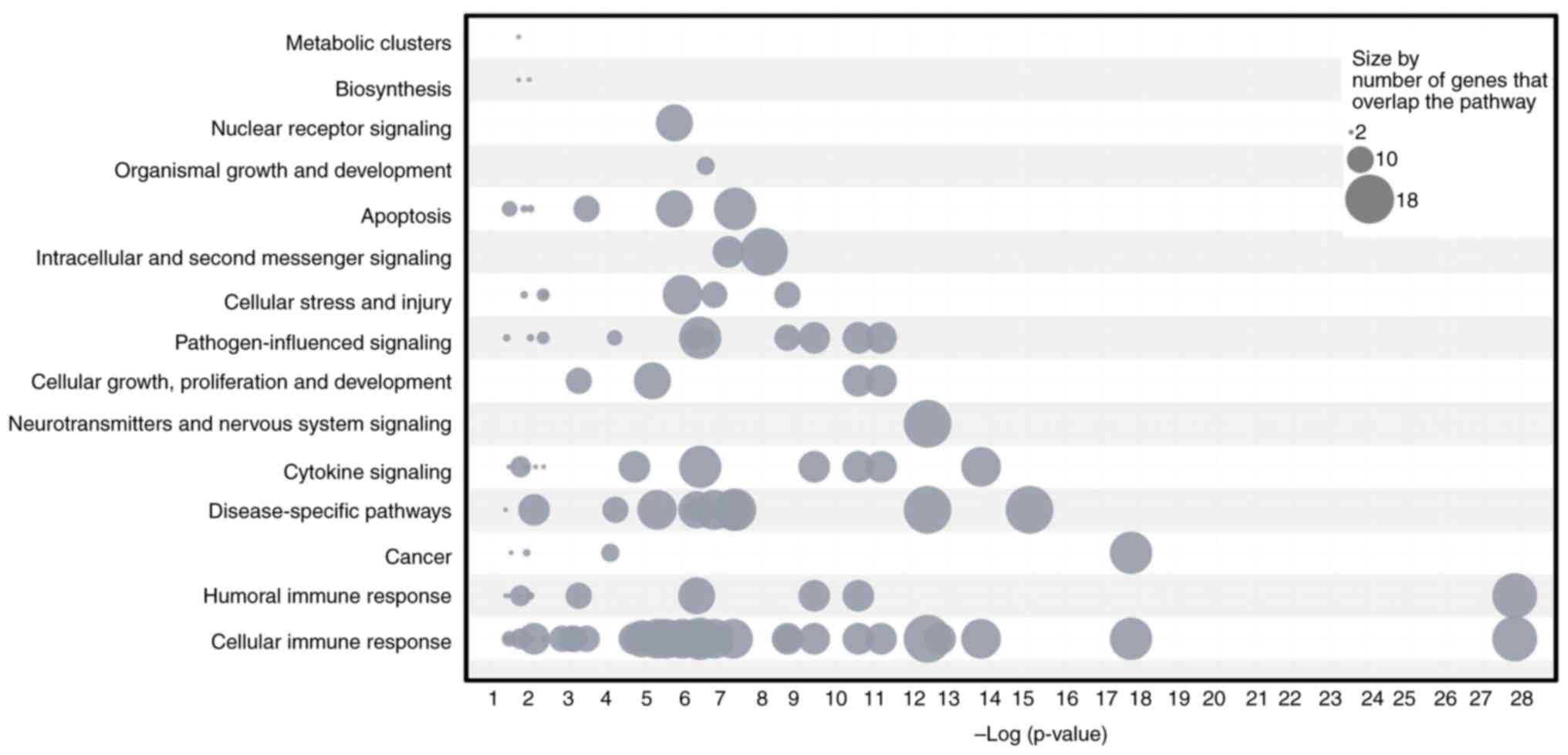
Table V. Furthermore, gene numbers
were cross-analyzed with pathways, resulting in the ranking of
various biological processes: Cellular immune response, humoral
immune response, cytokine signaling, cancer, disease-specific
pathways, neurotransmitters and nervous system signaling, cellular
growth, proliferation and development of neurotransmitters,
pathogen-influenced signaling, cellular stress and injury,
intracellular and second messenger signaling, apoptosis, organismal
growth and development, nuclear receptor signaling, biosynthesis,
and metabolic clusters (Fig. 3).
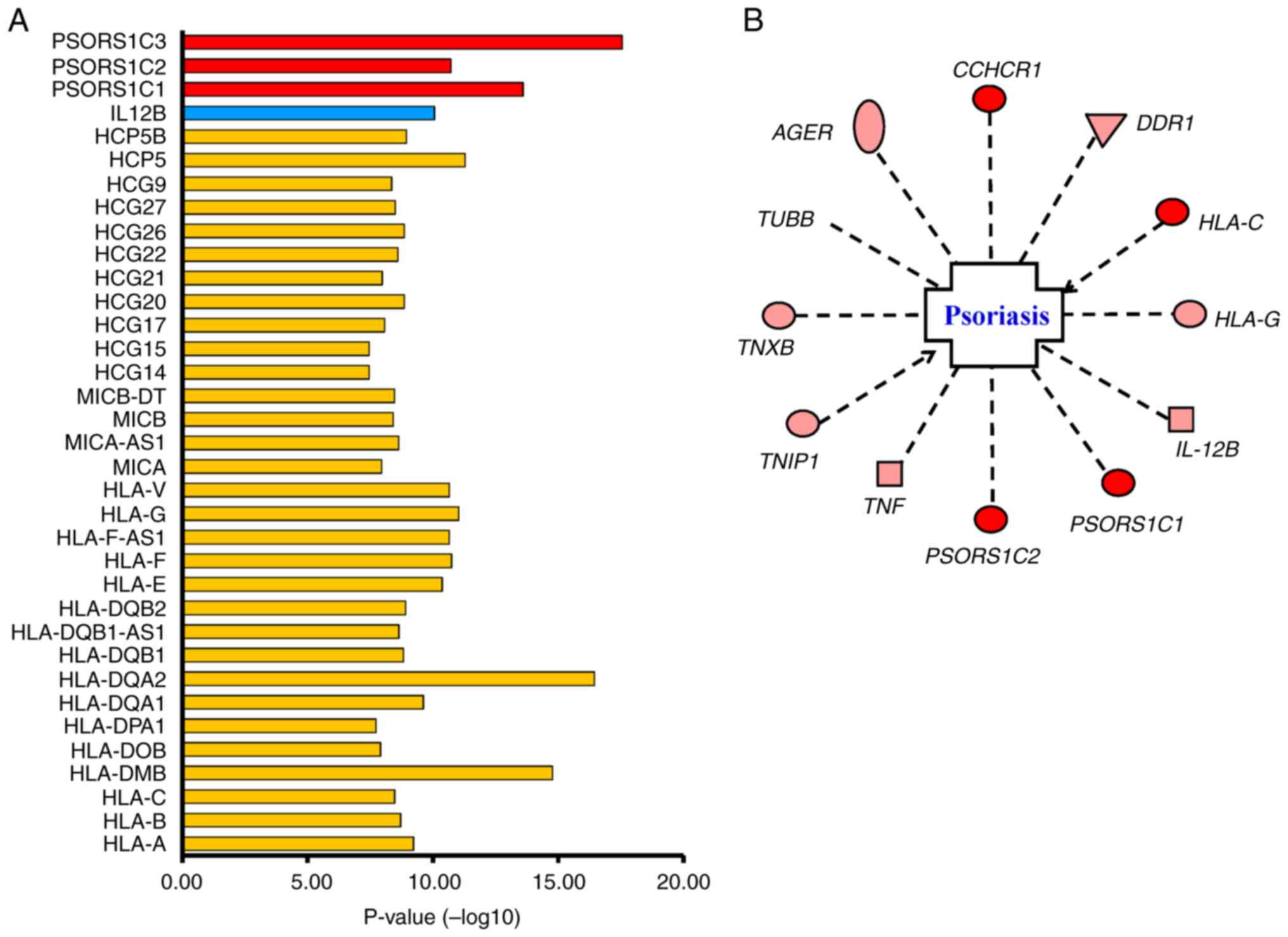
Our GWAS and network analysis findings suggested that the diversity
of specific genes, including HLA-A, HLA-C, HLA-DM, HLA-DP,
HLA-DQ, psoriasis susceptibility 1 (PSORS1), HLA complex
group on chromosome 6 and IL-12B on chromosome 5, play a
crucial role in the development of psoriasis (Fig. 4A). Additionally, our results showed
significant association with psoriasis between AGER,
discoidin domain receptor tyrosine kinase 1 (DDR1), TNF,
coiled-coil α-helical rod protein 1 (CCHCR1), tubulin β
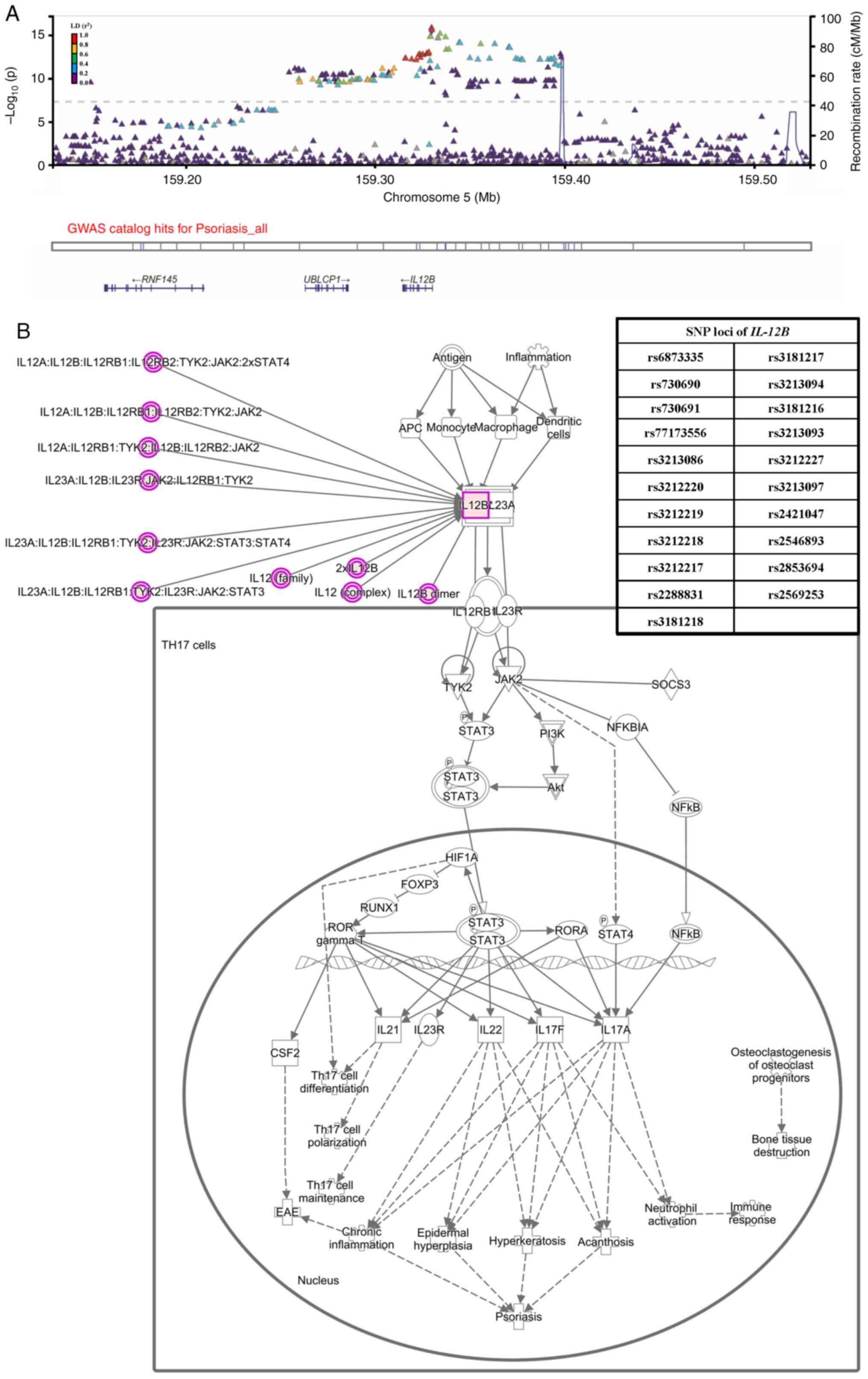
class I (TUBB) and TNXB genes (Fig. 4B). Furthermore, the regional
association plot (linkage disequilibrium score
r2>0.4) demonstrated a strong association between
variations in the IL12B gene and psoriasis (Fig. 5A). Finally, the psoriasis model
regulated by the IL12B/IL23 signaling pathway was analyzed using
the IPA database (Fig. 5B). The
analysis confirmed the presence of multiple highly correlated
IL12B genes and SNP sites, thereby providing substantial
evidence to support the existence of the IL-12 pathway.
To analyze the PRSs for psoriasis, the dataset was
divided into three sections: Base, target and validation, at a
ratio of 8:1:1. Each group was treated as a separate entity.
Summary statistics were calculated using data from the base group,
and a model was constructed using data from the target group. Data
from the validation group were used to assess the accuracy of the
model. A total of 1,684 SNPs with a significance level of
P<0.001 were chosen from a pool of 865 genes. The raw data for
the PRS models are presented in Table
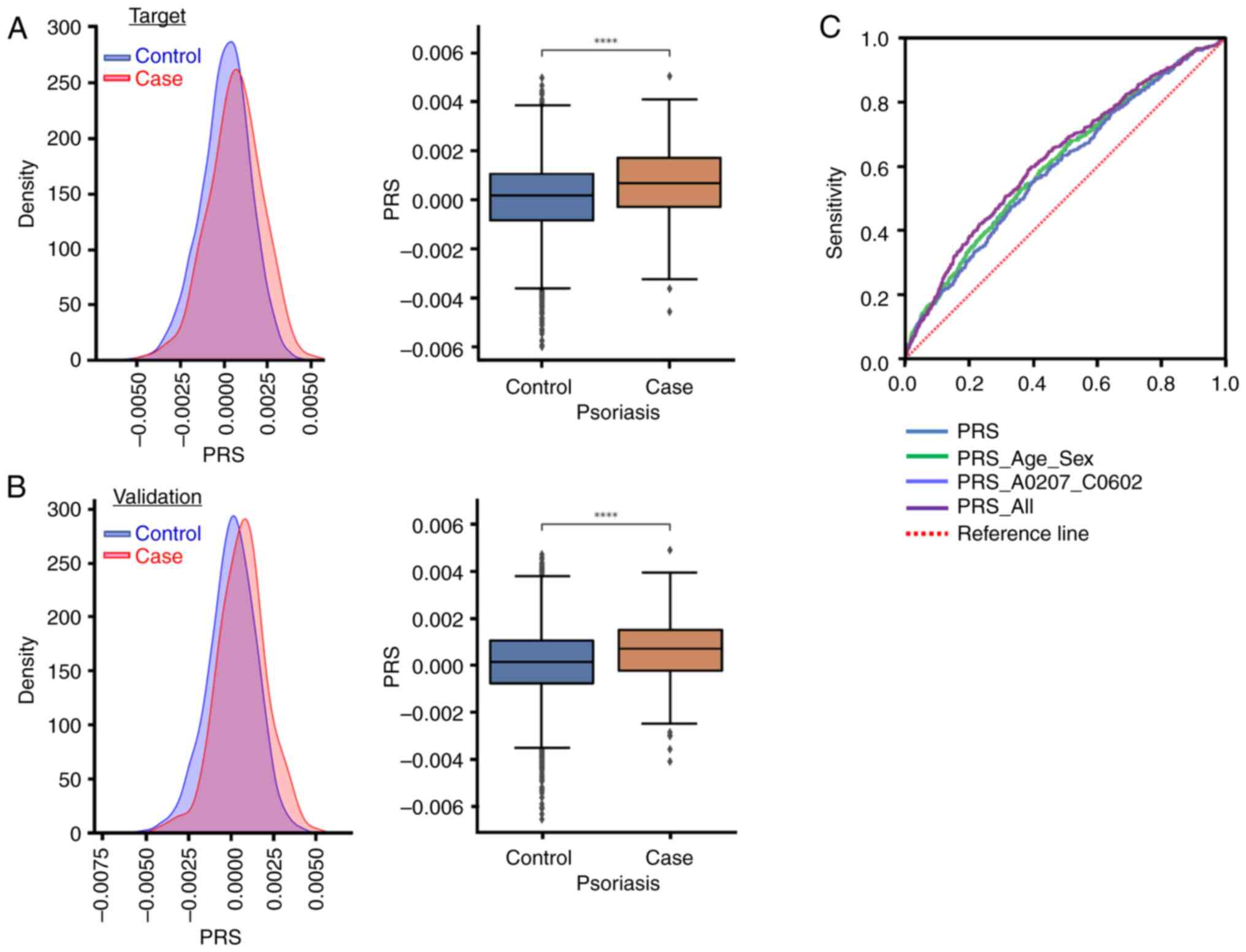
SIII. Fig. 6A and B illustrate
the PRS distribution and statistical findings applicable to the
target and validation groups. Fig.
6C shows the receiver operating characteristic curve of the PRS
for the prediction of psoriasis. The data indicated that patients
with psoriasis exhibited a significantly higher PRS than those in
the control group (P<0.001). Table
VI presents an analysis of the area under the curve (AUC) for
the PRS for psoriasis. The AUC of the PRS alone model was 0.611
[95% confidence interval (CI), 0.584–0.638], while that of the PRS
with age and sex combined was 0.611 (95% CI, 0.584–0.638). In
addition, the AUC of PRS with HLA-A*02:07 and
HLA-C*06:02 combined was 0.598 (95% CI, 0.571–0.624), while
that of PRS combined with age, sex, HLA-A*02:07 and
HLA-C*06:02 was 0.629 (95% CI, 0.602–0.656). It was
demonstrated that these SNPs collectively represent major risk
factors for the development of psoriasis. Furthermore,
HLA-A*02:07 and HLA-C*06:02 provided considerable
discriminatory ability, making a significant contribution to the
risk of psoriasis.
Finally, a meta-analysis was conducted in
collaboration with the BBJ to investigate the genetic relationships
within the East Asian population with respect to psoriasis. The
psoriasis GWAS from Biobank Japan (BBJ) was published previously
(29). The summary statistics of
psoriasis were downloaded from BBJ to replicate our results.
Table SIV presents unprocessed
results from the meta-analysis of SNPs. Upon completion of the
meta-analysis, the top 20 SNPs retained their genome-wide
significance (Table VIII). In
the BBJ cohort, results identified 438 significant single
nucleotide polymorphisms associated with psoriasis, with a
significance level of P<1×10−5. The majority of these
SNPs are located on chromosome 6. By conducting a meta-analysis of
the GWAS at key SNP sites within the BBJ Cohort, the persistent
significance of these SNPs wAS confirmed. This validates the
robustness and reliability of the present findings.
Psoriasis is a complex disease involving various
pathogenic and immunological mechanisms (17,96).
Although >100 psoriasis susceptibility loci have been identified
through GWAS in different countries and ethnicities (96–101), there have been limited studies on
genetic susceptibility loci in Taiwan (102,103). The present study conducted GWAS
and PRS to identify 92 novel genomic markers and identified 61
previously reported genomic markers associated with psoriasis.
Through network analysis, several biological processes were
discovered that contribute to psoriasis. These processes involve
factors such as HLA subtypes and genetic SNPs that affect
cytokines, and the PSORS1 locus. Additionally, it is
possible that transcriptional regulators and shared pathogenic
processes as well as other human diseases are involved in the
development of psoriasis.
The immune response plays an important role in the
development of psoriasis, and HLA polymorphisms have been
implicated in this process (104,105). Major histocompatibility complex
(MHC) molecules are present on every cell and provide peptide
antigens to CD4+ and CD8+ T cells (106,107). The length of these antigens is
usually 8–10 amino acids (108).
The presence of HLA polymorphisms leads to variable peptide-binding
grooves that contain two or three specific acceptor sites or
pockets (109). HLA molecules
present a variety of antigenic peptides by binding to specific
amino acid side chains (110).
The acceptor sites on HLA molecules differ from one another,
resulting in different peptide repertoires being presented for
different HLA molecules (109,110). The antigenic peptides contained
in each HLA molecule are unique, despite some overlap in binding
specificities (111,112). Certain HLA alleles, such as
HLA-A (61), HLA-B
(62,63), HLA-C (53,54),
HLA-DMB (64),
HLA-DQA1 (65),
HLA-DQA2 (66),
HLA-DQB1 (67),
HLA-DRB1 (67),
HLA-E (68), HLA-F
(69) and HLA-G (70), are more prevalent in patients with
psoriasis. The findings of the present study indicated a
significant association between the genetic variability of
HLA-related genes located on chromosome 6 and the
development of psoriasis. It was confirmed that certain HLA
genes, including HLA-DPA1, HLA-E, HLA-F, HLA-G, MICA and
MICB have amino acid mutations (68,113–115). The findings from the present
study indicated a notable association between HLA-A*02:07
and HLA-C*06:02 alleles and psoriasis in the Taiwanese
population. The HLA-C*06:02 allele is the most prominent
risk factor for psoriasis (116–119). The risk allele frequencies of
psoriasis have been reported to be 6–17% in Taiwan, 8–26% in Japan,
76.1% in Korea, 14% in Thailand and 46–67% in Caucasian populations
(120). Shen et al
demonstrated a significant association between psoriasis and
certain HLA alleles, including HLA-A*02:07 and
HLA-C*06:02, in Chinese patients with psoriasis in Singapore
(121). Furthermore, the
HLA-A*02:07 allele is significantly associated with the
development of psoriasis in individuals residing in southern China
(122). In Japan, psoriasis
vulgaris has been reported to be associated with HLA-A*02:07
and HLA-C*06:02 alleles (123). The HLA-Cw*06 allele is
associated with the highest degree of susceptibility to psoriasis
(116,124,125). It associates with the occurrence
of psoriasis as well as with improved treatment outcomes with
methotrexate, IL-12, IL-17 and IL-23 targeted therapeutic agents
(126). Furthermore,
HLA-C*06:02 has been identified as a biomarker for
predicting the response to biological treatment in patients with
psoriasis (116). The present
study also suggested that HLA-A*02:07 and HLA-C*06:02
significantly contribute to the risk of psoriasis, as demonstrated
by the PRS analysis.
Pro-inflammatory cytokines, including IL-12, IL-23
and TNF, play crucial roles in the development and progression of
psoriasis (72). IL-12B/IL-23 and
TNF are involved in inflammatory processes (72). IL-12B encodes for a subunit
of IL-12. IL-12 is a disulfide-linked heterodimer consisting of two
subunits: A 40 kD cytokine receptor-like subunit encoded by
IL-12B and a 35 kD subunit encoded by IL-12A
(127). IL-12 exerts its effects
on T and natural killer cells, leading to the activation of these
immune cell populations (128).
It is expressed by activated macrophages, which play a crucial role
in Th1 cell development. IL-12 is essential for maintaining an
adequate number of memory/effector Th1 cells to provide long-term
protection against intracellular pathogens (129). IL-23 cytokines are composed of
two subunits, IL-23A and IL-12B, both of which play significant
roles in various biological functions (127). IL-12B and IL-23 are particularly
important in the differentiation of T cells, specifically, the
generation of Th1 cells that produce IFN-γ and Th17 cells that
produce IL-17 (72). Previously,
it was observed that IL-12 is overexpressed on dendritic cells in
the skin lesions of patients with psoriasis (130). Moreover, ustekinumab, an
FDA-approved monoclonal antibody targeting IL-12/23p40, has shown
great efficacy in treating patients with psoriasis (99,131). In the present study a strong
association between variations in IL-12B levels was
identified in the psoriasis group. Previous studies identified
several SNPs associated with psoriasis in genes such as
IL-12 (rs3212220, rs3212217 and rs3212227),
IL-23/IL-23R and IL-17 (89,132). However, the present study did not
find any polymorphisms at the SNP loci of IL-23 or
IL-17. Overall, our findings suggested that IL-12B
and its associated SNPs on chromosome 5 are important in the
pathogenesis of psoriasis. Further investigation of these pathways
may provide valuable insights into the mechanisms underlying the
disease.
Previous studies have shown that HSPA1A/HSPA1B and
TRIM15 play roles in the pathogenesis of psoriasis (71,91).
HSPA1A and HSPA1B are heat shock proteins that regulate the
secretion of TNF-α, IL-1β and IL-10 from monocytes (133). TRIM15, on the other hand,
stimulates TNF-α and is involved in the TNF-α/NF-κB pathway,
contributing to the inflammatory response (91). In the present study psoriasis was
associated with SNPs in TNF (rs1800629) and TNIP1
(rs76956521, rs2233278, rs75851973 and rs8177833). Our study
established a connection between HSPA1A/HSPA1B, TRIM15 and
psoriasis. Additionally, several genes associated with enzymes and
kinases were identified including AGPAT1, ATAT1, BAG6, CARMIL1,
DHX16, DDAH2, GPX5, GPX6, NEU1, PGBD1, PPP1R11, SKIC2, VARS1
and VARS2, as SNP loci. Currently, there is no definitive
evidence linking enzymatic activity with psoriasis, which requires
further investigation. Future studies should, therefore, prioritize
research on these genes.
Familial recurrence of psoriasis has been
extensively studied, and it has been found that monozygotic twins
are more likely to have the disease than dizygotic twins (134). The main genetic factor
responsible for psoriasis is PSORS1, which is located within
the MHC on chromosome 6p21.3 (135,136). This region spans a range of
80–250 kb (137). The
PSORS1 locus contains several genes including HLA-C,
MICA, PSORS1C3 and CDSN. These candidate genes have
alleles within the PSORS1 locus and have been shown to be
expressed in skin cells (138).
HLA-C is responsible for presenting peptides to cytotoxic T cells
(CD8+T cells) by interacting with them on the cell
membrane, which activates the immune response and cytotoxicity of
cytotoxic T cells (139). MICA is
a cell-surface glycoprotein encoded by the MICA gene located
within the MHC locus. It is recognized by NK cells, γδ T cells and
CD8+ αβ T cells, which carry the NKG2D receptor on their
cell surfaces (140). CDSN is
primarily expressed in the upper layers of epidermis and hair
follicles. It contributes to keratinocyte cohesion, and is targeted
by proteases during epidermal desquamation (141). PSORS1C3 is a non-coding
gene and its RNA transcript is found in patients with psoriasis.
The functional role of PSORS1C3 is to modulate the
inflammatory response (142). An
association between PSORS1C3 polymorphisms and psoriasis has
been reported in various populations. Additionally, the specific
alleles of HLA-Cw*0602 and CDSN*5 consistently demonstrated a
significant association with psoriasis (142,143). Tawfik et al (144) conducted a study that showed an
association between psoriasis and three variants (rs10484554,
rs887466 and rs1062470), within the PSORS1 locus. These
variants are associated with LOC105375015, PSORS1C3 and
PSORS1C1 (137).
Additionally, GWAS identified 36 regions that contributed to
psoriasis susceptibility. However, >50% of the genetic variance
is attributed to a single MHC locus, specifically
PSORS1 (145). One of the
candidate genes in psoriasis is HLA-C, as demonstrated by a
GWAS performed using markers linked to HLA-Cw*0602 (145). However, some studies have
suggested that PSORS1 does not play a role in the
development of late-onset psoriasis (143,146,147). In the present study, a connection
between psoriasis and specific genetic markers was discovered,
namely, PSORS1C1, PSORS1C2, PSORS1C3, MICA and CDSN.
These genes have been linked to various factors, such as human
keratinocyte differentiation (53,148–150), the inflammatory response
(112,151–153) and in medication toxicity,
particularly the Stevens-Johnson syndrome associated with
allopurinol (142,154,155).
In Taiwan, few studies have been conducted on the
use of PRS for the treatment of psoriasis. Evidence suggests that
diabetes mellitus is increasingly prevalent among patients with
psoriasis (22,156,157). Eiris et al (158) showed a significant association
between three SNPs (rs6887695, rs3212227 and rs2201841) and
diabetes mellitus. Recent GWAS findings have indicated a genetic
association between psoriasis and autoimmune diseases (such as
multiple sclerosis, rheumatoid arthritis and autoimmune
hypothyroidism), neuromuscular diseases (including Alzheimer's and
Parkinson's diseases) (159–162), chronic inflammation (163) and skin diseases (164). Future investigations will aim to
conduct a transdisease meta-analysis and Mendelian randomization to
determine the causal relationship between psoriasis and the
aforementioned diseases. It is important to note that the present
study was subject to numerous limitations, including the risk of
false positives and false negatives. It cannot be confirmed whether
participants in the present were diagnosed with psoriasis or other
autoimmune diseases at different hospitals, leading to such
limitations in the present study. Additionally, due to the reliance
on retrospective medical record reviews for selecting the
experimental and control groups, accurate classification of the
severity of psoriasis could not be performed. It is also
hypothesized that the severity of psoriasis is highly associated
with genetics. Therefore, incorporating severity into future
studies will likely significantly enhance the efficacy of the PRS
model.
In conclusion, the present study has made
significant discoveries regarding psoriasis in Taiwan. This
confirms the influence of genetic variation and susceptibility on
the development of psoriasis. Loci such as the HLA region, PSORS1
and IL-12B were identified that are strongly associated with
psoriasis. The findings also showed an association between specific
alleles, such as HLA-A*02:07 and HLA-C*06:02, and HLA genotypes,
highlighting the role of genetic factors. Analysis of the PRS
indicates that the location of SNPs can accurately predict
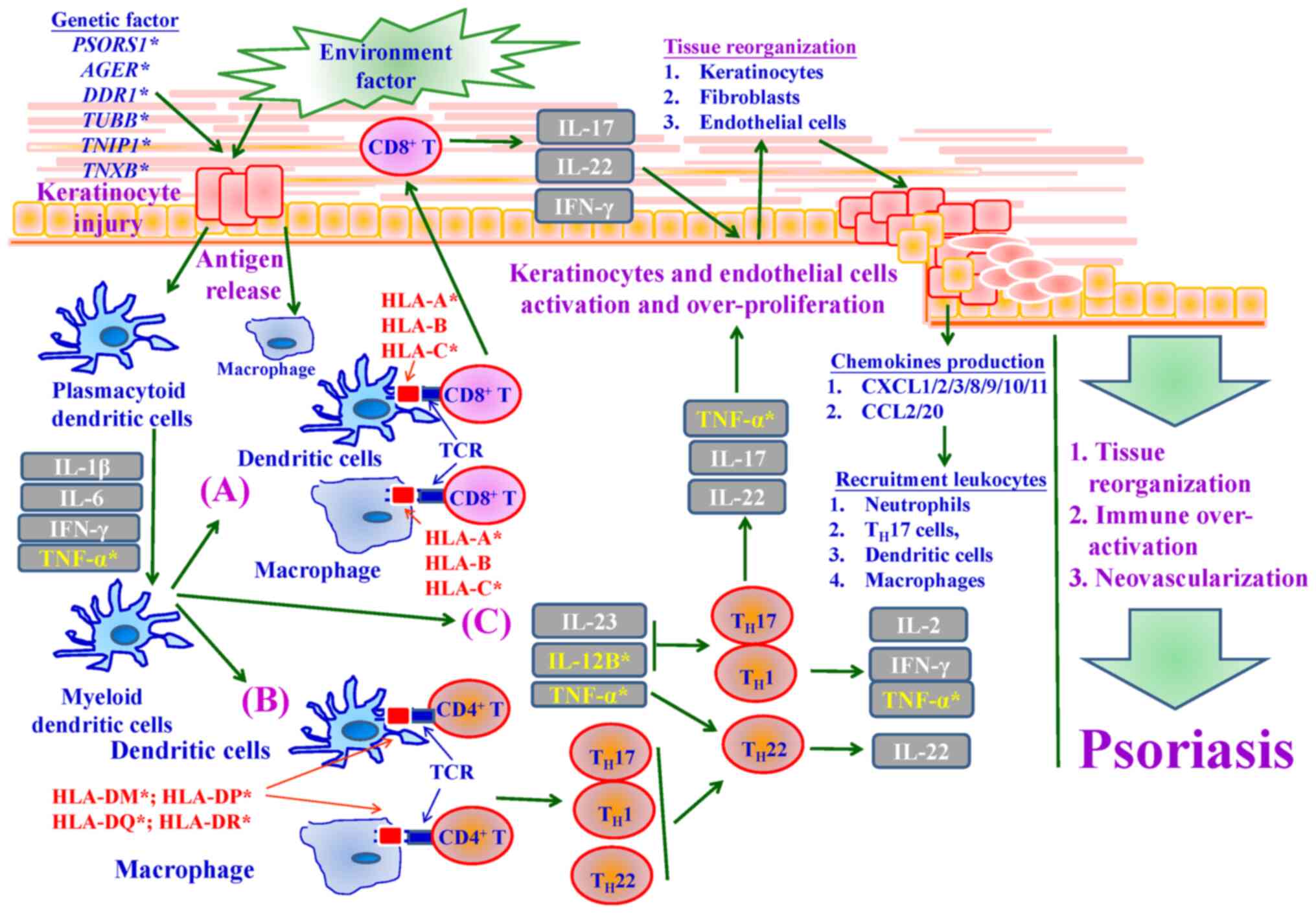
psoriasis occurrence. Fig. 8 shows
the importance of pathological mechanisms and signaling pathways in
psoriasis development, as shown by the GWAS results of the present
study. Importantly, the present study is the first to link multiple
genetic loci in Taiwanese individuals with the onset of psoriasis.
Overall, the present research emphasizes the critical role of
genetic factors and signaling pathways in psoriasis development,
providing valuable insights for future investigations and potential
therapeutic interventions.
The authors would like to thank the Office of
Research and Development, China Medical University (Taichung,
Taiwan) for providing Medical Research Core Facilities to perform
the experiments and data analysis. The authors also thank Dr
Kuan-Wen Chen and Dr Yao-Wei Jheng from GGA Corporation's Molecular
Science and Digital Innovation Center in Taipei, Taiwan, who made
contributions to the biological significance of the figures and
tables presented in this paper through their in-depth analysis and
interpretation. Their analysis was instrumental in accurately
deciphering the data.
This work was supported in part by China Medical University
Hospital, Taiwan (grant no. DMR-113-109) and The Ministry of
Science and Technology, Taiwan (grant no. MOST
111-2314-B-075-083-MY2).
JSY, TYL and FJT were responsible for the overall
conception and design. JSY, TYL and HFL performed the acquisition
of data. TYL, JSY and YWW performed the GWAS, PRS and PheWAS
analyses. TYL, HFL and WLL performed the interpretation of GWAS,
PRS and PheWAS results. SCT, JSY and YJC performed the analysis of
the bioinformatics network and interpreted the data. TYL and WLL
assessed the HLA diplotypes, and performed the allele frequency
analysis and interpretation of data. HFL and YWW performed the
meta-analysis and interpretation of data. YJC and WLL performed the
interpretation of clinical pathological mechanisms. JSY and FJT
confirm the authenticity of all the raw data. All authors have read
and approved the final version of the manuscript.
The study protocol was approved by the
Institutional Review Board of China Medical University Hospital and
categorized as the Precision Medicine Project (CMUHPMP) (IRB
number: CMUH110-REC3-005 and CMUH111-REC1-176). Patients have been
granted access to their medical records by the CMUH IRB. The CMUH
IRB also places considerable emphasis on ensuring patient
confidentiality. De-identified genetic and clinical data were
collected after obtaining informed consent from patients. The
present study complied with The Declaration of Helsinki.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Balda A, Wani I, Roohi TF, Suman, Krishna
KL, Mehdi S, Nadiga AP, Makkapati M and Baig MAI: Psoriasis and
skin cancer-Is there a link? Int Immunopharmacol. 121:1104642023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chi CC, Wu YW, Chao TH, Chen CC, Chen YJ,
Cheng HM, Chiu HY, Chiu YW, Chung WH, Hsieh TY, et al: 2022
Taiwanese Dermatological Association (TDA), Taiwanese Association
for Psoriasis and Skin Immunology (TAPSI), and Taiwan Society of
cardiology (TSOC) joint consensus recommendations for the
management of psoriatic disease with attention to cardiovascular
comorbidities. J Formos Med Assoc. 122:442–457. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Michalek IM, Loring B and John SM: A
systematic review of worldwide epidemiology of psoriasis. J Eur
Acad Dermatol Venereol. 31:205–212. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang TS, Hsieh CF and Tsai TF:
Epidemiology of psoriatic disease and current treatment patterns
from 2003 to 2013: A nationwide, population-based observational
study in Taiwan. J Dermatol Sci. 84:340–345. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kubota K, Kamijima Y, Sato T, Ooba N,
Koide D, Iizuka H and Nakagawa H: Epidemiology of psoriasis and
palmoplantar pustulosis: A nationwide study using the Japanese
national claims database. BMJ Open. 5:e0064502015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee JY, Kang S, Park JS and Jo SJ:
Prevalence of psoriasis in Korea: A population-based
epidemiological study using the Korean National Health Insurance
Database. Ann Dermatol. 29:761–767. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ding X, Wang T, Shen Y, Wang X, Zhou C,
Tian S, Liu Y, Peng G, Zhou J, Xue S, et al: Prevalence of
psoriasis in China: A population-based study in six cities. Eur J
Dermatol. 22:663–667. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang SF, Lin MH, Chou PC, Hu SK, Shih SY,
Yu HS and Yu S: Genetics of generalized pustular psoriasis: Current
understanding and implications for future therapeutics. Genes
(Basel). 14:12972023. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee LL, Huo AP and Chen SL: Experiences
and coping behaviors of patients with psoriasis: A qualitative
study. J Dermatolog Treat. 34:21936612023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang CY, Wang CW, Chen CB, Chen WT, Chang
YC, Hui RC and Chung WH: Pharmacogenomics on the treatment response
in patients with psoriasis: An updated review. Int J Mol Sci.
24:73292023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Honma M and Hayashi K: Psoriasis: Recent
progress in molecular-targeted therapies. J Dermatol. 48:761–777.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Furue M and Kadono T: Psoriasis: Behind
the scenes. J Dermatol. 43:4–8. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rendon A and Schakel K: Psoriasis
pathogenesis and treatment. Int J Mol Sci. 20:14752019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ohata C, Anezaki H, Kaneko S, Okazaki F,
Ito K, Matsuzaka Y, Kikuchi S, Koike Y, Murota H, Miyagi T, et al:
Clinical characteristics of patients with psoriasis with family
history: A multicenter observational study. J Dermatol. 50:746–752.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bayaraa B and Imafuku S: Relationship
between environmental factors, age of onset and familial history in
Japanese patients with psoriasis. J Dermatol. 45:715–718. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kaur I, Handa S and Kumar B: Natural
history of psoriasis: A study from the Indian subcontinent. J
Dermatol. 24:230–234. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kocaaga A and Kocaaga M: Psoriasis: An
Immunogenetic perspective. Glob Med Genet. 9:82–89. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Elyoussfi S, Rane SS, Eyre S and Warren
RB: TYK2 as a novel therapeutic target in psoriasis. Expert Rev
Clin Pharmacol. 16:549–558. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yarmolinsky J, Amos CI, Hung RJ, Moreno V,
Burrows K, Smith-Byrne K, Atkins JR, Brennan P; Colon Cancer Family
Registry (CCFR), Colorectal Cancer Transdisciplinary Study
(CORECT), ; et al: Association of germline TYK2 variation with lung
cancer and non-Hodgkin lymphoma risk. Int J Cancer. 151:2155–2160.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Enerback C, Sandin C, Lambert S,
Zawistowski M, Stuart PE, Verma D, Tsoi LC, Nair RP, Johnston A and
Elder JT: The psoriasis-protective TYK2 I684S variant impairs IL-12
stimulated pSTAT4 response in skin-homing CD4+ and CD8+ memory
T-cells. Sci Rep. 8:70432018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hao L, Kraft P, Berriz GF, Hynes ED, Koch
C, Korategere V Kumar P, Parpattedar SS, Steeves M, Yu W, et al:
Development of a clinical polygenic risk score assay and reporting
workflow. Nat Med. 28:1006–1013. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao WL, Huang YN, Chang YW, Liu TY, Lu
HF, Tiao ZY, Su PH, Wang CH and Tsai FJ: Combining polygenic risk
scores and human leukocyte antigen variants for personalized risk
assessment of type 1 diabetes in the Taiwanese population. Diabetes
Obes Metab. 25:2928–2936. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Janssens ACJW: Validity of polygenic risk
scores: Are we measuring what we think we are? Hum Mol Genet.
28:R143–R150. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cross B, Turner R and Pirmohamed M:
Polygenic risk scores: An overview from bench to bedside for
personalised medicine. Front Genet. 13:10006672022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dixon P, Keeney E, Taylor JC, Wordsworth S
and Martin RM: Can polygenic risk scores contribute to
cost-effective cancer screening? A systematic review. Genet Med.
24:1604–1617. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Byrne L and Toland AE: Polygenic risk
scores in prostate cancer risk assessment and screening. Urol Clin
North Am. 48:387–399. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lambert SA, Abraham G and Inouye M:
Towards clinical utility of polygenic risk scores. Hum Mol Genet.
28:R133–R142. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Johnson D, Wilke MAP, Lyle SM, Kowalec K,
Jorgensen A, Wright GEB and Drögemöller BI: A systematic review and
analysis of the use of polygenic scores in pharmacogenomics. Clin
Pharmacol Ther. 111:919–930. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sakaue S, Kanai M, Tanigawa Y, Karjalainen
J, Kurki M, Koshiba S, Narita A, Konuma T, Yamamoto K, Akiyama M,
et al: A cross-population atlas of genetic associations for 220
human phenotypes. Nat Genet. 53:1415–1424. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen TC, Wang TC, Yiu ZZN, Lee MS, Chen
LC, Chan KA, Griffiths CEM and Ashcroft DM; Global Psoriasis Atlas
(GPA), : Risk of serious infection and infection mortality in
patients with psoriasis: A nationwide cohort study using the Taiwan
National Health Insurance claims database. J Eur Acad Dermatol
Venereol. 38:136–144. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hsu MC, Wang CC, Huang LY, Lin CY, Lin FJ
and Toh S: Effect of ICD-9-CM to ICD-10-CM coding system transition
on identification of common conditions: An interrupted time series
analysis. Pharmacoepidemiol Drug Saf. 30:1653–1674. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang YH, Tang CH, Goh CH, Chang CL, Qiu
H, Yang YW, Saadoun C, Chang CL and Liu Y: Persistence and
adherence to biologics in patients with psoriasis in Taiwan: A new
biologics user cohort study. Front Pharmacol. 13:8809852022.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wei JC, Shi LH, Huang JY, Wu XF, Wu R and
Chiou JY: Epidemiology and medication pattern change of psoriatic
diseases in Taiwan from 2000 to 2013: A nationwide,
population-based cohort study. J Rheumatol. 45:385–392. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee WC, Wang LY and Cheng KF: An
easy-to-implement approach for analyzing case-control and case-only
studies assuming gene-environment independence and Hardy-Weinberg
equilibrium. Stat Med. 29:2557–2567. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chiou JS, Cheng CF, Liang WM, Chou CH,
Wang CH, Lin WD, Chiu ML, Cheng WC, Lin CW, Lin TH, et al: Your
height affects your health: genetic determinants and health-related
outcomes in Taiwan. BMC Med. 20:2502022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Genomes Project Consortium, . Auton A,
Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL,
McCarthy S, McVean GA and Abecasis GR: A global reference for human
genetic variation. Nature. 526:68–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu HF, Chou CH, Lin YJ, Uchiyama S, Terao
C, Wang YW, Yang JS, Liu TY, Wong HS, Chen SC and Tsai FJ: The
genome-wide association study of serum IgE levels demonstrated a
shared genetic background in allergic diseases. Clin Immunol.
260:1098972024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu TY, Lin CF, Wu HT, Wu YL, Chen YC,
Liao CC, Chou YP, Chao D, Chang YS, Lu HF, et al: Comparison of
multiple imputation algorithms and verification using whole-genome
sequencing in the CMUH genetic biobank. Biomedicine (Taipei).
11:57–65. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang JS, Liu TY, Chen YC, Tsai SC, Chiu
YJ, Liao CC and Tsai FJ: Genome-Wide association study of alopecia
areata in Taiwan: The conflict between individuals and hair
follicles. Clin Cosmet Investig Dermatol. 16:2597–2612. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu CY, Hu HY, Li CP, Chou YJ and Chang YT:
Comorbidity profiles of psoriasis in Taiwan: A latent class
analysis. PLoS One. 13:e01925372018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cheng YD, Lu CC, Hsu YM, Tsai FJ, Bau DT,
Tsai SC, Cheng CC, Lin JJ, Huang YY, Juan YN, et al: In Silico and
In Vitro studies of Taiwan Chingguan Yihau (NRICM101) on
TNF-α/IL-1β-induced human lung cells. Biomedicine (Taipei).
12:56–71. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang JS, Kang CY, Su CH, Chen CJ, Chiu YJ
and Hsu YM: Helicobacter pylori Targets in AGS Human gastric
adenocarcinoma: In situ proteomic profiling and systematic
analysis. Anticancer Res. 42:531–546. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liao WL, Liu TY, Cheng CF, Chou YP, Wang
TY, Chang YW, Chen SY and Tsai FJ: Analysis of HLA variants and
graves' disease and its comorbidities using a high resolution
imputation system to examine electronic medical health records.
Front Endocrinol (Lausanne). 13:8426732022. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu TY, Liao WL, Wang TY, Chan CJ, Chang
JG, Chen YC, Lu HF, Yang HH, Chen SY and Tsai FJ: Genome-wide
association study of hyperthyroidism based on electronic medical
record from Taiwan. Front Med (Lausanne). 9:8306212022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Willer CJ, Li Y and Abecasis GR: METAL:
Fast and efficient meta-analysis of genomewide association scans.
Bioinformatics. 26:2190–2191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chiu HY, Tsai SC, Tsai FJ, Lo YH, Cheng
CC, Liu TY, Jhan SR, Yang JS and Chiu YJ: Liraglutide With
Metformin Therapy Ameliorates Hepatic Steatosis and Liver Injury in
a Mouse Model of Non-alcoholic Steatohepatitis. In Vivo.
37:1037–1046. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Takenaka S, Itoh T and Fujiwara R:
Expression pattern of human ATP-binding cassette transporters in
skin. Pharmacol Res Perspect. 1:e000052013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Amagai R, Takahashi T, Terui H, Fujimura
T, Yamasaki K, Aiba S and Asano Y: The antimicrobial peptide
cathelicidin exerts immunomodulatory effects via scavenger
receptors. Int J Mol Sci. 24:8752023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Orsmark C, Skoog T, Jeskanen L, Kere J and
Saarialho-Kere U: Expression of allograft inflammatory factor-1 in
inflammatory skin disorders. Acta Derm Venereol. 87:223–227. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang F, Han L, Wang B, Huang Q, Yawalkar
N, Zhang Z and Yan K: Annexin A6 polymorphism is associated with
pro-atherogenic lipid profiles and with the downregulation of
methotrexate on anti-atherogenic lipid profiles in psoriasis. J
Clin Med. 11:70792022. View Article : Google Scholar
|
|
51
|
Slivka PF, Hsieh CL, Lipovsky A, Pratt SD,
Locklear J, Namovic MT, McDonald HA, Wetter J, Edelmayer R, Hu M,
et al: Small molecule and pooled CRISPR Screens Investigating IL17
Signaling Identify BRD2 as a novel contributor to keratinocyte
inflammatory responses. ACS Chem Biol. 14:857–872. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nakagawa H, Akazaki S, Asahina A, Tokunaga
K, Matsuki K, Kuwata S, Ishibashi Y and Juji T: Study of HLA class
I, class II and complement genes (C2, C4A, C4B and BF) in Japanese
psoriatics and analysis of a newly-found high-risk haplotype by
pulsed field gel electrophoresis. Arch Dermatol Res. 283:281–284.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Abbas Zadeh S, Mlitz V, Lachner J, Golabi
B, Mildner M, Pammer J, Tschachler E and Eckhart L: Phylogenetic
profiling and gene expression studies implicate a primary role of
PSORS1C2 in terminal differentiation of keratinocytes. Exp
Dermatol. 26:352–358. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sanchez F, Holm SJ, Mallbris L, O'Brien KP
and Stahle M: STG does not associate with psoriasis in the Swedish
population. Exp Dermatol. 13:413–418. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Acevedo F and Hammar H: Complement C3
proteins in psoriasis. Br J Dermatol. 121:329–335. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Fuentelsaz-Romero S, Cuervo A,
Estrada-Capetillo L, Celis R, García-Campos R, Ramírez J, Sastre S,
Samaniego R, Puig-Kröger A and Cañete JD: GM-CSF expression and
macrophage polarization in joints of undifferentiated arthritis
patients evolving to rheumatoid arthritis or psoriatic arthritis.
Front Immunol. 11:6139752021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nair RP, Stuart PE, Nistor I, Hiremagalore
R, Chia NVC, Jenisch S, Weichenthal M, Abecasis GR, Lim HW,
Christophers E, et al: Sequence and haplotype analysis supports
HLA-C as the psoriasis susceptibility 1 gene. Am J Hum Genet.
78:827–851. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
58
|
Knight J, Spain SL, Capon F, Hayday A,
Nestle FO, Clop A; Wellcome Trust Case Control Consortium; Genetic
Analysis of Psoriasis Consortium; I-chip for Psoriasis Consortium;
Barker JN, ; et al: Conditional analysis identifies three novel
major histocompatibility complex loci associated with psoriasis.
Hum Mol Genet. 21:5185–5192. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Rajesh D, Nagraj S, Kumar KSP, Kutty AVM
and Balakrishna S: Evaluation of HCP5 and Chemokine C Receptor type
5 gene polymorphisms in indian psoriatic patients. Indian J
Dermatol. 64:182–186. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li XL, Yu H and Wu GS: Investigating the
genetic association of HCP5, SPATA2, TNIP1, TNFAIP3 and COG6 with
psoriasis in Chinese population. Int J Immunogenet. 41:503–507.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Massy E, Pedini P, Pollet E, Martin M,
Roudier J, Picard C and Balandraud N: Association study between
HLA-A, -B, -C, -DRB1 alleles and Psoriatic arthritis in southern
France. Hum Immunol. 83:515–520. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cassia FF, Cardoso JF, Porto LC,
Ramos-E-Silva M and Carneiro S: Association of HLA Alleles and HLA
haplotypes with psoriasis, psoriatic arthritis and disease severity
in a miscegenated population. Psoriasis (Auckl). 11:41–51.
2021.PubMed/NCBI
|
|
63
|
Crivellato E and Zacchi T: The HLA system
and psoriasis: A family study. G Ital Dermatol Venereol.
120:247–254. 1985.(In Italian). PubMed/NCBI
|
|
64
|
Pyo CW, Hur SS, Kim YK, Kim TY and Kim TG:
Association of TAP and HLA-DM genes with psoriasis in Koreans. J
Invest Dermatol. 120:616–622. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Zhou X, He Y, Kuang Y, Chen W and Zhu W:
HLA-DQA1 and DQB1 alleles are associated with acitretin response in
patients with psoriasis. Front Biosci (Landmark Ed). 27:2662022.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhu KJ, Lv YM, Yin XY, Wang ZX, Sun LD, He
SM, Cheng H, Hu DY, Zhang Z, Li Y, et al: Psoriasis regression
analysis of MHC loci identifies shared genetic variants with
vitiligo. PLoS One. 6:e230892011. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Shawkatova I, Javor J, Parnicka Z, Kozub
P, Zilínková M, Frey P, Ferenčík S and Buc M: HLA-C, DRB1 and DQB1
alleles involved in genetic predisposition to psoriasis vulgaris in
the Slovak population. Folia Microbiol (Praha). 58:319–324. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Patel F, Marusina AI, Duong C, Adamopoulos
IE and Maverakis E: NKG2C, HLA-E and their association with
psoriasis. Exp Dermatol. 22:797–799. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tamiya G, Shiina T, Oka A, Tomizawa M, Ota
M, Katsuyama Y, Yoshitome M, Makino S, Kimura M and Inoko H: New
polymorphic microsatellite markers in the human MHC class I region.
Tissue Antigens. 54:221–228. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Aractingi S, Briand N, Le Danff C, Viguier
M, Bachelez H, Michel L, Dubertret L and Carosella ED: HLA-G and NK
receptor are expressed in psoriatic skin: A possible pathway for
regulating infiltrating T cells? Am J Pathol. 159:71–77. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lamore SD and Wondrak GT: Zinc pyrithione
impairs zinc homeostasis and upregulates stress response gene
expression in reconstructed human epidermis. Biometals. 24:875–890.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Vincken NLA, Welsing PMJ, Silva-Cardoso
SC, Bekker CPJ, Lopes AP, Olde Nordkamp M, Leijten EFA, Radstake
TRDJ and Angiolilli C: Suppression of IL-12/IL-23 p40 subunit in
the skin and blood of psoriasis patients by Tofacitinib is
dependent on active interferon-ү signaling in dendritic cells:
Implications for the treatment of psoriasis and interferon-driven
diseases. Exp Dermatol. 31:962–969. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Bojko A, Ostasz R, Bialecka M, Klimowicz
A, Malinowski D, Budawski R, Bojko P, Droździk M and Kurzawski M:
IL12B, IL23A, IL23R and HLA-C*06 genetic variants in psoriasis
susceptibility and response to treatment. Hum Immunol. 79:213–217.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Woo J and Lee C: Identification of
functional haplotypes in the promoter region of the LST1 gene.
Biochem Genet. 52:365–371. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Morelli M, Galluzzo M, Scarponi C, Madonna
S, Scaglione GL, Girolomoni G, Talamonti M, Bianchi L and Albanesi
C: Allelic Variants of HLA-C Upstream Region, PSORS1C3, MICA, TNFA
and genes involved in epidermal homeostasis and barrier function
influence the clinical response to Anti-IL-12/IL-23 treatment of
patients with psoriasis. Vaccines (Basel). 10:19772022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Morelli M, Galluzzo M, Madonna S, Scarponi
C, Scaglione GL, Galluccio T, Andreani M, Pallotta S, Girolomoni G,
Bianchi L, et al: HLA-Cw6 and other HLA-C alleles, as well as
MICB-DT, DDX58, and TYK2 genetic variants associate with optimal
response to anti-IL-17A treatment in patients with psoriasis.
Expert Opin Biol Ther. 21:259–270. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Lommers E, Depierreux F, Hansen I, Dive D
and Maquet P: NMOSD with anti-MOG antibodies following anti-TNFα
therapy: A case report. Mult Scler Relat Disord. 26:37–39. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Setsirichok D, Tienboon P, Jaroonruang N,
Kittichaijaroen S, Wongseree W, Piroonratana T, Usavanarong T,
Limwongse C, Aporntewan C, Phadoongsidhi M and Chaiyaratana N: An
omnibus permutation test on ensembles of two-locus analyses can
detect pure epistasis and genetic heterogeneity in genome-wide
association studies. Springerplus. 2:2302013. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Wu X, Dong H, Luo L, Zhu Y, Peng G,
Reveille JD and Xiong M: A novel statistic for genome-wide
interaction analysis. PLoS Genet. 6:e10011312010. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Frew JW: The contradictory inefficacy of
methotrexate in hidradenitis suppurativa: A need to revise
pathogenesis or acknowledge disease heterogeneity? J Dermatolog
Treat. 31:422–423. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Cai M, Huang H, Hu Z, Yuan T, Li W, Liu Y,
Zheng L, Zhang Y, Sheng Y and Zhang X: Two variants in the NOTCH4
and HLA-C genes contribute to familial clustering of psoriasis. Int
J Genomics. 2020:69073782020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Michailidis C, Karpouzis A, Kourmouli N,
Tripsianis G, Diplas A and Veletza S: Notch2, notch4 gene
polymorphisms in psoriasis vulgaris. Eur J Dermatol. 23:146–153.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Gao W, Sweeney C, Walsh C, Rooney P,
McCormick J, Veale DJ and Fearon U: Notch signalling pathways
mediate synovial angiogenesis in response to vascular endothelial
growth factor and angiopoietin 2. Ann Rheum Dis. 72:1080–1088.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Mehta K, Jaiswal P, Briggs F, Faubion WA,
Tabibian JH, Cominelli F and Dave M: In-patient outcomes of
hematopoietic stem cell transplantation in patients with immune
mediated inflammatory diseases: A nationwide study. Sci Rep.
8:68252018. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Chang YT, Liu HN, Shiao YM, Lin MW, Lee
DD, Liu MT, Wang WJ, Wu S, Lai CY and Tsai SF: A study of PSORS1C1
gene polymorphisms in Chinese patients with psoriasis. Br J
Dermatol. 153:90–96. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Skibola CF, Bracci PM, Halperin E, Conde
L, Craig DW, Agana L, Iyadurai K, Becker N, Brooks-Wilson A, Curry
JD, et al: Genetic variants at 6p21.33 are associated with
susceptibility to follicular lymphoma. Nat Genet. 41:873–875. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Rahman P, Roslin NM, Pellett FJ, Lemire M,
Greenwood CM, Beyene J, Pope A, Peddle L, Paterson AD, Uddin M and
Gladman DD: High resolution mapping in the major histocompatibility
complex region identifies multiple independent novel loci for
psoriatic arthritis. Ann Rheum Dis. 70:690–694. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Gregory AP, Dendrou CA, Attfield KE,
Haghikia A, Xifara DK, Butter F, Poschmann G, Kaur G, Lambert L,
Leach OA, et al: TNF receptor 1 genetic risk mirrors outcome of
anti-TNF therapy in multiple sclerosis. Nature. 488:508–511. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Nair RP, Duffin KC, Helms C, Ding J,
Stuart PE, Goldgar D, Gudjonsson JE, Li Y, Tejasvi T, Feng BJ, et
al: Genome-wide scan reveals association of psoriasis with IL-23
and NF-kappaB pathways. Nat Genet. 41:199–204. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Witoelar A, Jansen IE, Wang Y, Desikan RS,
Gibbs JR, Blauwendraat C, Thompson WK, Hernandez DG, Djurovic S,
Schork AJ, et al: Genome-wide Pleiotropy Between Parkinson Disease
and Autoimmune Diseases. JAMA Neurol. 74:780–792. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Roy M, Singh K, Shinde A, Singh J, Mane M,
Bedekar S, Tailor Y, Gohel D, Vasiyani H, Currim F and Singh R:
TNF-α-induced E3 ligase, TRIM15 inhibits TNF-alpha-regulated NF-κB
pathway by promoting turnover of K63 linked ubiquitination of TAK1.
Cell Signal. 91:1102102022. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Miao X, Xiang Y, Mao W, Chen Y, Li Q and
Fan B: TRIM27 promotes IL-6-induced proliferation and inflammation
factor production by activating STAT3 signaling in HaCaT cells. Am
J Physiol Cell Physiol. 318:C272–C281. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
He P, Cao RR, Deng FY and Lei SF:
Identification of potential pleiotropic genes for immune and
skeletal diseases using multivariate MetaCCA analysis. Curr
Genomics. 22:596–606. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Burkhart JG, Wu G, Song X, Raimondi F,
McWeeney S, Wong MH and Deng Y: Biology-inspired graph neural
network encodes reactome and reveals biochemical reactions of
disease. Patterns (NY). 4:1007582023. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Zhen Q, Yang Z, Wang W, Li B, Bai M, Wu J,
Ge H, Dong Z, Shen J, Tang H, et al: Genetic study on small
insertions and deletions in psoriasis reveals a role in complex
human diseases. J Invest Dermatol. 139:2302–2312 e14. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Gupta R, Debbaneh MG and Liao W: Genetic
epidemiology of psoriasis. Curr Dermatol Rep. 3:61–78. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Oh SM, Kim SK, Ahn HJ and Jeong KH: A
pilot genome-wide association study identifies novel markers of
metabolic syndrome in patients with psoriasis. Ann Dermatol.
35:285–292. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Chen W, Wang W, Yong L, Zhen Q, Yu Y, Ge
H, Mao Y, Cao L, Zhang R, Hu X, et al: Genome-wide meta-analysis
identifies ten new psoriasis susceptibility loci in the Chinese
population. J Genet Genomics. 49:177–180. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Connell WT, Hong J and Liao W: Genome-Wide
association study of ustekinumab response in psoriasis. Front
Immunol. 12:8151212022. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Ren Y, Wang L, Dai H, Qiu G, Liu J, Yu D,
Liu J, Lyu CZ, Liu L and Zheng M: Genome-wide association analysis
of anti-TNF-α treatment response in Chinese patients with
psoriasis. Front Pharmacol. 13:9689352022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Kisielnicka A, Sobalska-Kwapis M,
Purzycka-Bohdan D, Nedoszytko B, Zabłotna M, Seweryn M, Strapagiel
D, Nowicki RJ, Reich A, Samotij D, et al: The analysis of a
genome-wide association study (GWAS) of overweight and obesity in
psoriasis. Int J Mol Sci. 23:73962022. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Huang YH, See LC, Chang YC, Chung WH,
Chang LC, Yang SF and Su SC: Impact of ABCG2 gene polymorphism on
the predisposition to psoriasis. Genes (Basel). 12:16012021.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Chang YC, Wu WM, Huang YH, Chung WH, Tsai
HY and Hsu LA: The (CCTTT) n pentanucleotide repeat polymorphism in
the inducible nitric oxide synthase gene promoter and the risk of
psoriasis in Taiwanese. Arch Dermatol Res. 307:425–432. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Chandran V and Raychaudhuri SP:
Geoepidemiology and environmental factors of psoriasis and
psoriatic arthritis. J Autoimmun. 34:J314–J321. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Elder JT, Bruce AT, Gudjonsson JE,
Johnston A, Stuart PE, Tejasvi T, Voorhees JJ, Abecasis GR and Nair
RP: Molecular dissection of psoriasis: Integrating genetics and
biology. J Invest Dermatol. 130:1213–1226. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Sanchez-Mazas A: A review of HLA allele
and SNP associations with highly prevalent infectious diseases in
human populations. Swiss Med Wkly. 150:W202142020.PubMed/NCBI
|
|
107
|
Buhler S and Sanchez-Mazas A: HLA DNA
sequence variation among human populations: Molecular signatures of
demographic and selective events. PLoS One. 6:e146432011.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Melief CJ and Kast WM: Cytotoxic T
lymphocyte therapy of cancer and tumor escape mechanisms. Semin
Cancer Biol. 2:347–354. 1991.PubMed/NCBI
|
|
109
|
Neefjes J, Jongsma ML, Paul P and Bakke O:
Towards a systems understanding of MHC class I and MHC class II
antigen presentation. Nat Rev Immunol. 11:823–836. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Rock KL, Reits E and Neefjes J: Present
Yourself! By MHC Class I and MHC Class II Molecules. Trends
Immunol. 37:724–737. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Gottlieb AB: Immunologic mechanisms in
psoriasis. J Invest Dermatol. 95 (5 Suppl):18S–19S. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Harden JL, Krueger JG and Bowcock AM: The
immunogenetics of psoriasis: A comprehensive review. J Autoimmun.
64:66–73. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Bartosinska J, Michalak-Stoma A, Kowal M,
Raczkiewicz D, Krasowska D, Chodorowska G and Giannopoulos K:
Analysis of circulating soluble programmed death 1 (PD-1),
neuropilin 1 (NRP-1) and human leukocyte antigen-G (HLA-G) in
psoriatic patients. Postepy Dermatol Alergol. 36:167–172. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Lee YH, Choi SJ, Ji JD and Song GG:
Genome-wide pathway analysis of a genome-wide association study on
psoriasis and Behcet's disease. Mol Biol Rep. 39:5953–5959. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Sokolik R, Gebura K, Iwaszko M, Świerkot
J, Korman L, Wiland P and Bogunia-Kubik K: Significance of
association of HLA-C and HLA-E with psoriatic arthritis. Hum
Immunol. 75:1188–1191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Dand N, Duckworth M, Baudry D, Russell A,
Curtis CJ, Lee SH, Evans I, Mason KJ, Alsharqi A, Becher G, et al:
HLA-C*06:02 genotype is a predictive biomarker of biologic
treatment response in psoriasis. J Allergy Clin Immunol.
143:2120–2130. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Talamonti M, Galluzzo M, Zangrilli A,
Papoutsaki M, Egan CG, Bavetta M, Tambone S, Fargnoli MC and
Bianchi L: HLA-C*06:02 does not predispose to clinical response
following long-term adalimumab treatment in psoriatic patients: A
retrospective cohort study. Mol Diagn Ther. 21:295–301. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Stuart PE, Tejasvi T, Shaiq PA,
Kullavanijaya P, Qamar R, Raja GK, Li Y, Voorhees JJ, Abecasis GR,
Elder JT and Nair RP: A single SNP surrogate for genotyping
HLA-C*06:02 in diverse populations. J Invest Dermatol.
135:1177–1180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Nikamo P and Stahle M: Cost-effective
HLA-Cw06:02 typing in a Caucasian population. Exp Dermatol.
21:221–223. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Chiu HY, Huang PY, Jee SH, Hu CY, Chou CT,
Chang YT, Hwang CY and Tsai TF: HLA polymorphism among Chinese
patients with chronic plaque psoriasis: subgroup analysis. Br J
Dermatol. 166:288–297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Shen M, Lim SWD, Tan ES, Oon HH and Ren
EC: HLA correlations with clinical phenotypes and risk of metabolic
comorbidities in Singapore Chinese psoriasis patients. Mol Diagn
Ther. 23:751–760. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Cai M, Huang H, Ran D, Zheng X, Wen L, Zhu
Z, Liu L, Zhang C, Hong X, Hong J, et al: HLA-C*01:02 and
HLA-A*02:07 confer risk specific for psoriatic patients in Southern
China. J Invest Dermatol. 139:2045–2048. e42019. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Hirata J, Hirota T, Ozeki T, Kanai M, Sudo
T, Tanaka T, Hizawa N, Nakagawa H, Sato S, Mushiroda T, et al:
Variants at HLA-A, HLA-C, and HLA-DQB1 Confer Risk of Psoriasis
Vulgaris in Japanese. J Invest Dermatol. 138:542–548. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
124
|
van Vugt LJ, van den Reek JMPA, Hannink G,
Coenen MJH and de Jong EMGJ: Association of HLA-C*06:02 Status With
Differential Response to Ustekinumab in Patients With Psoriasis: A
systematic review and meta-analysis. JAMA Dermatol. 155:708–715.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Wei P, Yang Y, Liu Z, Luo Z, Tu W, Han J,
Deng Y and Yin L: Characterization of autoantigen presentation by
HLA-C*06:02 in psoriasis. J Invest Dermatol. 137:2238–2241. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Huang YW and Tsai TF: HLA-Cw1 and
psoriasis. Am J Clin Dermatol. 22:339–347. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Teng MW, Bowman EP, McElwee JJ, Smyth MJ,
Casanova JL, Cooper AM and Cua DJ: IL-12 and IL-23 cytokines: from
discovery to targeted therapies for immune-mediated inflammatory
diseases. Nat Med. 21:719–729. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Mirlekar B and Pylayeva-Gupta Y: IL-12
family cytokines in cancer and immunotherapy. Cancers (Basel).
13:1672021. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Gee K, Guzzo C, Che Mat NF, Ma W and Kumar
A: The IL-12 family of cytokines in infection, inflammation and
autoimmune disorders. Inflamm Allergy Drug Targets. 8:40–52. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Ek WE, Karlsson T, Hoglund J,
Rask-Andersen M and Johansson A: Causal effects of inflammatory
protein biomarkers on inflammatory diseases. Sci Adv.
7:eabl43592021. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Lowes MA, Russell CB, Martin DA, Towne JE
and Krueger JG: The IL-23/T17 pathogenic axis in psoriasis is
amplified by keratinocyte responses. Trends Immunol. 34:174–181.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Magyari L, Varszegi D, Sarlos P, Jaromi L,
Melegh BI, Duga B, Kisfali P, Kovesdi E, Matyas P, Szabo A, et al:
Marked differences of haplotype tagging SNP distribution, linkage,
and haplotype profile of IL23 receptor gene in Roma and Hungarian
population samples. Cytokine. 65:148–152. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Ogbodo E, Michelangeli F and Williams JHH:
Exogenous heat shock proteins HSPA1A and HSPB1 regulate TNF-α,
IL-1β and IL-10 secretion from monocytic cells. FEBS Open Bio.
13:1922–1940. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Generali E, Ceribelli A, Stazi MA and
Selmi C: Lessons learned from twins in autoimmune and chronic
inflammatory diseases. J Autoimmun. 83:51–61. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Valdimarsson H: The genetic basis of
psoriasis. Clin Dermatol. 25:563–567. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Kere J: Mapping and identifying genes for
asthma and psoriasis. Philos Trans R Soc Lond B Biol Sci.
360:1551–1561. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Feng BJ, Sun LD, Soltani-Arabshahi R,
Bowcock AM, Nair RP, Stuart P, Elder JT, Schrodi SJ, Begovich AB,
Abecasis GR, et al: Multiple Loci within the major
histocompatibility complex confer risk of psoriasis. PLoS Genet.
5:e10006062009. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Fan X, Yang S, Huang W, Wang ZM, Sun LD,
Liang YH, Gao M, Ren YQ, Zhang KY, Du WH, et al: Fine mapping of
the psoriasis susceptibility locus PSORS1 supports HLA-C as the
susceptibility gene in the Han Chinese population. PLoS Genet.
4:e10000382008. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Velastegui E, Vera E, Vanden Berghe W,
Munoz MS and Orellana-Manzano A: ‘HLA-C: Evolution, epigenetics,
and pathological implications in the major histocompatibility
complex’. Front Genet. 14:12060342023. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Bauer S, Groh V, Wu J, Steinle A, Phillips
JH, Lanier LL and Spies T: Activation of NK cells and T cells by
NKG2D, a receptor for stress-inducible MICA. Science. 285:727–729.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Kowalczyk AP and Green KJ: Structure,
function, and regulation of desmosomes. Prog Mol Biol Transl Sci.
116:95–118. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Linh NTT, Giang NH, Lien NTK, Trang BK,
Trang DT, Ngoc NT, Nghia VX, My LT, Mao CV, Hoang NH and Xuan NT:
Association of PSORS1C3, CARD14 and TLR4 genotypes and haplotypes
with psoriasis susceptibility. Genet Mol Biol. 45:e202200992022.
View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Allen MH, Ameen H, Veal C, Evans J,
Ramrakha-Jones VS, Marsland AM, Burden AD, Griffiths CE, Trembath
RC and Barker JN: The major psoriasis susceptibility locus PSORS1
is not a risk factor for late-onset psoriasis. J Invest Dermatol.
124:103–106. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Tawfik NZ, Abdallah HY, Hassan R, Hosny A,
Ghanem DE, Adel A and Atwa MA: PSORS1 locus genotyping profile in
psoriasis: A pilot case-control study. Diagnostics (Basel).
12:10352022. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Clop A, Bertoni A, Spain SL, Simpson MA,
Pullabhatla V, Tonda R, Hundhausen C, Di Meglio P, De Jong P,
Hayday AC, et al: An in-depth characterization of the major
psoriasis susceptibility locus identifies candidate susceptibility
alleles within an HLA-C enhancer element. PLoS One. 8:e716902013.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Wisniewski A, Matusiak L,
Szczerkowska-Dobosz A, Nowak I and Kusnierczyk P:
HLA-C*06:02-independent, gender-related association of PSORS1C3 and
PSORS1C1/CDSN single-nucleotide polymorphisms with risk and
severity of psoriasis. Mol Genet Genomics. 293:957–966. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Ameen M, Allen MH, Fisher SA, Lewis CM,
Cuthbert A, Kondeatis E, Vaughan RW, Murakami H, Nakagawa H and
Barker JN: Corneodesmosin (CDSN) gene association with psoriasis
vulgaris in Caucasian but not in Japanese populations. Clin Exp
Dermatol. 30:414–418. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Allen M, Ishida-Yamamoto A, McGrath J,
Davison S, Iizuka H, Simon M, Guerrin M, Hayday A, Vaughan R, Serre
G, et al: Corneodesmosin expression in psoriasis vulgaris differs
from normal skin and other inflammatory skin disorders. Lab Invest.
81:969–976. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Tazi Ahnini R, Camp NJ, Cork MJ, Mee JB,
Keohane SG, Duff GW and di Giovine FS: Novel genetic association
between the corneodesmosin (MHC S) gene and susceptibility to
psoriasis. Hum Mol Genet. 8:1135–1140. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Zwirner NW, Dole K and Stastny P:
Differential surface expression of MICA by endothelial cells,
fibroblasts, keratinocytes, and monocytes. Hum Immunol. 60:323–330.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Carlen L, Sakuraba K, Stahle M and Sanchez
F: HLA-C expression pattern is spatially different between
psoriasis and eczema skin lesions. J Invest Dermatol. 127:342–348.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Schon MP, Boehncke WH and Brocker EB:
Psoriasis: Clinical manifestations, pathogenesis and therapeutic
perspectives. Discov Med. 5:253–258. 2005.PubMed/NCBI
|
|
153
|
Telem DF, Israeli S, Sarig O and Sprecher
E: Inflammatory peeling skin syndrome caused a novel mutation in
CDSN. Arch Dermatol Res. 304:251–255. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Maekawa K, Nishikawa J, Kaniwa N, Sugiyama
E, Koizumi T, Kurose K, Tohkin M and Saito Y: Development of a
rapid and inexpensive assay for detecting a surrogate genetic
polymorphism of HLA-B*58:01: A partially predictive but useful
biomarker for allopurinol-related Stevens-Johnson syndrome/toxic
epidermal necrolysis in Japanese. Drug Metab Pharmacokinet.
27:447–450. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Lu HF, Liu TY, Chou YP, Chang SS, Hsieh
YW, Chang JG and Tsai FJ: Comprehensive characterization of
pharmacogenes in a Taiwanese Han population. Front Genet.
13:9486162022. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Patrick MT, Stuart PE, Zhang H, Zhao Q,
Yin X, He K, Zhou XJ, Mehta NN, Voorhees JJ, Boehnke M, et al:
Causal relationship and shared genetic loci between psoriasis and
type 2 diabetes through trans-disease meta-analysis. J Invest
Dermatol. 141:1493–1502. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Abramczyk R, Queller JN, Rachfal AW and
Schwartz SS: Diabetes and psoriasis: Different sides of the same
prism. Diabetes Metab Syndr Obes. 13:3571–3577. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Eiris N, Gonzalez-Lara L, Santos-Juanes J,
Queiro R, Coto E and Coto-Segura P: Genetic variation at IL12B,
IL23R and IL23A is associated with psoriasis severity, psoriatic
arthritis and type 2 diabetes mellitus. J Dermatol Sci. 75:167–172.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Patrick MT, Nair RP, He K, Stuart PE,
Billi AC, Zhou X, Gudjonsson JE, Oksenberg JR, Elder JT and Tsoi
LC: Shared genetic risk factors for multiple sclerosis/psoriasis
suggest involvement of interleukin-17 and janus kinase-signal
transducers and activators of transcription signaling. Ann Neurol.
94:384–397. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Tang Y, Liu W, Kong W, Zhang S and Zhu T:
Multisite chronic pain and the risk of autoimmune diseases: A
Mendelian randomization study. Front Immunol. 14:10770882023.
View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Huang J, Su B, Karhunen V, Gill D, Zuber
V, Ahola-Olli A, Palaniswamy S, Auvinen J, Herzig KH,
Keinänen-Kiukaanniemi S, et al: Inflammatory diseases, inflammatory
biomarkers, and Alzheimer disease: An observational analysis and
mendelian randomization. Neurology. 100:e568–e581. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Li C, Li X, Lin J, Cui Y and Shang H:
Psoriasis and progression of Parkinson's disease: a Mendelian
randomization study. J Eur Acad Dermatol Venereol. 36:2401–2405.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Ellinghaus D, Jostins L, Spain SL, Cortes
A, Bethune J, Han B, Park YR, Raychaudhuri S, Pouget JG, Hübenthal
M, et al: Analysis of five chronic inflammatory diseases identifies
27 new associations and highlights disease-specific patterns at
shared loci. Nat Genet. 48:510–518. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Yin X, Low HQ, Wang L, Li Y, Ellinghaus E,
Han J, Estivill X, Sun L, Zuo X, Shen C, et al: Genome-wide
meta-analysis identifies multiple novel associations and ethnic
heterogeneity of psoriasis susceptibility. Nat Commun. 6:69162015.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Gandhi G, Buttar BS, Albert L, Hasan Q and
Aggarwal RK: Psoriasis-associated genetic polymorphism in North
Indian population in the CCHCR1 gene and in a genomic segment
flanking the HLA-C region. Dis Markers. 31:361–370. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Chang YT, Hsu CY, Chou CT, Lin MW, Shiao
YM, Tsai CY, Yu CW, Shiue JJ, Lee YF, Huang CH, et al: The genetic
polymorphisms of POU5F1 gene are associated with psoriasis vulgaris
in Chinese. J Dermatol Sci. 46:153–156. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Chang YT, Tsai SF, Lee DD, Shiao YM, Huang
CY, Liu HN, Wang WJ and Wong CK: A study of candidate genes for
psoriasis near HLA-C in Chinese patients with psoriasis. Br J
Dermatol. 148:418–423. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Xia X, Yu H, Li Y, Liang Y, Li G and Huang
F: transcriptome analysis identifies biomarkers for the diagnosis
and management of psoriasis complicated with depression. Clin
Cosmet Investig Dermatol. 16:1287–1301. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Chantarangsu S, Mushiroda T,
Mahasirimongkol S, Kiertiburanakul S, Sungkanuparph S, Manosuthi W,
Tantisiriwat W, Charoenyingwattana A, Sura T, Takahashi A, et al:
Genome-wide association study identifies variations in 6p21.3
associated with nevirapine-induced rash. Clin Infect Dis.
53:341–348. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Sanchez FO, Linga Reddy MV, Mallbris L,
Sakuraba K, Stahle M and Alarcon-Riquelme ME: IFN-regulatory factor
5 gene variants interact with the class I MHC locus in the Swedish
psoriasis population. J Invest Dermatol. 128:1704–1709. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Tervaniemi MH, Siitonen HA, Soderhall C,
Minhas G, Vuola J, Tiala I, Sormunen R, Samuelsson L, Suomela S,
Kere J and Elomaa O: Centrosomal localization of the psoriasis
candidate gene product, CCHCR1, supports a role in cytoskeletal
organization. PLoS One. 7:e499202012. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Ishida-Yamamoto A, Furio L, Igawa S, Honma
M, Tron E, Malan V, Murakami M and Hovnanian A: Inflammatory
peeling skin syndrome caused by homozygous genomic deletion in the
PSORS1 region encompassing the CDSN gene. Exp Dermatol. 23:60–63.
2014. View Article : Google Scholar : PubMed/NCBI
|