Introduction
Chronic kidney disease (CKD), a slowly progressive
and irreversible kidney function failure, affects ~10% of the
global population (1).
Accumulating clinical evidence suggests that excessive sodium
intake accelerates the progression of CKD and its related
hypertension (2,3). Additionally, a high level of sodium
has been proven to induce kidney cytokine expression as well as
tissue regeneration and fibrosis by activating Wnt/β-catenin and
TGF-β signaling (4–6). Consequently, excessive sodium intake
is an important risk factor for CKD progression. Therefore, besides
advocating salt reduction, it is critical to explore remedies to
reconcile the adverse impact of excessive sodium consumption on
CKD.
The intestine has a rich and structurally diverse
microbiome that is engaged in multiple interactions affecting host
health by influencing the intrinsic immunity and metabolism.
Moreover, the composition and function of the gut microbiota is
dynamic and easily affected by diet. There is an effect of dietary
habits on host-microbe interactions, and nutrition can either
maintain homeostasis or lead to disease susceptibility (7). Although salt is one of the most
essential dietary components, excessive salt intake has been
extensively studied and proven to cause a wide range of diseases
through its effects on the gut microbiome (8). Previous studies have shown that high
salt (HS) consumption results in alteration of the gut microbiota
structure that may be pro-inflammatory, which exacerbates colitis
and hypertension (9,10). In addition, HS-induced gut barrier
disruption triggers renal function injury (11). These data indicated that excessive
salt intake could cause susceptibility to a wide range of diseases
through the gut microbiome. Moreover, depletion of the gut
microbiome or administering probiotics can ease the progression of
numerous diseases (11,12). Taken together, ideas were provided
to reverse the progression of HS-related CKD through regulation of
the gut microbiome.
A previous review advocated non-pharmacological
strategies such as dietary and lifestyle modulation as well as
kidney disease-specific pharmacological interventions to achieve
kidney function preservation (13). Puerariae lobatae Radix (PLR)
is rich in nutrients, including flavonoids, starch, saponins and
amino acids, and has always been used as a medicine and food
homologous herb to relieve gastrointestinal and cardiovascular
diseases (14). Additionally, PLR
is consistent with the principles of the CKD management guidelines
that advocate dietary control (15). Multiple studies have indicated that
PLR and its active ingredient might alleviate diabetes-related
kidney damage and non-alcoholic fatty liver disease through
modulating the gut microbiota (16,17).
Previously, it was also confirmed that PLR promotes gut microbiota
homeostasis, increases the relative abundance of beneficial
bacteria, and protects the structural and functional integrity of
the gut barrier, blood-brain barrier and placental barrier, thus
affecting the symptoms of vascular anomaly-related diseases
including ischemic stroke and pre-eclampsia (14,18).
CKD is a disease closely related to host metabolic disorder and
vascular dysfunction and bioflavonoids and other bioactive
compounds have been verified to relieve CKD-associated biochemical
abnormalities and kidney inflammation (19). However, few studies have focused on
whether PLR exerts a protective effect on these conditions.
Because excessive sodium intake is a risk factor for
the development of CKD, dietary preferences have great potential in
influencing host health (11). In
the present study, a HS-induced CKD murine model was constructed
and the renoprotective effects of PLR were investigated.
Materials and methods
Source and preparation of PLR
decoction
Medicinal slices form of PLR were purchased from
Guangzhou Weida Company. Standard substance of puerarin (purity
≥99.0%), daidzin (purity ≥98.0%) and daidzein (purity ≥98.0%) were
purchased from Chengdu Must Bio-Technology Co., Ltd. PLR decoction
preparation was conducted as previously described (14,20,21).
In brief, a certain quality of PLR was received and extracted in
boiling distilled water at a ratio of 1:10 (w/v) for 1 h; the
filtrate was collected and the residue was boiled with distilled
water (1:6, w/v) for another 1 h. The aforementioned filtrates were
concentrated and the content of puerarin, daidzin and daidzein was
quantified by using ultra-performance liquid chromatography
(specific method is listed in Table
SI and Fig. S1). Briefly, the
column was an Agilent ZORBAX SB-C18 (3.0×100 mm, 1.8-Micron). The
mobile phase comprised methanol and water with a flow rate of 0.3
ml/min. Changes in the proportion of the mobile phase are shown in
Table SI. Puerarin, daidzin and
daidzein were detected at a UV wavelength of 250, 270 and 260 nm,
respectively. Testosterone (240 nm) acted as the internal standard.
The retention times of puerarin, daidzin and daidzein were 3.82,
5.19 and 9.55 min, respectively. Organic phase methanol was
obtained from Tianjin Damao Chemical Reagent Factory. The
respective concentrations of these three compounds were 10.9, 17.5
and 0.3 mg/ml.
Animals and experimental design
All animal experimental procedures were conducted in
strict accordance with the National Institute of Health guidelines
and were approved (approval no. L-2019216) by the Institutional
Animal Ethical Care Committee of Southern Medical University
Experimental Animal Center (Guangzhou, China) (22). Specific pathogen-free male C57BL/6
mice (6–8 week, weighing 20–26 g) were purchased from SPF
Biotechnology Co., Ltd. The total number purchased was 55. All mice
were housed under standard conditions of temperature (22±1°C) and
humidity (50±5%) with a 12/12-h light-dark cycle and free access to
food and water. After 1 week of acclimatization, HS-feeding mice
were administered drinking water containing 2% (w/v) NaCl for 8
weeks as previously described (11) (HS, n=6), The amount of drinking
water provided minus the amount of water remaining indicated the
amount of water consumed during the day. while PLR-intervention
mice received Pueraria decoction (containing 18, 36 and 72
mg/kg Puerarin, respectively) by oral gavage once a day along with
8 weeks HS feeding (PLR-L, PLR-M and PLR-H, n=6 per group). Control
mice received normal drinking water during the experiment (CON,
n=10). There were no differences in baseline covariates among
groups. The feces and urine were collected every week for further
examination. After an 8-week period of modeling, cervical
dislocation was performed under complete anesthesia with an
intraperitoneal injection of 1.5% sodium pentobarbital at a
concentration of 40 mg/kg. Death of mice was verified by cardiac
arrest, a reduction in body temperature, and a lack of response to
strong stimuli. Sampling operation time was ~10–15 min per mouse.
Serum, kidney, liver, spleen, colon and cecal contents samples were
harvested in a sterile manner for further analysis. Calculation of
organ index was performed using organ weight/body weight ratio.
After separation of the colon, the length was measured from the
junction of the cecum and colon to the anal canal, which was
recorded as the length of the colon. Blood collection (0.5–0.8 ml)
was performed by open laparotomy through the abdominal aorta with a
fine needle after euthanasia under complete anesthesia.
For blood pressure measurement, systolic blood
pressure (SBP) and mean blood pressure (MBP) were measured every
week via non-invasive tail cuff method, using a BP-2010A instrument
(Beijing Softron Biotechnology Co., Ltd.). All measurements were
operated between 8 to 12 am. At least six continuous stable results
of each mouse were obtained and the average value was calculated as
the final result.
The mice were weighed at regular times each day,
their bedding was changed and their health was assessed. Humane
endpoints were as follows: The body weight of mice being 20% lower
than before the experiment, mice being unable to feed or drink, or
not responding to gentle stimulation. In the aforementioned
circumstances, mice would be considered unsuitable for further
experimentation and would be euthanized by neck dissection after
complete anesthesia using the anesthesia method and dosage as
aforementioned. In the present study, none of the mice reached
these humane endpoints.
Fecal microbiota transplantation (FMT)
experiment
FMT experiment was performed based on the modified
method previously described (11,23).
In brief, feces from the donor mice (HS and PLR-H group mice) were
collected in aseptic centrifugal tubes respectively and resuspended
in PBS at 125 mg/ml, the mixtures were centrifuged at 1,000 × g for
1 min at 37°C and the supernatants were collected and saved in
separate 1.5 ml tubes for subsequently microbiota transplantation.
Before FMT experiment, the acceptor male C57BL/6 mice (6–8 weeks)
were given antibiotics (vancomycin, 100 mg/kg; neomycin sulfate 200
mg/kg; metronidazole 200 mg/kg; and ampicillin 200 mg/kg)
intragastrically once a day for 1 w to deplete the gut microbiota.
All the mice then received drinking water containing 2% (w/v) NaCl
for 8 weeks as aforementioned. A volume of 150 µl of the fecal
microbiota solution was simultaneously orally gavaged to mice once
a day in the first 2 weeks, every other day in the following 2
weeks and twice each week in the last 4 weeks. The mice that
received fecal microbiota solution from HS mice or PLR-H mice were
referred to as HS-FMT group or PLR-FMT group (n=8 per group).
Control mice received normal drinking water during the experiment
(CON, n=5). The total number of mice purchased in this part of the
experiment was 21. Blood pressures were measured as aforementioned
every week. All the mice had free access to food and water, and the
feces and urine were collected before sacrifice. Mice were
sacrificed under anesthesia and then blood, kidney, liver, spleen,
colon and cecal contents samples were harvested in a sterile manner
for further analysis.
16S rDNA gene sequencing and microbe
analysis
Feces were collected in sterilized 1.5 ml tubes and
frozen at −80°C before DNA extraction. Microbial DNA from fecal
samples were extracted as previously described (24,25).
V3-V4 region of 16S rDNA was amplified by PCR using the following
primers: 341F, 5′-CCTACGGGNGGCWGCAG-3′ and 806R,
5′-GGACTACHVGGGTATCTAAT-3′. Amplicons were purified using the
AxyPrep DNA Gel Extraction Kit (cat. no. AP-GX-250; Axygen;
Corning, Inc.). Equimolar pooling of purified amplicons was
performed and paired end sequenced (PE250) was conducted on an
Illumina platform according to the standard protocols. Alpha
diversity were calculated in QIIME (26) (version 1.9.1). The abundance
statistics of each taxonomy was visualized using Krona (27) (version 2.6). Principal coordinate
analysis (PCoA) and non-metric multidimensional scaling (NMDS) of
weighted unifrac distances were generated in R project Vegan
package (version 2.5.3). Biomarker features in each group were
screened by LEfSe software (28)
(version 1.0). Microbial dysbiosis index (MDI) was calculated as
follows: MDI=log10[(total abundance in genera increased
in disease group)/(total abundance in genera decreased in disease
group)]. The Kyoto Encylopedia of Genes and Genomes (KEGG) pathway
analysis of the operational taxonomic units was inferred using
Tax4Fun (29) (version 1.0) and
was generated using Omicsmart, a dynamic real-time interactive
online platform for data analysis (http://www.omicsmart.com). Spearman correlation
coefficient between environmental factors and species was
calculated in R project psych package (version 1.8.4) then
generated using the Wekemo Bioincloud (https://www.bioincloud.tech). The calculated P-value
was conducted through false discovery rate (FDR) correction, taking
FDR ≤0.05 as a threshold.
RNA-seq analysis
Kidney samples were collected in sterilized 1.5-ml
tubes and frozen at −80°C before DNA extraction. Total RNA was
extracted using the Eastep™ Super Total RNA Extraction Kit (cat.
no. LS1040; Promega Corporation) according to the manufacturer's
instructions. After total RNA was extracted, eukaryotic mRNA was
enriched by Oligo(dT) beads. Then fragments were transcribed into
cDNA by using NEBNext Ultra RNA Library Prep Kit for Illumina (cat.
no. 7530; New England Biolabs, Inc.). The ligation reaction was
purified, and PCR amplified. The resulting cDNA library was
sequenced using Illumina Novaseq6000 by Gene Denovo Biotechnology
Co. Differential expression analysis of RNAs was performed by
DESeq2 (30) software between two
different groups. The transcripts with the parameter of FDR below
0.05 and absolute fold change ≥2 were considered differentially
expressed transcripts. Gene set enrichment analysis (GSEA) was
performed using software GSEA (31) and MSigDB (31) to identify whether a set of genes in
specific KEGG pathway shows significant differences in two
groups.
Gene expression analysis
Total RNA was extracted from kidney and colon tissue
using the Animal Total RNA Isolation Kit (cat. no. RE-03011/03014;
Foregene Co., Ltd,) according to the manufacturer's instructions. A
reverse transcription enzyme, HiScript III RT SuperMix for qPCR
(+gDNA wiper) (Vazyme Biotech Co., Ltd.), was applied to obtain
cDNA (temperature and duration: 50°C for 15 min; 85°C for 5 sec).
The quantitative PCR reactions (initial denaturation: 95°C for 30
sec; 40 cycles of amplification at 95°C for 10 sec and 60°C for 30
sec; followed by melting curve analysis at 95°C for 15 sec, 60°C
for 1 min and 95°C for 15 sec) were performed on the LightCycler480
(Roche Diagnostics) using a ChamQ SYBR qPCR Master Mix (Vazyme
Biotech Co., Ltd.). Relative quantification of target genes were
calculated using the 2−ΔΔCq method (32). GAPDH was used as a control gene.
All the target gene primer sequences are listed in Table SII.
Protein expression and biochemical
analysis
The kidney and colon tissue protein were extracted
with a commercial RIPA lysis buffer (cat. no. PC101; Epizyme
Biomedical Technology Co., Ltd.) supplemented with 1% cocktail
protease inhibitor (cat. no. MB2678; Dalian Meilun Biology
Technology Co., Ltd.). Protein samples were quantified using a BCA
Protein Assay Kit (cat. no. ZJ102; Epizyme Biomedical Technology
Co., Ltd.) and 40 µg protein was loaded per lane. Samples were
separated by SDS-PAGE on 10% gels and transferred to a PVDF
membrane. After blocking with 5% non-fat milk for 1 h at 37°C,
samples were incubated overnight at 4°C with the following primary
antibodies: Rabbit anti-Occludin (1:2,000; cat. no. ab216327;
Abcam), rabbit anti-ZO-1 (1:2,000; cat. no. ab216880; Abcam),
rabbit anti-Claudin-1 (1:2,000; cat. no. YT0942; ImmunoWay
Biotechnology Company), mouse anti-TNF-α (1:1,000; cat. no. YM3477;
ImmunoWay Biotechnology Company), rabbit anti-IL-1β (1:1,000; cat.
no. YT5201; ImmunoWay Biotechnology Company), rabbit anti-β-catenin
(1:5,000; cat. no. 51067-2-AP; Proteintech Group, Inc.) and mouse
anti-β-actin (1:5,000; cat. no. RM2001; RayBiotech, Inc.).
Subsequently, they were incubated with horseradish
peroxidase-conjugated goat anti-mouse/rabbit secondary antibody at
37°C for 1 h (1:5,000; cat. no. LF101/LF102; Epizyme Biomedical
Technology Co., Ltd.). Membranes were visualized with a Femto Light
Chemiluminescence Kit (cat. no. SQ201; Epizyme Biomedical
Technology Co., Ltd.) and imaged with a gel image processing system
(cat. no. 92-15313-00; FluorChem R System; Bio-Techne). The band
intensity was assessed using ImageJ software (version 1.53K;
National Institutes of Health).
Serum creatinine was determined by using a
Creatinine Assay kit (sarcosine oxidase) (cat. no. C011-2-1;
Nanjing Jiancheng Bioengineering Institute). The urinary protein,
serum lipopolysaccharide (LPS) and serum TNF-α level were detected
manually with commercial ELISA Kits (cat. nos. MM-44286M2,
MM-0634M2 and MM-0132M2; Shanghai Meimian Biotechnology, Co., Ltd.)
according to the manufacturer's protocols. Serum and urine
K+, Ca2+ and Na+ concentrations
were determined on an automatic biomedical analyzer (Roche
Diagnostics).
FD-4 permeability experiment
FITC Dextran 4-KD (FD-4; Sigma-Adrich; Merck KGaA)
was used to detect intestinal permeability in vivo. FD-4 was
orally administered to mice at a dose of 0.6 mg/kg 4 h before serum
was harvested. The FD-4 level in serum was detected in the dark,
based on a standard curve and spectro-fluorometrically with an
excitation wavelength of 485 nm and an emission wavelength of 530
nm in a microplate fluorescence reader (Infinite® M1000
PRO; Tecan Group, Ltd.).
Imaging of intestinal inflammation in
vivo
L-012 (FUJIFILM Wako Pure Chemical Corporation) was
used to confirm intestinal inflammation in vivo (33). Mice were anesthetized with 3–4%
isoflurane induction by inhalation and maintained with 1–1.5%
isoflurane and injected with 100 µl of a 20-mmol L-012 solution.
After 1 min, IVIS Spectrum CT system was used to acquire
bioluminescent images. For post-acquisition analysis, the Living
Image software was used to quantify the bioluminescent signals at
standardized regions of interest (ROIs) defined on the abdomen.
Histological staining and
analysis
The left kidney and colon tissue of mice were
collected and fixed in 4% paraformaldehyde for 3 days at room
temperature. The samples were then embedded in paraffin, sectioned
at 4-µm thickness and stained with hematoxylin and eosin (H&E)
(hematoxylin for 4 min and eosin for 20 sec) or Masson's trichrome
(MASSON) (Weigert's iron hematoxylin stain for 8 min, ponceau S for
5 min and aniline blue for 2 min) at room temperature. The kidney
tissue histological damage was assessed according to a previously
described scoring method. Briefly, the extent of damage was
assessed based on interstitial inflammatory cell infiltration,
tubular pattern of renal tubules and loss of brush border. The
lesion score was quantified as follows: score 0=0%; score 1=1–10%;
score 2=11–25%; score 3=26–75%; score 4=76–100% (34). The kidney fibrosis area was
selected randomly from at least 5 points of cortical fields and
quantified using ImageJ software (version 1.53K; National
Institutes of Health). Immunohistochemistry was performed using the
primary antibodies for Occludin, ZO-1 and Claudin-1. Quantification
of the average optical density was performed by automated image
analysis in five randomly chosen fields of each sample
(magnification, ×200). All images were scanned by a NanoZoomer
Digital slide scanner and captured at ×200 or ×400 magnification
with an NDP. View2 Plus Image viewing software (Hamamatsu Photonics
K.K.) was used.
Immunofluorescence
Colon paraffin section samples were generated as
aforementioned. TBS solution containing 0.3% Triton X-100 (cat. no.
9002-93-1; Beijing Solarbio Science & Technology Co., Ltd.),
0.25% protein free rapid blocking buffer (cat. no. PS108P; Epizyme
Biomedical Technology Co., Ltd.) and 5% goat serum (cat. no.
BL210A; Biosharp Life Sciences) was used to block samples for 1 h
at room temperature. The samples were then incubated at 4°C with
rabbit anti-β-catenin antibody (1:50; cat. no. 51067-2-AP;
Proteintech Group, Inc.) overnight. Subsequently, they were
incubated with Alexa Fluor 488-labeled goat anti-rabbit secondary
antibody (1:500; cat. no. A0423; Beyotime Institute of
Biotechnology) at 37°C for 1 h. Then the slices were washed and
stained with DAPI (0.5 µg/ml) for 10 min. Images were captured with
fluorescent microscopy (Olympus Corporation). Quantification of the
fluorescence intensity was calculated by automated image analysis
in five randomly chosen fields of each sample (magnification,
×200).
Statistical analysis
Results were represented as the mean ± standard
error of the mean (SEM) and performed using GraphPad Prism software
(version 8.0) (GraphPad Software; Dotmatics). When comparing the
difference in means between two samples, the Mann-Whitney test was
used for non-normal distributions; when comparing the difference in
means between multiple samples, the one-way ANOVA was used if the
data were normally distributed and the variance was homogeneous,
and the Kruskal-Wallis test was used for non-normal distributions.
Comparisons between groups involving the same time point and
between groups at different time points within the same group were
made using two-way repeated-measures ANOVA. Dunn's post hoc test
was used after Kruskal-Wallis test, and Bonferroni's post hoc test
or Tukey's HSD post hoc test was used after ANOVA. Adonis analysis
and anosim analyses were used for PCoA. P<0.05 was considered to
indicate a statistically significant difference.
Results
PLR relieves elevated SBP and slow
weight gain induced by an HS diet
There were no differences in the baseline weight and
blood pressure among the groups (Fig.
S2A-C); however, after consuming the same amount of sodium,
there was a significant difference in SBP among the groups
(Fig. 1A). Compared with the CON
group, the SBP of the HS group showed significant elevation from
the 5th week. By contrast, the SBP of the PLR-H group maintained no
difference from the CON group during the whole experiment. SBP in
the PLR-L and PLR-M groups were lower than the HS group; however,
this reduction was not significant at the end of the experiment
(Fig. 1A). Additively, changes in
MBP were not significantly different among the groups (Fig. S2D). These results suggested that
high-dose PLR supplementation could effectively reduce SBP in CKD.
Besides, the weight change was also recorded; HS group gained
weight more slowly and exhibited significant different in weight
compared with those in the CON group from the 4th week onwards. PLR
intervention could maintain a normal rate of weight gain to some
extent (Fig. S2E).
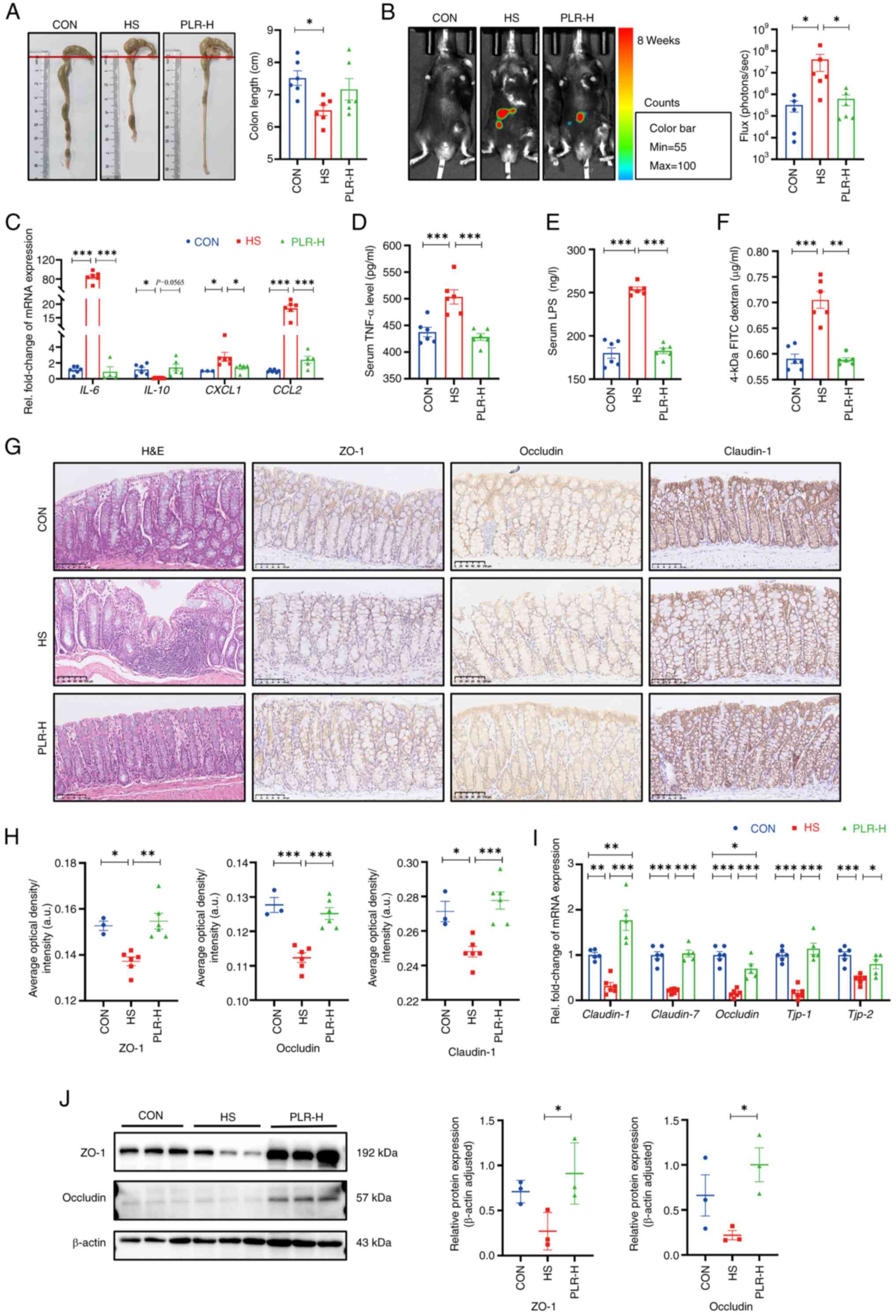
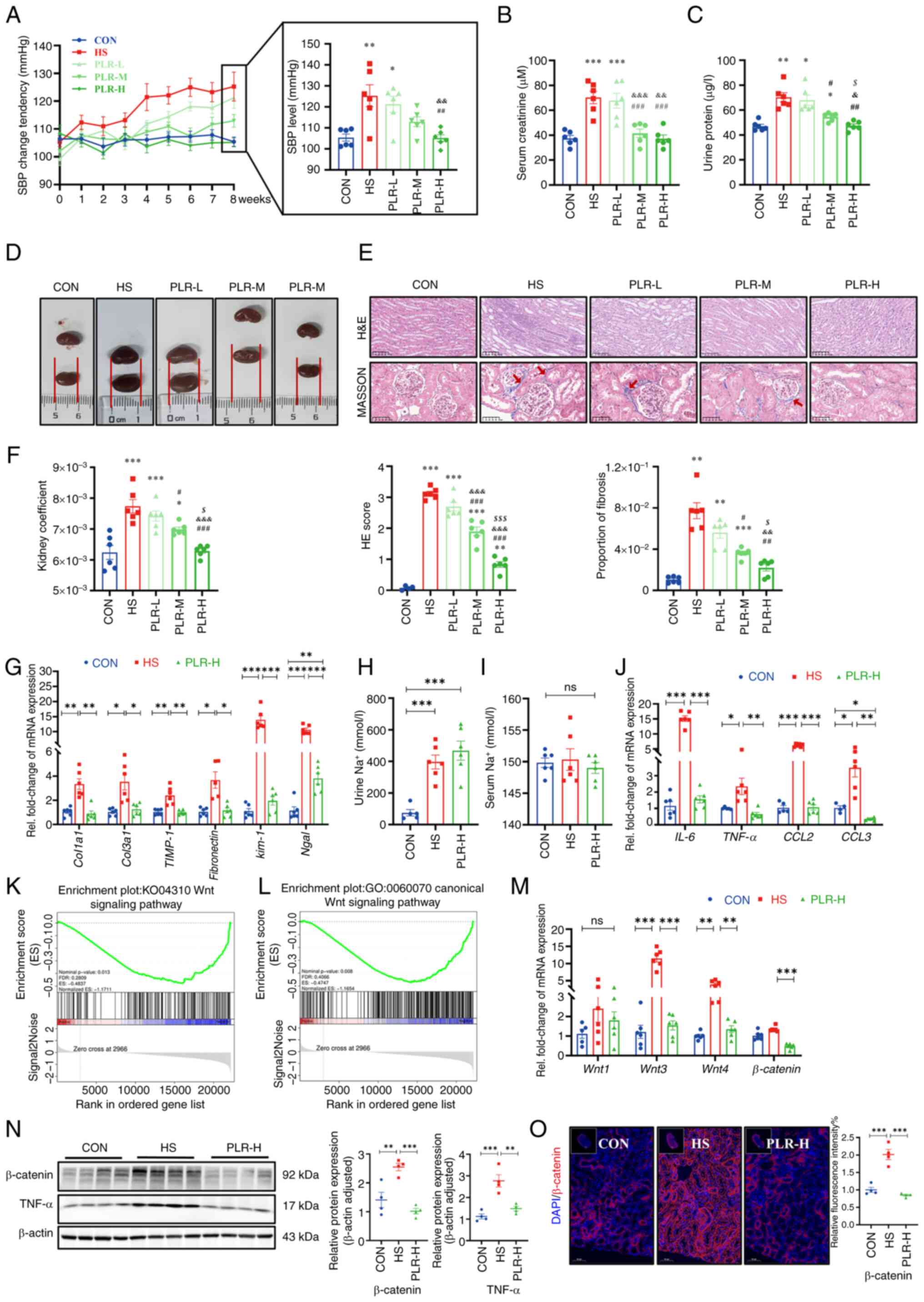 | Figure 1.PLR alleviates kidney injuries
induced by HS diet and downregulates the Wnt/β-catenin pathway in
kidney of HS mice. (A) SBP change of groups were evaluated
non-invasively in the course of experiment (n=6). (B) Serum
creatinine after 8 weeks (n=6). (C) Urine protein after 8 weeks
(n=6). (D) Representative gross anatomy images of the kidney (n=6).
(E) H&E and MASSON staining of the kidney and the pathology
scores and histological analysis. Fibrosis in the glomeruli and
renal interstitium is indicated with red arrows. H&E staining
scale bar, 100 µm; MASSON staining scale bar, 50 µm (n=6). (F)
Kidney coefficient (n=6). (G) RT-qPCR analysis of kidney
injuries-related genes (n=6). (H) Urine Na+
concentration (n=6). (I) Serum Na+ concentration (n=6).
(J) RT-qPCR analysis of inflammatory factors-related genes in
kidney (n=6). (K) Gene-set enrichment analyses based on Kyoto
Encyclopedia of Genes and Genomes database performed on the RNA
sequencing data (n=3). (L) Gene-set enrichment analyses based on
Gene Ontology database performed on the RNA sequencing data (n=3).
(M) RT-qPCR analysis of Wnt1, Wnt3, Wnt4 and β-catenin genes in
kidney (n=5-6). (N) β-catenin and TNF-α protein levels in the
kidney (n=4). (O) Representative in situ detection of
β-catenin in renal cortex was measured by immunofluorescent
staining. Kidney tissue sections were stained with DAPI (blue) and
probed with β-catenin (red). Scale bar, 50 µm (n=4). Results are
expressed as the mean ± SEM. *P<0.05, **P<0.01 and
***P<0.001 vs. the CON group; #P<0.05,
##P<0.01 and ###P<0.001 vs. the HS
group; &P<0.05, &&P<0.01
and &&&P<0.001 vs. the PLR-L group;
$P<0.05, and $$$P<0.001 vs. the PLR-M
group in A-E. ***P<0.001, **P<0.01 and *P<0.05 were
determined by one-way ANOVA with Bonferroni's post hoc test in G-J
and M-O. PLR, Puerariae lobatae Radix; HS, high salt; SBP,
systolic blood pressure; H&E, hematoxylin-eosin; RT-qPCR,
reverse transcription-quantitative PCR; ns, not significant. |
PLR alleviates CKD and renal fibrosis
induced by an HS diet
In the 8th week, serum creatinine, urine protein and
kidney histologic stains were used to assess the extent of kidney
damage as well as the efficiency of PLR on kidney protection.
Compared with the CON group, the HS group revealed significantly
aggravated serum creatinine and urine protein levels (Fig. 1B and C); however, these biochemical
parameters were significantly improved in the PLR-M and PLR-H
groups. Moreover, the macroscopic images of the kidneys of the
PLR-M and PLR-H groups exhibited markedly less hyperemia and
swelling compared with those of the HS and PLR-L groups (Fig. 1D). Consistent with this appearance,
the kidney coefficient and the histopathological injury observed by
H&E staining and the proportion of fibrosis observed by MASSON
staining were significantly lower in the PLR-M and PLR-H groups
than those in the HS group (Fig. 1E
and F). Such histological coefficient differences were not
indicated in the liver and spleen among the groups (Fig. S2F and G), however. Notably,
middle-and high-dose PLR supplementation significantly alleviated
CKD. PLR-H more efficiently reduced the degree of fibrosis and
renal H&E scores compared with PLR-M. Taken together, the high
PLR dose was considered as the most effective for CKD protection
and was evaluated in the following experiment.
Compared with the CON group, the HS mice
demonstrated significantly upregulated mRNA levels of Col1a1,
Col3a1, TIMP-1 and Fibronectin (Fig. 1G), which indicated fibrosis in the
kidneys of the HS group. High-dose PLR supplementation
significantly downregulated these fibrosis-related gene expression
levels. In addition, the mRNA levels of Kim-1 and
Ngal, which indicated renal tubular injury, were
significantly higher in the HS group than in the PLR-H group.
Considering that the amount of salt intake could influence kidney
injury, the total water intake was compared and it was found that
the HS and PLR (all doses) groups consumed similar amounts of salt
in the whole experiment (Fig.
S2H). Additionally, both HS intervention and administation of
high-dose PLR did not alter serum sodium loading, but they did
increase urinary sodium excretion, as measured by urine and serum
Na+ (Fig. 1H and I).
Serum concentrations of the other major ions also had no
differences among the groups (Fig.
S2I and J).
PLR mitigates the inflammatory
response and downregulates the Wnt/β-catenin pathway in the
kidney
To investigate the mechanism by which high-dose PLR
exerted its renoprotective effects, RNA-seq was performed to assess
the gene expression profile of the kidneys in the HS group vs. the
PLR-H group. Compared with the HS group, administration of
high-dose PLR altered the expression of 532 genes in the kidneys,
with 53 transcripts being significantly upregulated and 479 being
downregulated (Fig. S2K). GSEA
based on the Gene Ontology (GO) and KEGG databases were performed
on the RNA-seq data and revealed that a high PLR dose substantially
decreased the expression of genes related to the canonical Wnt
signaling pathway (Fig. 1K and L).
Furthermore, it was found that HS intake induced a significant
increase in the mRNA levels of IL-6, TNF-α, CCL2 and CCL3 in the
kidney (Fig. 1J), and this effect
was reversed in the PLR-H group. To further verify whether the
protective effect of high-dose PLR is related to the Wnt/β-catenin
pathway, the mRNA levels of molecules in this pathway were next
analyzed. As presented in Fig. 1M,
HS intake significantly increased the Wnt3 and Wnt4 mRNA levels.
Furthermore, the protein level of β-catenin was measured (Fig. 1N and O), the expression level of
which was significantly increased in the HS mice compared with the
CON group, demonstrating that HS intake could activate the
canonical Wnt signaling pathway. While high-dose PLR administration
could significantly downregulate the mRNA and protein level of
β-catenin, it also downregulated the mRNA levels of Wnt3 and Wnt4.
In addition, TNF-α, which was able to induce β-catenin activation,
was also found to have an elevated protein level in the kidney
tissue in the HS group and was reduced by high-dose PLR
administration. Overall, it was found that high-dose PLR could
exert nephroprotective effects by reducing the activation of the
Wnt/β-catenin pathway.
PLR reduces intestinal inflammation
and protects against intestinal barrier damage
Maternal intestinal damage, including inflammation
and concomitant increased gut permeability, are involved in
numerous extraintestinal diseases including CKD (35). Hence, the intestinal alterations
were examined, as shown in Fig.
2A. Compared with the HS group, colon shortening was relieved
to certain extent in the PLR-H group, although no statistical
difference was observed. To further assess the inflammation level,
pro-inflammatory cytokines in the colonic tissues were measured
(Fig. 2C). The mRNA levels of
IL-6, CXCL1 and CCL2 in the HS group were significantly higher,
whereas the mRNA level of inflammatory protective factor IL-10 was
reduced compared with those in the CON group. However, high-dose
PLR supplementation significantly inhibited the expression of
pro-inflammatory factors. In Fig.
2B, an L-012 chemiluminescent probe was used to assess
intestinal reactive oxygen species (ROS) in vivo (33). The images indicated that chronic HS
intervention markedly elevated the ROS concentration in the colon
of mice, with a significantly increased total flux in photons, and
this elevation was significantly dampened by high-dose PLR
administration.
To further demonstrate the role of PLR
administration in intestinal protection, intestinal tight junction
markers in the colon were measured. Compared with the CON group,
the HS group revealed significantly downregulated mRNA levels of
tight junction proteins (Fig. 2I);
whereas the serum TNF-α level, LPS level and FD-4 permeability were
elevated (Fig. 2D-F). Moreover,
the HS mice presented significantly deteriorative histopathological
injury in the colon as observed by H&E staining, as well as
decreased mRNA and protein levels of ZO-1, Occludin and Claudin-1
observed by immunohistochemical staining and western blotting
(Fig. 2G-J). However, all the
aforementioned damages induced by HS were largely attenuated by
high-dose PLR administration. Collectively, the aforementioned
results demonstrated that therapeutic high-dose PLR administration
alleviated intestinal inflammation and reinforced the gut
barrier.
PLR reverses gut microbial dysbiosis
and increases the relative abundance of beneficial bacteria
A pathophysiological colon change is always
associated with enteric dysbiosis (36,37).
The impact of high-dose PLR on the gut microbiota composition and
equilibrium was further examined via 16S rDNA sequencing. The
relative abundance of Firmicutes as well as the
Firmicutes/Bacteroidetes ratio in the cecal content of HS mice were
lower than those in the cecal content of CON mice, whereas the
Bacteroidetes level was significantly higher in the HS group
(Fig. S3A). High-dose PLR
intervention reversed the dominant position of Bacteroidetes and
upregulated the Firmicutes/Bacteroidetes ratio. In addition, the
bacterial composition in the cecal content samples from the three
groups in terms of bacterial phyla and genera were significantly
different (Figs. 3A and S3B). At the phylum level,
Verrucomicrobia was the predominant phylum in the PLR-H group
compared with that of the HS group. At the genus level, the fecal
microbiota was dominated by Lachnospiraceae_NK4A136_group,
Akkermansia and Lactobacillus. Besides, different
abundances of fecal bacterial taxa in the three groups were
identified by LEfSe analysis (Fig.
3B), which indicated that seven bacterial genera including
short-chain fatty acid (SCFA)-producing bacteria, such as
Akkermansia and Bifidobacterium, were enriched by
PLR-H, while the other eight taxa, including Rikenella,
Prevotellaceae_UCG-001 and Lachnoclostridium were
enriched in the HS group (LDA score >3). Moreover, indicator
species analysis showed a statistically significant higher relative
abundance of Faecalibaculum and Akkermansia at the
genus level in the PLR-H group than in the HS group (Fig. 3C). It was also found that
Lactobacillus and Bifidobacterium showed an increasing trend after
high-dose PLR administration compared with the HS group.
Additionally, the MDI revealed an increased value in HS mice
(Fig. 3D), which was reversed by
high-dose PLR intervention, indicating a decrease in the level of
intestinal flora disruption.
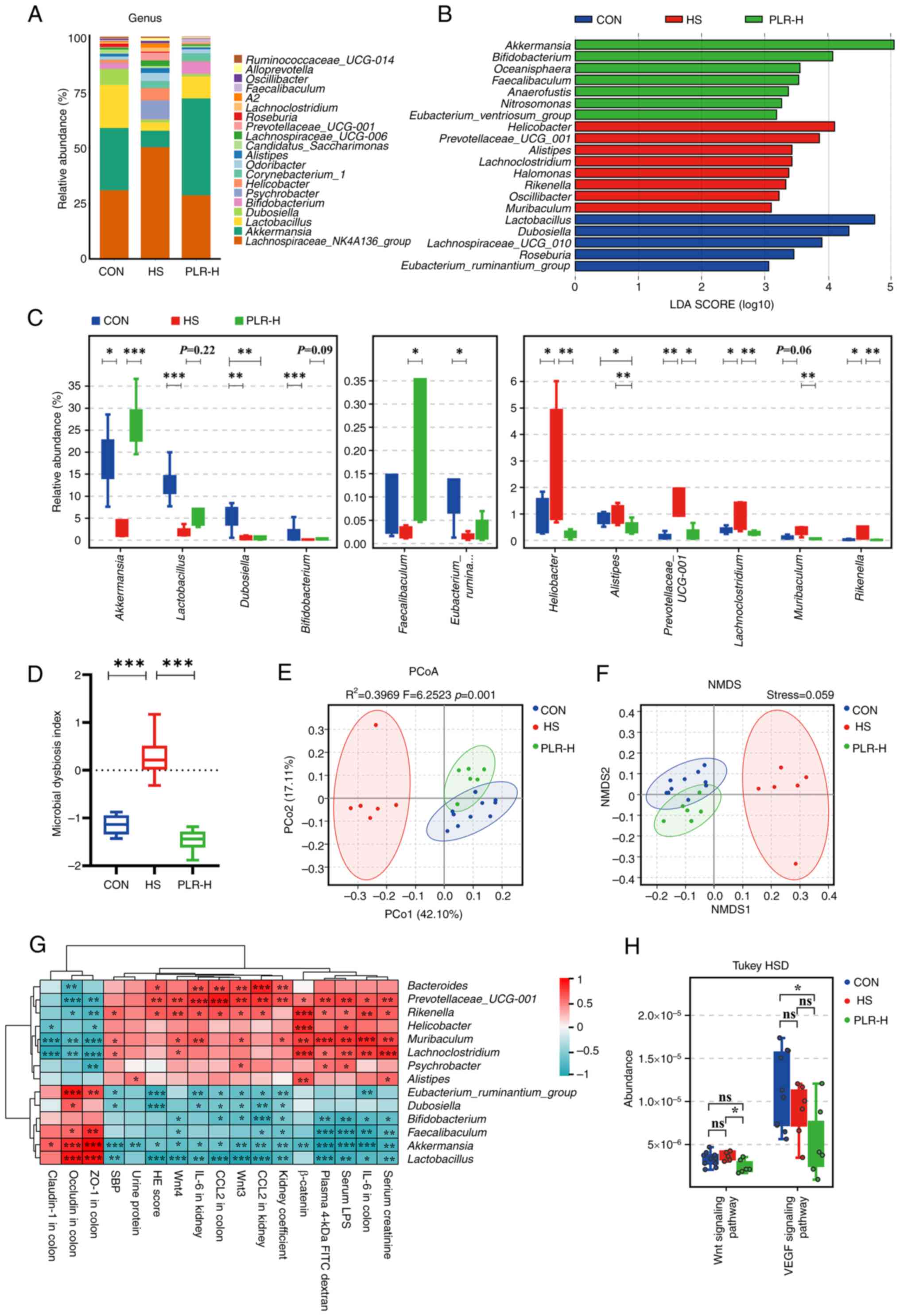 | Figure 3.PLR reverses intestinal microbial
dysbiosis in HS mice and remodels gut microbiota by increasing the
relative abundance of probiotics. (A) Relative bacterial abundance
at the genus level in the feces of mice. (B) Histogram of the LDA
score showing the biomarker at the genus level of each group. (C)
Relative abundance of indicator species at the genus level showing
the enriched bacteria in the gut microbiome among groups. (D)
Microbial Dysbiosis index of each group. (E) PCoA based on the
weighted UniFrac analysis of operational taxonomic units. (F) NMDS
based on the weighted UniFrac analysis of operational taxonomic
units. (G) Correlation heatmap of major indicator species and
biomarkers based on LDA score and major injury indicators, scale
shows correlation coefficient. (H) Kyoto Encyclopedia of Genes and
Genomes pathway analysis of function distribution and difference
analysis based on Tax4Fun prediction results. Results are expressed
as the mean ± SEM (n=6–10 for each group). ***P<0.001,
**P<0.01 and *P<0.05 were determined by one-way ANOVA with
Bonferroni's post hoc test or Kruskal-Wallis test with Dunn's post
hoc test in C and D, adonis analysis and anosim analysis in E,
Spearman analysis in G and ANOVA test with Tukey's HSD test in H.
PLR, Puerariae lobatae Radix; HS, high salt; LDA, linear
discriminant analysis; PCoA, principal coordinates analysis; NMDS,
non-metric multidimensional scaling; ns, not significant. |
In the present study, the alpha diversity was
impacted by neither HS nor PLR-H administration (Fig. S3C). However, NMDS and PCoA
demonstrated that the overall structure of the gut microbiota was
significantly different among the three groups (Fig. 3E and F), indicating that the gut
microbiota structure in HS mice was markedly influenced and shifted
closer towards that of the CON group by PLR intervention.
To further the present investigation, Spearman
correlation analysis was performed (Figs. 3G and S3D). The relative abundances of
Akkermansia, Lactobacillus and Bifidobacterium were
negatively correlated with kidney injury-related parameters and the
Wnt/β-catenin signaling pathway transcripts, while positively
correlated with tight junction proteins in the colon. The relative
abundance of these beneficial bacteria was also negatively
correlated with inflammatory factors not only in the colon but also
in the kidney. By contrast, the relative abundance of dominant
bacteria (such as Rikenella, Prevotellaceae_UCG-001 and
Lachnoclostridium) in the HS group had a strong positive
correlation with injury severity in the colon and kidney.
Tax4Fun prediction analysis was then used to
annotate the 16S rDNA data with metabolic pathways from the KEGG
database (Fig. 3H). The relative
abundance of the metabolism-associated signal transduction pathway
showed a significant downregulation of the Wnt signaling pathway in
the PLR-H group compared with that of the HS group, which was
consistent with the GSEA results obtained from RNA-seq about the
protective role of high-dose PLR in kidney injury. In addition, the
vascular endothelial growth factor (VEGF) signaling pathway, which
was proven by previous studies to promote fibrosis in CKD (38), was lower in the PLR-H group than
that in the HS group, suggesting that fibrotic signaling pathways
were suppressed by high-dose PLR supplementation.
Collectively, PLR administration could rectify gut
microbiota dysbiosis and enhance the relative abundances of
beneficial bacteria in mice, which were significantly negatively
correlated with CKD severity.
Gut microbiota reestablished by PLR
reduce renal tissue fibrosis and intestinal epithelial barrier
impairment
Next, to confirm whether the protective effect of
PLR against CKD was dependent on the gut microbiome, the gut
microbiota derived from mice treated with PLR-H were transplanted
to CKD mice (Fig. 4A). Baseline
weight and blood pressure had no differences among the groups
(Fig. S4A-C). Compared with the
HS-FMT group, SBP, serum creatinine and urine protein were
significantly reduced in the PLR-FMT group (Fig. 4B-D). As presented in Fig. 4E and F, the PLR-FMT group
demonstrated slight macroscopic injury and a lower kidney
coefficient than that of the HS-FMT group; however, no statistical
differences were exhibited in the liver and spleen coefficient
among the three groups (Fig.
S4D). Additionally, the kidney histopathological injury and
fibrosis proportion were also reduced in the PLR-FMT group compared
with those of the HS-FMT group (Fig.
4G). Furthermore, the mRNA levels of the inflammatory factors
and chemokines, along with the mRNA levels of renal tubular-related
injury and interstitial fibrosis, were also decreased in the
PLR-FMT group compared with those of the HS-FMT group (Figs. 4H, I and S4E).
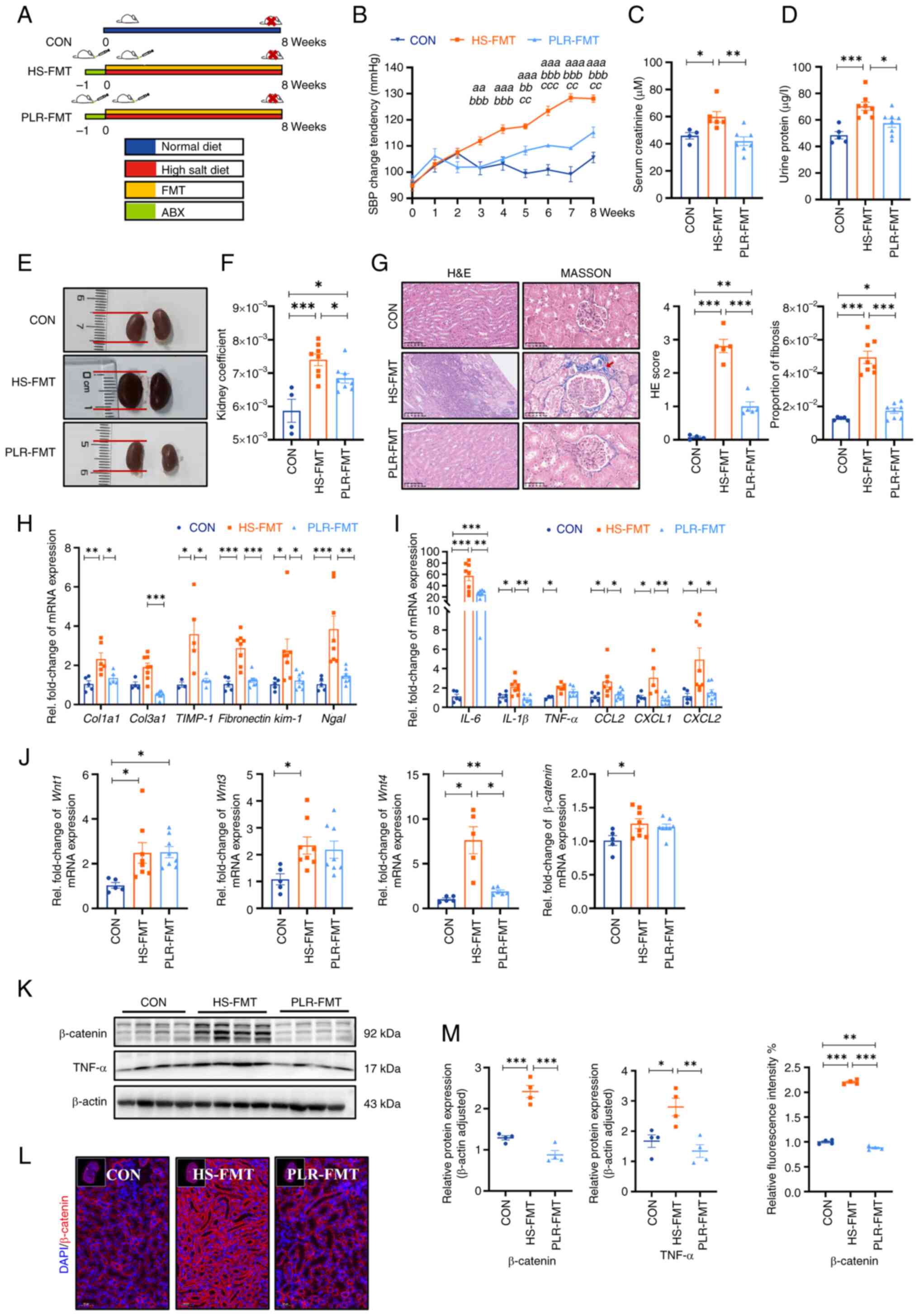 | Figure 4.Gut microbiota from mice treated with
PLR improve kidney tissue damage induced by HS diet and
downregulate the Wnt/β-catenin pathway. (A) Flow chart of FMT
experimental design. 8-week-old male C57BL/6 mice were administered
drinking water containing 2% w/v NaCl for 8 weeks. (B) SBP change
was evaluated non-invasively during experiment.
aaP<0.01 and aaaP<0.001 for the CON
group vs. the HS-FMT group on the corresponding week;
bbP<0.01 and bbbP<0.001 for the PLR-FMT
group vs. the HS-FMT group on the corresponding week;
ccP<0.01 and cccP<0.001 for the PLR-FMT
group vs. the CON group on the corresponding week. Two-way
repeated-measures ANOVA with Bonferroni's post hoc test was used
for statistical analysis. (C) Serum creatinine after 8 weeks. (D)
Urine protein after 8 weeks. (E) Representative gross anatomy
pictures of the kidney. (F) Kidney coefficient. (G) H&E and
MASSON staining of the kidney and the pathology scores and
histological analysis., Fibrosis in the glomeruli and renal
interstitium is indicated with a red arrow. H&E staining scale
bar, 100 µm; MASSON staining scale bar, 50 µm (n=4-8). (H) RT-qPCR
analysis of kidney injuries-related genes. (I) RT-qPCR analysis of
inflammatory factors' genes in kidney. (J) RT-qPCR analysis of
Wnt1, Wnt3, Wnt4 and β-catenin genes in kidney
(n=5-8). (K) β-catenin and TNF-α protein levels in the kidney. (L)
Representative in situ detection of β-catenin in renal
cortex was measured by immunofluorescent staining. Kidney tissue
sections were stained with DAPI (blue) and probed with β-catenin
(red). Scale bar, 50 µm (n=4). (M) Quantification of western blot
and immunofluorescence. Results are expressed as the mean ± SEM
(n=5–8 for each group). ***P<0.001, **P<0.01 and *P<0.05
were determined by one-way ANOVA with Bonferroni's post hoc test in
C, D and F-I. PLR, Puerariae lobatae Radix; HS, high salt;
FMT, fecal microbiota transplantation; ABX, antibiotic; SBP,
systolic blood pressure; HE, hematoxylin-eosin; RT-qPCR, reverse
transcription-quantitative PCR. |
In addition, the activation of the Wnt/β-catenin
pathway in the kidney was also explored. As shown in Fig. 4J, the mRNA levels of Wnt1, Wnt3,
Wnt4 and β-catenin of the PLR-FMT group were lower than
those in the HS-FMT group. Moreover, compared with the HS-FMT
group, the protein levels of β-catenin and TNF-α were significantly
reduced in the PLR-FMT group (Fig.
4K-M). Taken together, these results suggested that the gut
microbiota remodeled by PLR could protect kidney function, the
effect of which is probably associated with the downregulation of
the Wnt/β-catenin pathway.
Afterwards, it was investigated whether the gut
microbiota derived from the PLR group was effective in alleviating
the intestine inflammatory response and protecting intestinal
barrier function. Colonic length was significantly reduced in the
HS-FMT group compared with the CON group; however, this reduction
was significantly mitigated by the intervention of PLR-derived gut
microbiota and was restored to a length that was not significantly
different from that of the CON group (Fig. 5A). Decreased mRNA levels of
IL-6, CXCL1 and CCL2, as well as increased
IL-10 mRNA expression in the colon were observed in the
PLR-FMT group (Fig. 5C) compared
with those of the HS-FMT group, which indicated a lower colonic
inflammatory environment generated by PLR-derived gut microbiota.
Furthermore, the bioluminescence imaging system exhibited images of
less ROS in the colonic tissues of the PLR-FMT group than that of
the HS-FMT group (Fig. 5B).
Furthermore, H&E staining of the PLR-FMT colon revealed less
abscission and epithelial cell necrosis, and less inflammatory cell
infiltration compared with that of the HS-FMT group. As presented
in Fig. 5G-J, the mRNA and protein
levels of ZO-1, Occludin and Claudin-1 in the colon were
significantly higher; while the serum TNF-α level, LPS level and
FD-4 permeability were significantly lower in the PLR-FMT group
than those in the HS-FMT group (Fig.
5D-F). These results suggested that the HS-derived gut
microbiota triggered gut permeability by deteriorating intestinal
barrier function, while the microbiota remodeled by PLR protected
against colonic inflammation and intestinal barrier disruption,
which reduced translocation of bacterial products and inflammatory
cytokines into the serum. In addition, the aforementioned results
indicated that the gut microbiota reestablished by PLR could reduce
renal tissue fibrosis and colonic epithelial barrier damage.
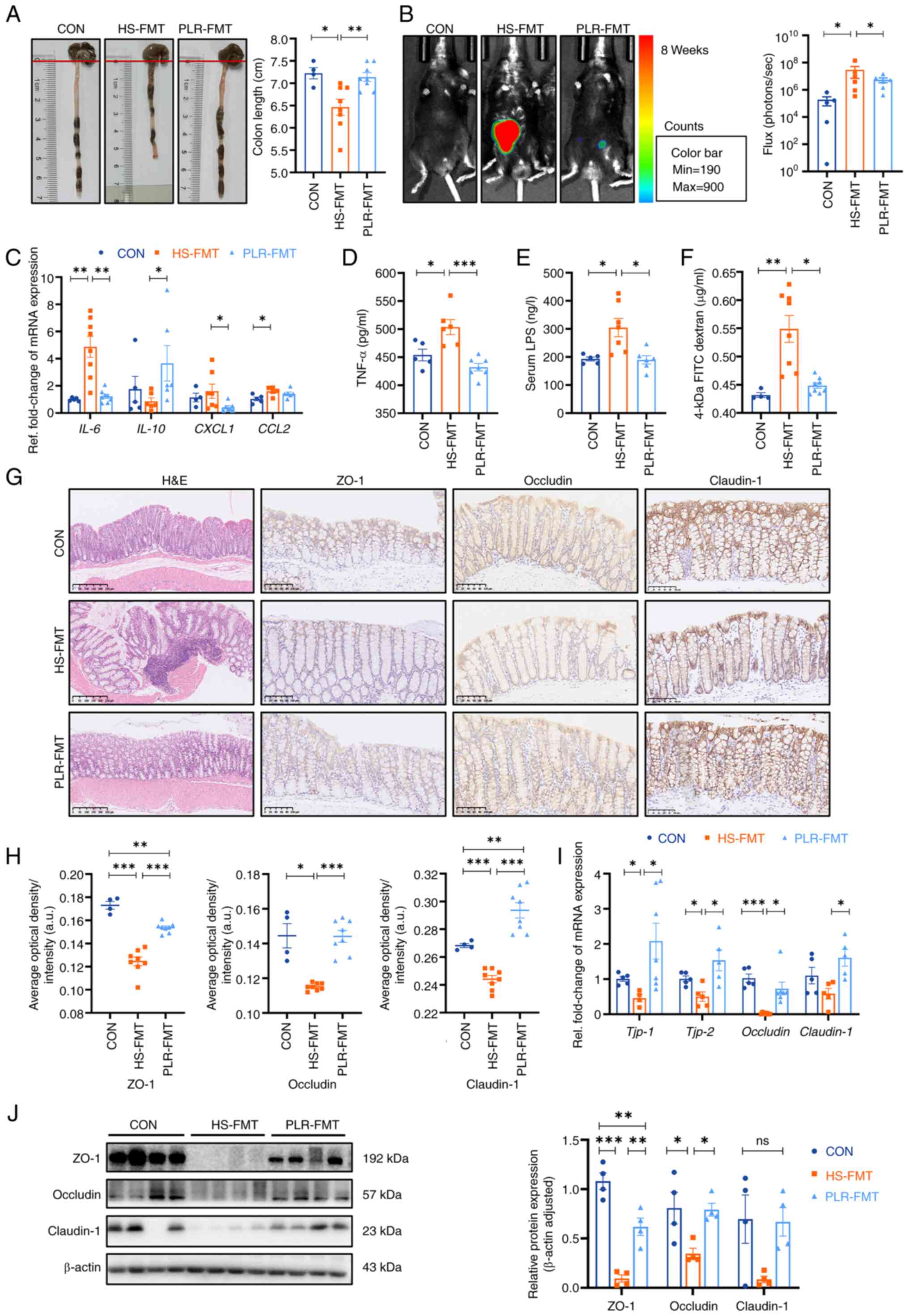 | Figure 5.FMT from mice treated with PLR reduce
intestinal inflammation and protect intestinal barrier function.
(A) Representative gross anatomy pictures of the colon and the
colon length measurement. (B) Representative L-012 fluorescent
staining and animal fluorescence imaging (n=5-6). (C) RT-qPCR
analysis of inflammatory factors' genes in colon. (D) Relative
serum TNF-αlevels. (E) Relative serum LPS levels. (F) FD-4 levels
in the plasma. (G) H&E staining and ZO-1, Occludin and
Claudin-1 immunohistochemical staining in colon tissues. Scale bar,
100 or 250 µm (n=3-6). (H) Quantification of immunohistochemistry.
(I) RT-qPCR analysis of tight junction proteins genes in colon
(n=5-6). (J) ZO-1, Occludin and Claudin-1 protein levels in the
colon (n=4). Results are expressed as the mean ± SEM (n=4–8 for
each group). ***P<0.001, **P<0.01 and *P<0.05 were
determined by one-way ANOVA with Bonferroni's post hoc test. FMT,
fecal microbiota transplantation; PLR, Puerariae lobatae
Radix; HS, high salt; CON, control; LPS, lipopolysaccharide; FD-4,
FITC-dextran 4-KD; H&E, hematoxylin-eosin; ns, not significant;
RT-qPCR, reverse transcription-quantitative PCR; ZO-1, zonula
occludens 1. |
Gut microbiota rebuilt by PLR promote
intestinal homeostasis in CKD
To investigate the gut microbiota characteristic
that exerted renoprotective effects in the FMT intervention, the
microbial compositions from the HS-FMT and PLR-FMT groups were
further analyzed. The main composition in either the phylum or
genus level was similar among the groups (Figs. 6A and S5A). Notably, LEfSe and indicator
species analysis indicated that Lactobacillus and
Ruminococcaceae_UCG_014 at the genus level were
significantly enriched by PLR-FMT (LDA score >3.6, Fig. 6B and C), while higher relative
abundances of Akkermansia and Bifidobacterium were
also found in the PLR-FMT group. As shown in Fig. S6A, the main composition in the
genus level was similar between the PLR-H and the PLR-FMT groups.
Indicator species analysis revealed 111 shared genera species in
the PLR-H and PLR-FMT groups (Fig.
S6B). There were no significant differences in the relative
abundance of major genera. The relative abundance of
Akkermansia and Alistipes in the PLR-FMT group was
significantly lower than that of the PLR-H group, but the relative
abundance of Lactobacillus and Lachnoclostridium was
significantly higher than that of the PLR-H group (Fig. S6C). In addition, the increased MDI
value in the HS-FMT mice was reversed by PLR-FMT intervention
(Fig. 6D). Although alpha
diversity was not impacted by either HS-FMT or PLR-FMT
administration (Fig. S5B), PCoA
analysis and NMDS showed that the gut microbiota structure was
completely clustered (Fig. 6E and
F). Furthermore, Spearman correlation analysis revealed that
Lactobacillus, Akkermansia and
Ruminococcaceae_UCG_014, which were enriched in the PLR-FMT
group, were significantly positively correlated with the gut
barrier-related tight junction protein, while negatively correlated
with damage-related parameters and the Wnt signaling
pathway-related genes in kidney (Fig.
6G). Finally, different gut microbial metabolic functions were
explored via Tax4Fun prediction analysis from the KEGG data
(Fig. 6H). The VEGF and Wnt
signaling pathways were significantly downregulated in PLR-FMT mice
compared with the HS-FMT group.
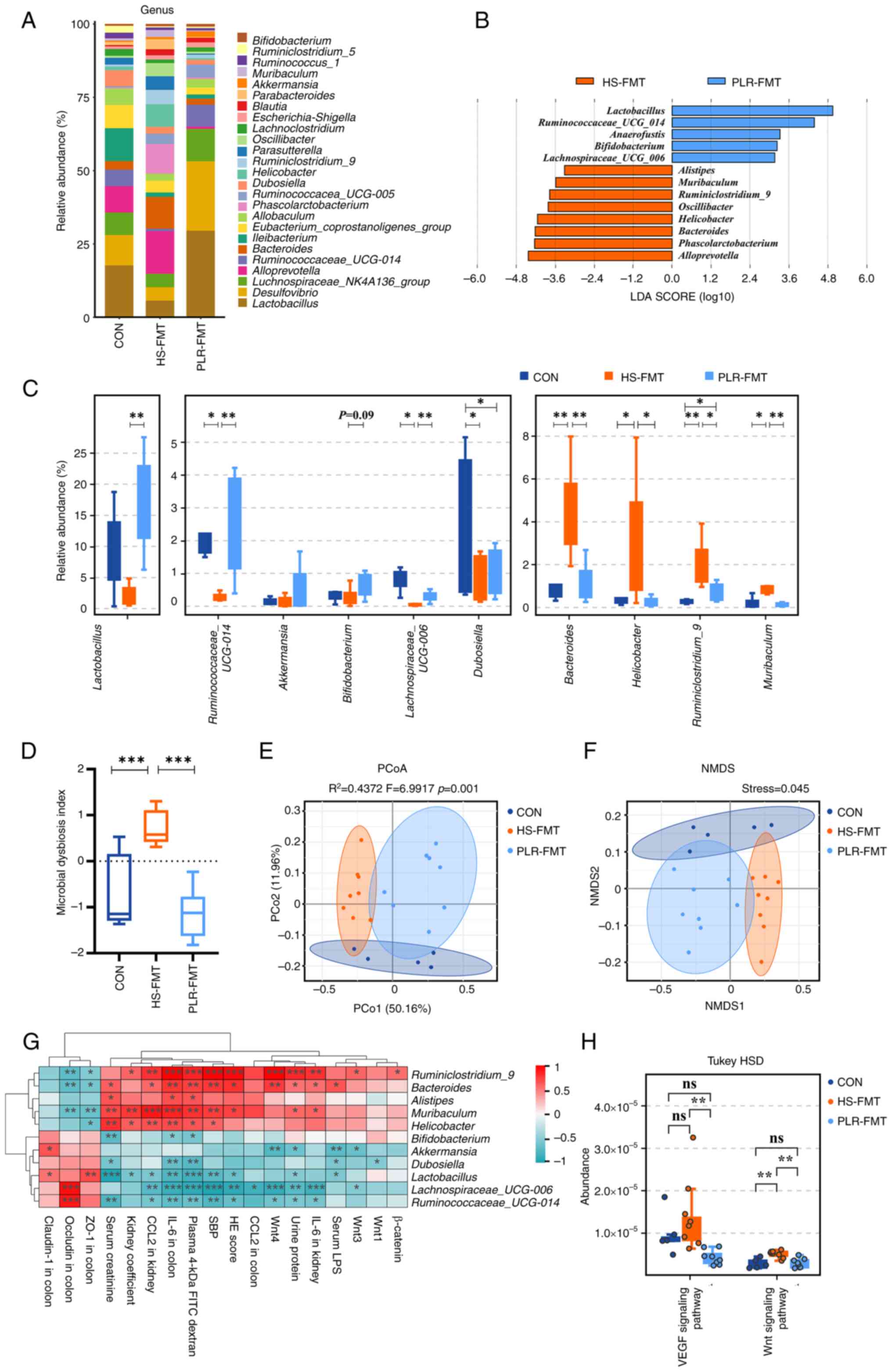 | Figure 6.FMT from mice treated with PLR
relieve intestinal microbial dysbiosis and rebuild healthy
microbiota environment. (A) Relative bacterial abundance at the
genus level in the feces of mice. (B) Histogram of the LDA score
showing the biomarker at the genus level between the HS-FMT group
and the PLR-FMT group. (C) Relative abundance of indicator species
at the genus level showing the enriched bacteria in the gut
microbiome among groups. (D) Microbial Dysbiosis index of each
group. (E) PCoA based on the weighted UniFrac analysis of
operational taxonomic units. (F) NMDS based on the weighted UniFrac
analysis of operational taxonomic units. (G) Correlation heatmap of
major indicator species and biomarkers based on LDA score and major
injury indicators, scale shows correlation coefficient. (H) Kyoto
Encyclopedia of Genes and Genomes pathway analysis of function
distribution and difference analysis based on Tax4Fun prediction
results. Results are expressed as the mean ± SEM (n=5–8 for each
group). ***P<0.001, **P<0.01 and *P<0.05 were determined
by one-way ANOVA with Bonferroni's post hoc test or Kruskal-Wallis
test with Dunn's post hoc test in C and D, adonis analysis and
anosim analysis in E, Spearman analysis in G, ANOVA test with
Tukey's HSD test in H. FMT, fecal microbiota transplantation; HS,
high salt; PLR, Puerariae lobatae Radix; LDA, linear
discriminant analysis; PCoA, principal coordinates analysis; NMDS,
non-metric multidimensional scaling. |
Overall, the aforementioned results indicated that
FMT intervention could alter the structure of the gut microbiota,
and the supplementary microbiota remodeled by high-dose PLR exerted
a similar renoprotective effect as the PLR-H group. Further,
Lactobacillus, Akkermansia and Bifidobacterium might
be the dominating beneficial bacteria that induced CKD alleviation
in both the PLR-H and PLR-FMT groups.
Discussion
PLR is a traditional Chinese medicine according to
the Chinese pharmacopoeia, and its main components are puerarin,
daidzin and daidzein. These three main components are flavonoid
natural drug compounds, which are characterized by low solubility,
poor absorption and rapid metabolism, which to a moderate extent
limit their application in diseases. However, PLR is not a simple
superimposition of the aforementioned three individual ingredients.
It has been previously shown that compared with the administration
of pure daidzin alone, administration of the same dose of crude
daidzin contained in a methanol extract of Radix Puerariae
increased bioavailability of daidzin by 10-fold (39). It can be observed that PLR solves
the dilemma of low oral bioavailability but high bioactivity of
monomer drugs. In the present study, for the first time, to the
best of our knowledge, it was found that oral PLR administration
could alleviate CKD. On the other hand, from the point of view of
intestinal flora, it is more likely to be affected by PLR compared
with monomer components. The results of preclinical and clinical
studies on herb-microbiota interactions suggest that traditional
herbs can exert health-promoting and disease-preventing effects by
influencing the structure of the intestinal flora (40). In the present study, the protective
effect of PLR was achieved, at least partly, by remodeling the gut
microbiota.
Salt is one of the most common dietary elements and
plays an important role in maintaining the water-salt balance of
the body. In the present study, a prolonged excessive HS diet led
to gut microbiota disruption, intestinal inflammation and
permeability increasement. It was also identified that an HS diet
induced both inflammatory and fibrotic damage to the kidneys,
probably because of harmful bacterial metabolites, such as LPS and
inflammatory factors, including TNF-α, originating from the gut
that then enters the circulation and induces damage to distant
organs. However, the therapeutic intervention of PLR could protect
against both intestinal and renal injury, which may be a result of
its ability to reverse disturbances in the gut microbiota. It was
also found that PLR intervention increased Akkermansia,
Lactobacillus and Bifidobacterium and decreased
Rikenella, Prevotellaceae_UCG-001 and
Lachnoclostridium. Previously, numerous clinical studies
have found that the gut microbiota can mediate associations between
the gut and other organs, such as via the ‘gut-brain axis’ and
‘gut-kidney axis’ (41,42). This suggests that the gut
microbiota may be a potential target for intervention in abenteric
disease progression. Indeed, it was confirmed that PLR exerted
efficacy, at least partially, through the gut microbiota in the FMT
experiment. Akkermansia muciniphila (A. muciniphila)
is considered one of the key players in colonic mucus-associated
microbiota and is necessary for the gut to produce mucus to
maintain a healthy mucus layer and intestinal wall thickness
(43,44). A recent study demonstrated that
increased colonization of A. muciniphila in the colon of
mice mitigated gut barrier leakage and blood endotoxemia in
experimental colitis (45).
Furthermore, previous clinical research revealed that the relative
abundance of Akkermansia in patients with CKD was
significantly lower than that in the healthy group (46). Oral gavage of mice with A.
muciniphila protected against HFD/CCl4-induced liver
and kidney fibrosis by modulating the inflammatory response
(47). Moreover, A.
muciniphila administration suppressed epithelial-mesenchymal
transition and reduced renal interstitial fibrosis in 5/6
nephrectomy rats (48). This
suggested that besides repairing the intestinal barrier,
Akkermansia was also closely associated with alleviating the
damage of CKD and renal fibrosis. Similarly, in the present study,
the results also revealed that the relative abundances of
Akkermansia were negatively correlated with the nephritic
histopathological fibrotic degree and biochemical indexes of kidney
injury. Therefore, it was hypothesized that the alleviating effect
of PLR on CKD and renal fibrosis may be associated with increased
colonization of Akkermansia in the intestine.
The gut microbiota is a complex ecosystem in which
various bacteria strains can interact with each other through
resource competition and nutrient symbiosis (14). The protective effect of a single
bacterium, the symbiotic relationship between beneficial bacteria
in intestinal homeostasis maintenance and disease protection should
not be overlooked (49). In the
present study, it was found that PLR also increased the relative
abundances of Bifidobacterium and Lactobacillus,
which are capable of enhancing the intestinal mucus layer and
goblet cell function, thus protecting intestinal barrier integrity
(50,51). Additionally, in vitro
experiments demonstrated that A. muciniphila can stimulate
the growth and change the gene expression profile of
Lactobacillus (52,53). Studies have also shown that
Bifidobacterium and Lactobacillus have potential
benefits in reducing uremic toxin levels and protecting against CKD
(54–56). Regarding PLR, it is rich in
macromolecules such as starch, cellulose and lignin (49,57).
These macromolecules reach the colon and provide energy for the
synergistic growth of carbohydrate-utilizing bacteria such as
Bifidobacterium and Lactobacillus (9,14,58).
In addition, the aforementioned probiotics, together with
Faecalibaculum, which could be enriched by PLR intervention
in the present study, have been reported as SCFA-producing bacteria
(59–61), capable of increasing SCFA
production and thus creating an anti-inflammatory and
anti-oxidative environment in the intestine. Moreover, a
non-inflammatory stable state reduced gut barrier damage and
permeation of harmful metabolites into the plasma, thus decreasing
the adverse impact on abenteric organs. Taken together, the gut
microbiota reshaped by PLR and characterized by Akkermansia,
Bifidobacterium and Lactobacillus is a key factor in its
enteroprotective and nephroprotective effects.
A continuous intake of HS necessitates salt to be
excreted in the urine to maintain the water-salt balance in the
plasma, and this sustained kidney stimulation in urine production
may cause pathological damage and kidney fibrosis. Currently, the
Wnt signaling pathway is more accepted to influence HS-related CKD
(62). Previous literature
indicated that transient activation of Wnt/β-catenin facilitates
kidney tissue generation after acute kidney injury, whereas
sustained activation stimulates kidney fibrosis in CKD (63). With the presence of a HS load, mice
presented with more fibrosis and upregulated Wnt/β-catenin
signaling in the heart and kidney (64,65).
In the present study, PLR intervention significantly alleviated the
degree of renal fibrosis in HS-induced CKD. More importantly, GSEA
showed the downregulation of the canonical Wnt signaling pathway in
the PLR group. This might explain a potential pathway by which PLR
protects the kidney from fibrosis. In the present study, the
Tax4Fun prediction analysis from the KEGG database was inspected
and it was found that the Wnt signaling pathway was correlated with
the gut microbial metabolism. Besides, the current results showed
that the activation of the Wnt/β-catenin pathway and the level of
pathological damage in the kidney were significantly negatively
correlated with the relative abundance of beneficial bacteria, such
as Akkermansia, Lactobacillus and Bifidobacterium,
increased by PLR intervention, which led the authors to hypothesize
that the gut microbiota may be effective in regulating the Wnt
signaling pathway expression and subsequently decelerating renal
fibrosis progression. Numerous studies have found that the Wnt
signaling pathway can be regulated by the gut microbiota (66,67).
Treatment of mice with A. muciniphila is reported to
significantly suppress epithelial-mesenchymal transition and reduce
renal interstitial fibrosis (48).
Furthermore, treatment of Bifidobacterium bifidum and
Lactobacillus gasseri with quercetin could inhibit the
canonical Wnt/β-catenin signaling pathway to protect against
colorectal cancer in mice (68).
Both PLR and PLR-FMT interventions could significantly downregulate
the Wnt signaling pathway. From this, it was hypothesized by the
authors that alterations in the Wnt signaling pathway and
alleviation of renal fibrosis by PLR intervention was associated
with the gut microbiota and its metabolism.
Inflammation and other signaling pathways can also
contribute to CKD. In the present study, a significantly elevated
TNF-α protein level was found in the serum and kidneys of CKD mice.
Increased TNF-α is one of the upstream targets for triggering
β-catenin activation in the kidney (69,70),
and the reduction of β-catenin has been shown to result in a lower
expression of fibrosis markers including fibronectin, Col1a1 and
Col3a1 (71–74). However, the elevation of TNF-α and
β-catenin was significantly suppressed by PLR intervention in the
present study, which added to the possible targets of the
nephroprotective effects of PLR. In addition, the VEGF signaling
pathway could also be downregulated by PLR intervention in the
current study. Previous studies have shown that numerous therapies
targeted the HIF-1α/VEGF signaling pathway to relieve liver
fibrosis (75,76). The VEGF signaling pathway was also
reported to have a synergistic effect with the Wnt/β-catenin
signaling pathway in angiogenesis and fibrosis (77,78).
In the present study, these two pathways may have corporate effects
on renal fibrosis after PLR intervention.
In the present study, the protective effect of PLR
against elevated blood pressure induced by HSD was found. Sodium
excretion and diuresis using diuretics is one of the clinical
strategies to reduce blood pressure. In the present study, there
was a trend toward higher urinary sodium excretion in the PLR group
compared with the HS group, but the difference was not significant.
To the best of our knowledge, no previous studies have identified a
diuretic effect of PLR or its main components. Therefore, further
exploration is needed regarding whether PLR can promote urinary
sodium excretion.
PLR, a traditional Chinese medicine and food
homologous herb, has multi-target and multi-pathway pharmacodynamic
routes of action. In the present study, it was confirmed that PLR
could alleviate CKD by remodeling the gut microbiota, repairing the
intestinal epithelial barrier, and downregulating the Wnt signaling
pathway in the kidney. However, there are certain limitations to
the present study; although a previous study suggested that removal
of gut flora could attenuate HS diet-induced kidney injury
(11), in the present study, this
finding was not validated and the FMT experiment was directly
performed. Furthermore, the present study focused on gut microbiota
entirely and its beneficial effect on CKD, and single strains of
bacteria were not accessed. In addition, although focus was
addressed on Wnt1, Wnt3 and Wnt4 in the canonical Wnt/β-catenin
signaling pathway, this does not mean that other Wnt genes have no
influence (79). It has been
previously revealed that upregulation of the Wnt4 and Wnt5 genes
could activate the non-canonical Wnt pathway in hepatic stellate
cells of fibrotic livers (80).
Moreover, it was apparent that the beneficial pharmaceutical
effects of PLR were mediated through several mechanisms. Previous
studies have shown that mTOR and AMPK pathways are also involved in
the regulation and progression of CKD (81,82).
In the future, more in depth understanding about the remodeling
ability of PLR towards the gut microbiota should be further
explored and mTOR and AMPK pathways could be potential targets for
subsequent research. Overall, the present study provided evidence
for a new function of PLR regarding kidney protection and a novel
direction for the treatment of kidney disease.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant nos. 82173304 and 82370782) and the
Natural Science Foundation of Guangdong (grant no.
2023A1515012565).
Availability of data and materials
The data generated in the present study may be
found in the Genome Sequence Archive (Genomics, Proteomics &
Bioinformatics 2021), National Genomics Data Center (Nucleic Acids
Res 2022), China National Center for Bioinformation/Beijing
Institute of Genomics, Chinese Academy of Sciences under accession
numbers CRA015711, CRA015797 and CRA015712 or at the following URL:
https://ngdc.cncb.ac.cn/gsa.
Authors' contributions
PW, JZ and JWX conceived, designed and interpreted
the study. JWX, ZRZ, QS and YY undertook the data acquisition and
analysis. MX, ZRZ, BLW, PCH and ZYF performed the experiments. JZ
and JWX were major contributors in writing the manuscript. JZ and
PW contributed to the review of data and the final manuscript. PW
and JWX confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The animal study protocol was approved (approval
no. L-2019216) by the Institutional Animal Ethical Care Committee
of Southern Medical University Experimental Animal Center
(Guangzhou, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Cockwell P and Fisher LA: The global
burden of chronic kidney disease. Lancet. 395:662–664. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Garofalo C, Borrelli S, Provenzano M, De
Stefano T, Vita C, Chiodini P, Minutolo R, De Nicola L and Conte G:
Dietary salt restriction in chronic kidney disease: A meta-analysis
of randomized clinical trials. Nutrients. 10:7322018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kim SM and Jung JY: Nutritional management
in patients with chronic kidney disease. Korean J Intern Med.
35:1279–1290. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oppelaar JJ and Vogt L: Body
Fluid-Independent effects of dietary salt consumption in chronic
kidney disease. Nutrients. 11:27792019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yu HC, Burrell LM, Black MJ, Wu LL, Dilley
RJ, Cooper ME and Johnston CI: Salt induces myocardial and renal
fibrosis in normotensive and hypertensive rats. Circulation.
98:2621–2628. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou B, Liu Y, Kahn M, Ann DK, Han A, Wang
H, Nguyen C, Flodby P, Zhong Q, Krishnaveni MS, et al: Interactions
between β-catenin and transforming growth factor-β signaling
pathways mediate epithelial-mesenchymal transition and are
dependent on the transcriptional co-activator cAMP-response
element-binding protein (CREB)-binding protein (CBP). J Biol Chem.
287:7026–7038. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zmora N, Suez J and Elinav E: You are what
you eat: Diet, health and the gut microbiota. Nat Rev Gastroenterol
Hepatol. 16:35–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He FJ, Tan M, Ma Y and MacGregor GA: Salt
reduction to prevent hypertension and cardiovascular disease: JACC
State-of-the-Art review. J Am Coll Cardiol. 75:632–647. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Miranda PM, De Palma G, Serkis V, Lu J,
Louis-Auguste MP, McCarville JL, Verdu EF, Collins SM and Bercik P:
High salt diet exacerbates colitis in mice by decreasing
Lactobacillus levels and butyrate production. Microbiome. 6:572018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan X, Jin J, Su X, Yin X, Gao J, Wang X,
Zhang S, Bu P, Wang M, Zhang Y, et al: Intestinal flora modulates
blood pressure by regulating the synthesis of Intestinal-derived
corticosterone in high salt-induced hypertension. Circ Res.
126:839–853. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu J, Luo H, Wang J, Tang W, Lu J, Wu S,
Xiong Z, Yang G, Chen Z, Lan T, et al: Enteric dysbiosis-linked gut
barrier disruption triggers early renal injury induced by chronic
high salt feeding in mice. Exp Mol Med. 49:e370. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li HB, Xu ML, Xu XD, Tang YY, Jiang HL, Li
L, Xia WJ, Cui N, Bai J, Dai ZM, et al: Faecalibacterium
prausnitzii attenuates CKD via Butyrate-Renal GPR43 axis. Circ Res.
131:e120–e134. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kalantar-Zadeh K, Jafar TH, Nitsch D,
Neuen BL and Perkovic V: Chronic kidney disease. Lancet.
398:786–802. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang L, Liu Z, Wu P, Yue X, Lian Z, He P,
Liu Y, Zhou R and Zhao J: Puerariae lobatae radix alleviates
Pre-Eclampsia by remodeling gut microbiota and protecting the gut
and placental barriers. Nutrients. 14:50252022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Inker LA, Astor BC, Fox CH, Isakova T,
Lash JP, Peralta CA, Kurella Tamura M and Feldman HI: KDOQI US
commentary on the 2012 KDIGO clinical practice guideline for the
evaluation and management of CKD. Am J Kidney Dis. 63:713–735.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Q, Liu W, Feng Y, Hou H, Zhang Z, Yu Q,
Zhou Y, Luo Q, Luo Y, Ouyang H, et al: Radix Puerariae thomsonii
polysaccharide (RPP) improves inflammation and lipid peroxidation
in alcohol and high-fat diet mice by regulating gut microbiota. Int
J Biol Macromol. 209:858–870. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang SS, Zhang NN, Guo S, Liu SJ, Hou YF,
Li S, Ho CT and Bai NS: Glycosides and flavonoids from the extract
of Pueraria thomsonii Benth leaf alleviate type 2 diabetes in
high-fat diet plus streptozotocin-induced mice by modulating the
gut microbiota. Food Funct. 13:3931–3945. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen R, Wu P, Cai Z, Fang Y, Zhou H,
Lasanajak Y, Tang L, Ye L, Hou C and Zhao J: Puerariae
lobatae Radix with chuanxiong Rhizoma for treatment of cerebral
ischemic stroke by remodeling gut microbiota to regulate the
brain-gut barriers. J Nutr Biochem. 65:101–114. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Avila-Carrasco L, García-Mayorga EA,
Díaz-Avila DL, Garza-Veloz I, Martinez-Fierro ML and González-Mateo
GT: Potential therapeutic effects of natural plant compounds in
kidney disease. Molecules. 26:60962021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang W, Sheng L, Chen Y, Li Z, Wu H, Ma J,
Zhang D, Chen X and Zhang S: Total coumarin derivates from
Hydrangea paniculata attenuate renal injuries in cationized-BSA
induced membranous nephropathy by inhibiting complement activation
and interleukin 10-mediated interstitial fibrosis. Phytomedicine.
96:1538862022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma L, Huang M, Sun G, Lin Y, Lu D and Wu
B: Puerariae lobatae radix protects against UVB-induced skin
aging via antagonism of REV-ERBα in mice. Front Pharmacol.
13:10882942022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
National Research Council (US) Committee
for the Update of the Guide for the care use of Laboratory Animals:
Guide for the Care and Use of Laboratory Animals. 8th edition.
National Academies Press (US); Washington (DC): 2011
|
|
23
|
Li Z, Zhang X, Wu H, Ma Z, Liu X, Ma J,
Zhang D, Sheng L, Chen X and Zhang S: Hydrangea paniculata
coumarins attenuate experimental membranous nephritis by
bidirectional interactions with the gut microbiota. Commun Biol.
6:11892023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou Z, Qiu Y, Li K, Sun Q, Xie M, Huang
P, Yu Y, Wang B, Xue J, Zhu Z, et al: Unraveling the impact of
Lactobacillus spp. and other urinary microorganisms on the efficacy
of mirabegron in female patients with overactive bladder. Front
Cell Infect Microbiol. 12:10303152022. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qiu Y, Gao Y, Chen C, Xie M, Huang P, Sun
Q, Zhou Z, Li B, Zhao J and Wu P: Deciphering the influence of
urinary microbiota on FoxP3+ regulatory T cell infiltration and
prognosis in Chinese patients with non-muscle-invasive bladder
cancer. Human Cell. 35:511–521. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Caporaso JG, Kuczynski J, Stombaugh J,
Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich
JK, Gordon JI, et al: QIIME allows analysis of high-throughput
community sequencing data. Nat Methods. 7:335–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ondov BD, Bergman NH and Phillippy AM:
Interactive metagenomic visualization in a Web browser. BMC
Bioinformatics. 12:3852011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Segata N, Izard J, Waldron L, Gevers D,
Miropolsky L, Garrett WS and Huttenhower C: Metagenomic biomarker
discovery and explanation. Genome Biol. 12:R602011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aßhauer KP, Wemheuer B, Daniel R and
Meinicke P: Tax4Fun: Predicting functional profiles from
metagenomic 16S rRNA data. Bioinformatics. 31:2882–2884. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Love MI, Huber W and Anders S: Moderated
estimation of fold change and dispersion for RNA-seq data with
DESeq2. Genome Biol. 15:5502014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wirtz S, Popp V, Kindermann M, Gerlach K,
Weigmann B, Fichtner-Feigl S and Neurath MF: Chemically induced
mouse models of acute and chronic intestinal inflammation. Nat
Protoc. 12:1295–1309. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Manabe E, Ito S, Ohno Y, Tanaka T, Naito
Y, Sasaki N, Asakura M, Masuyama T, Ishihara M and Tsujino T:
Reduced lifespan of erythrocytes in Dahl/Salt sensitive rats is the
cause of the renal proximal tubule damage. Sci Rep. 10:220232020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gunathilake M, Lee J, Choi IJ, Kim YI,
Yoon J, Sul WJ, Kim JF and Kim J: Alterations in gastric microbial
communities are associated with risk of gastric cancer in a Korean
population: A Case-Control study. Cancers. 12:26192020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yan AW, Fouts DE, Brandl J, Stärkel P,
Torralba M, Schott E, Tsukamoto H, Nelson KE, Brenner DA and
Schnabl B: Enteric dysbiosis associated with a mouse model of
alcoholic liver disease. Hepatology. 53:96–105. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu LC, Shih YA, Wu LL, Lin YD, Kuo WT,
Peng WH, Lu KS, Wei SC, Turner JR and Ni YH: Enteric dysbiosis
promotes antibiotic-resistant bacterial infection: Systemic
dissemination of resistant and commensal bacteria through
epithelial transcytosis. Am J Physiol Gastrointest Liver Physiol.
307:G824–G835. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Miao C, Zhu X, Wei X, Long M, Jiang L, Li
C, Jin D and Du Y: Pro- and anti-fibrotic effects of vascular
endothelial growth factor in chronic kidney diseases. Ren Fail.
44:881–892. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Keung WM, Lazo O, Kunze L and Vallee BL:
Potentiation of the bioavailability of daidzin by an extract of
Radix puerariae. Proc Natl Acad Sci USA. 93:4284–4288. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen F, Wen Q, Jiang J, Li HL, Tan YF, Li
YH and Zeng NK: Could the gut microbiota reconcile the oral
bioavailability conundrum of traditional herbs? J Ethnopharmacol.
179:253–264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang HX and Wang YP: Gut Microbiota-brain
Axis. Chin Med J (Engl). 129:2373–2380. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yang T, Richards EM, Pepine CJ and Raizada
MK: The gut microbiota and the brain-gut-kidney axis in
hypertension and chronic kidney disease. Nat Rev Nephrol.
14:442–456. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Belzer C and de Vos WM: Microbes
inside-from diversity to function: The case of Akkermansia. ISME J.
6:1449–1458. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lopez-Siles M, Enrich-Capó N, Aldeguer X,
Sabat-Mir M, Duncan SH, Garcia-Gil LJ and Martinez-Medina M:
Alterations in the abundance and Co-occurrence of Akkermansia
muciniphila and Faecalibacterium prausnitzii in the colonic
mucosa of inflammatory bowel disease subjects. Front Cell Infect
Microbiol. 8:2812018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wade H, Pan K, Duan Q, Kaluzny S, Pandey
E, Fatumoju L, Saraswathi V, Wu R, Harris EN and Su Q:
Akkermansia muciniphila and its membrane protein Ameliorates
intestinal inflammatory stress and promotes epithelial wound
healing via CREBH and miR-143/145. J Biomed Sci. 30:382023.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Li F, Wang M, Wang J, Li R and Zhang Y:
Alterations to the gut microbiota and their correlation with
inflammatory factors in chronic kidney disease. Front Cell Infect
Microbiol. 9:2062019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Keshavarz Azizi Raftar S, Ashrafian F,
Yadegar A, Lari A, Moradi HR, Shahriary A, Azimirad M, Alavifard H,
Mohsenifar Z, Davari M, et al: The protective effects of live and
pasteurized Akkermansia muciniphila and its extracellular
vesicles against HFD/CCl4-induced liver injury. Microbiol Spectr.
9:e00484212021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pei T, Hu R, Wang F, Yang S, Feng H, Li Q,
Zhang J, Yan S, Ju L, He Z, et al: Akkermansia muciniphila
ameliorates chronic kidney disease interstitial fibrosis via the
gut-renal axis. Microb Pathog. 174:1058912023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lian Z, Xu Y, Wang C, Chen Y, Yuan L, Liu
Z, Liu Y, He P, Cai Z and Zhao J: Gut microbiota-derived melatonin
from Puerariae lobatae Radix-resistant starch
supplementation attenuates ischemic stroke injury via a positive
microbial co-occurrence pattern. Pharmacol Res. 190:1067142023.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Engevik MA, Luk B, Chang-Graham AL, Hall
A, Herrmann B, Ruan W, Endres BT, Shi Z, Garey KW, Hyser JM and
Versalovic J: Bifidobacterium dentium fortifies the intestinal
mucus layer via autophagy and calcium signaling pathways. mBio.
10:e01087–19. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang J, Ji H, Wang S, Liu H, Zhang W,
Zhang D and Wang Y: Probiotic lactobacillus plantarum promotes
intestinal barrier function by strengthening the epithelium and
modulating gut microbiota. Front Microbiol. 9:19532018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang B, Chen X, Chen Z, Xiao H, Dong J, Li
Y, Zeng X, Liu J, Wan G, Fan S and Cui M: Stable colonization of
Akkermansia muciniphila educates host intestinal
microecology and immunity to battle against inflammatory intestinal
diseases. Exp Mol Med. 55:55–68. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cozzolino A, Vergalito F, Tremonte P,
Iorizzo M, Lombardi SJ, Sorrentino E, Luongo D, Coppola R, Di Marco
R and Succi M: Preliminary evaluation of the safety and probiotic
potential of Akkermansia muciniphila DSM 22959 in comparison
with lactobacillus rhamnosus GG. Microorganisms. 8:1892020.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang X, Yang S, Li S, Zhao L, Hao Y, Qin
J, Zhang L, Zhang C, Bian W, Zuo L, et al: Aberrant gut microbiota
alters host metabolome and impacts renal failure in humans and
rodents. Gut. 69:2131–2142. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Tian N, Li L, Ng JKC and Li PKT: The
potential benefits and controversies of probiotics use in patients
at different stages of chronic kidney disease. Nutrients.
14:40442022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mitrović M, Stanković-Popović V, Tolinački
M, Golić N, Soković Bajić S, Veljović K, Nastasijević B, Soldatović
I, Svorcan P and Dimković N: The Impact of synbiotic treatment on
the levels of gut-derived uremic toxins, inflammation, and gut
microbiome of chronic kidney disease Patients-A randomized trial. J
Ren Nutr. 33:278–288. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Yue SY, Zhou RR, Nan TG, Huang LQ and Yuan
Y: Comparison of major chemical components in puerariae thomsonii
radix and Puerariae lobatae radix. Zhongguo Zhong Yao Za
Zhi. 47:2689–2697. 2022.(In Chinese). PubMed/NCBI
|
|
58
|
Luo Y, Xiao Y, Zhao J, Zhang H, Chen W and
Zhai Q: The role of mucin and oligosaccharides via cross-feeding
activities by Bifidobacterium: A review. Int J Biol Macromol.
167:1329–1337. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wu Z, Huang S, Li T, Li N, Han D, Zhang B,
Xu ZZ, Zhang S, Pang J, Wang S, et al: Gut microbiota from green
tea polyphenol-dosed mice improves intestinal epithelial
homeostasis and ameliorates experimental colitis. Microbiome.
9:1842021. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Chen R, Xu Y, Wu P, Zhou H, Lasanajak Y,
Fang Y, Tang L, Ye L, Li X, Cai Z and Zhao J: Transplantation of
fecal microbiota rich in short chain fatty acids and butyric acid
treat cerebral ischemic stroke by regulating gut microbiota.
Pharmacol Res. 148:1044032019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lee J, d'Aigle J, Atadja L, Quaicoe V,
Honarpisheh P, Ganesh BP, Hassan A, Graf J, Petrosino J, Putluri N,
et al: Gut Microbiota-derived Short-chain fatty acids promote
poststroke recovery in aged mice. Circ Res. 127:453–465. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Feng Y, Ren J, Gui Y, Wei W, Shu B, Lu Q,
Xue X, Sun X, He W, Yang J and Dai C: Wnt/β-Catenin-Promoted
macrophage alternative activation contributes to kidney fibrosis. J
Am Soc Nephrol. 29:182–193. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Schunk SJ, Floege J, Fliser D and Speer T:
WNT-β-catenin signalling-a versatile player in kidney injury and
repair. Nat Rev Nephrol. 17:172–184. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yamamoto M, Takahashi-Yanaga F, Arioka M,
Igawa K, Tomooka K, Yamaura K and Sasaguri T: Cardiac and renal
protective effects of 2,5-dimethylcelecoxib in angiotensin II and
high-salt-induced hypertension model mice. J Hypertens. 39:892–903.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Manning JA, Shah SS, Nikolic A, Henshall
TL, Khew-Goodall Y and Kumar S: The ubiquitin ligase NEDD4-2/NEDD4L
regulates both sodium homeostasis and fibrotic signaling to prevent
end-stage renal disease. Cell Death Dis. 12:3982021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Feng Q, Chen WD and Wang YD: Gut
Microbiota: An integral moderator in health and disease. Front
Microbiol. 9:1512018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Chen J, Zhao K-N and Vitetta L: Effects of
intestinal microbial−elaborated butyrate on oncogenic
signaling pathways. Nutrients. 11:10262019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Benito I, Encío IJ, Milagro FI, Alfaro M,
Martínez-Peñuela A, Barajas M and Marzo F: Microencapsulated
bifidobacterium bifidum and lactobacillus gasseri in combination
with quercetin inhibit colorectal cancer development in ApcMin/+
mice. Int J Mol Sci. 22:49062021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Hiyama A, Yokoyama K, Nukaga T, Sakai D
and Mochida J: A complex interaction between Wnt signaling and
TNF-α in nucleus pulposus cells. Arthritis Res Ther. 15:R1892013.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zhao Y, Wang C, Hong X, Miao J, Liao Y,
Hou FF, Zhou L and Liu Y: Wnt/β-catenin signaling mediates both
heart and kidney injury in type 2 cardiorenal syndrome. Kidney Int.
95:815–829. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Pan B, Zhang H, Hong Y, Ma M, Wan X and
Cao C: Indoleamine-2,3-dioxygenase activates Wnt/β-catenin inducing
kidney fibrosis after acute kidney injury. Gerontology. 67:611–619.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Gelfand BD, Meller J, Pryor AW, Kahn M,
Bortz PDS, Wamhoff BR and Blackman BR: Hemodynamic activation of
beta-catenin and T-cell-specific transcription factor signaling in
vascular endothelium regulates fibronectin expression. Arterioscler
Thromb Vasc Biol. 31:1625–1633. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Xiang FL, Fang M and Yutzey KE: Loss of
β-catenin in resident cardiac fibroblasts attenuates fibrosis
induced by pressure overload in mice. Nat Commun. 8:7122017.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Xiao L, Xu B, Zhou L, Tan RJ, Zhou D, Fu
H, Li A, Hou FF and Liu Y: Wnt/β-catenin regulates blood pressure
and kidney injury in rats. Biochim Biophys Acta Mol Basis Dis.
1865:1313–1322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lin Y, Dong MQ, Liu ZM, Xu M, Huang ZH,
Liu HJ, Gao Y and Zhou WJ: A strategy of vascular-targeted therapy
for liver fibrosis. Hepatology. 76:660–675. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Sun J, Shi L, Xiao T, Xue J, Li J, Wang P,
Wu L, Dai X, Ni X and Liu Q: microRNA-21, via the HIF-1α/VEGF
signaling pathway, is involved in arsenite-induced hepatic fibrosis
through aberrant cross-talk of hepatocytes and hepatic stellate
cells. Chemosphere. 266:1291772021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Jiang L, Yin M, Wei X, Liu J, Wang X, Niu
C, Kang X, Xu J, Zhou Z, Sun S, et al: Bach1 represses
Wnt/β-Catenin signaling and angiogenesis. Circ Res. 117:364–375.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hung SW, Zhang R, Tan Z, Chung JPW, Zhang
T and Wang CC: Pharmaceuticals targeting signaling pathways of
endometriosis as potential new medical treatment: A review. Med Res
Rev. 41:2489–2564. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Hong F, Hong J, Wang L, Zhou Y, Liu D, Xu
B, Yu X and Sheng L: Chronic exposure to nanoparticulate TiO2
causes renal fibrosis involving activation of the Wnt pathway in
mouse kidney. J Agric Food Chem. 63:1639–1647. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Jiang F, Parsons CJ and Stefanovic B: Gene
expression profile of quiescent and activated rat hepatic stellate
cells implicates Wnt signaling pathway in activation. J Hepatol.
45:401–409. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Yoo EJ, Oh KH, Piao H, Kang HJ, Jeong GW,
Park H, Lee CJ, Ryu H, Yang SH, Kim MG, et al: Macrophage
transcription factor TonEBP promotes systemic lupus erythematosus
and kidney injury via damage-induced signaling pathways. Kidney
Int. 104:163–180. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Huynh C, Ryu J, Lee J, Inoki A and Inoki
K: Nutrient-sensing mTORC1 and AMPK pathways in chronic kidney
diseases. Nat Rev Nephrol. 19:102–122. 2023. View Article : Google Scholar : PubMed/NCBI
|