Introduction
Sarcopenia refers to the gradual loss of muscle mass
and muscle strength, and the main symptoms are muscle loss and
weakness (1,2). Although sarcopenia is mainly a
disease of elderly individuals, it can be associated with diseases
that are not confined to the elderly population. Muscle reduction
syndrome is characterized by a gradual overall loss of skeletal
muscle mass and muscle strength, and is closely associated with
physical disability and death (3,4).
However, there is currently no treatment for sarcopenia and its
diagnosis is complex (5,6). A microRNA (miRNA/miR) is a non-coding
RNA ~22 nucleotides long, which serves an important role in
regulating mRNA expression at the post-transcriptional level by
degrading and/or inhibiting the translation of mRNAs with sequences
complementary to the miRNA (7–9). In
most cases, primary miRNAs are transcribed from miRNA-encoding DNA,
processed into precursor miRNAs, transported to the cytoplasm and
then processed into miRNAs (10).
Numerous studies have aimed to identify miRNAs that may serve as
biomarkers or treatments of muscular diseases (11–13).
However, the levels of precursor miRNAs do not exhibit clear
positive correlations with those of mature miRNAs (14). A previous study demonstrated that
the expression of miR-886 is increased in prostate cancer (15), whereas another study revealed that
the expression of precursor miR-886 is decreased (16). These findings suggested that
precursor miRNA levels may not be positively correlated with mature
miRNAs, even during disease. The role of precursor miRNAs in muscle
atrophy remains largely unknown, suggesting that the levels of
mature miRNAs associated with various muscle diseases may differ
from those of their precursors. Overexpression of PHD finger
protein 20 (PHF20) has been shown to inhibit skeletal muscle
differentiation in mice, and to reduce the expression of proteins
that regulate myogenesis in such mice (17), indicating that PHF20 serves
negative roles in myogenesis and injury-induced muscle regeneration
in vivo. These findings suggested that the PHF20 transgenic
(TG) mouse may serve as a useful animal model when studying muscle
atrophy.
The present study comprehensively analyzed the miRNA
precursor expression profiles in exosomes from human skeletal
muscle. The findings revealed a previously unidentified muscle
atrophy-associated miRNA precursor, the miR-206 precursor.
Furthermore, miR-6516 was revealed to inhibit muscle atrophy.
Notably, expression of the miR-206 precursor was upregulated during
muscle atrophy, whereas miR-6516 was shown to serve a pivotal role
in the regulation of muscle atrophy by modulating the expression of
cyclin-dependent kinase inhibitor 1b (Cdkn1b).
Materials and methods
Plasma samples from patients and
normal subjects
For experiments using human derivatives, materials,
methods, ethical considerations and reasons for exemption from
research subject consent were reviewed by the institutional review
board of Chungnam National University Hospital, and exemption from
review was approved on the condition that human derivatives were
provided from a biobank (approval no. CNUH 2022-11-087; Daejeon,
South Korea). All human plasma samples were provided by the biobank
of Chungbuk National University Hospital. All human plasma samples
were collected and stored in advance by the biobank with informed
patient consent. Normal and patient plasma samples were obtained
from individuals with disease codes Z00 (people without symptom
complaints or reported diagnoses) and N18.5 (stage 5 chronic kidney
disease), respectively, according to the Korean Standard
Classification of Diseases (www.kcdcode.kr). The patient group was further
adjusted to those with a history of diabetes. In addition, clinical
information on the age, disease code, and additional diseases of
the normal and patient groups was provided by the biobank. A total
of 1 ml plasma was received from each of the 20 normal subjects
(average age, 23.4 years; 10 men and 10 women) and 19 patients
(average age, 64.8 years; 9 men and 10 women).
Animals
The Institutional Animal Care and Use Committee of
Chungnam National University approved all animal management and
experiment protocols (approval nos. 202305A-CNU-083 and
202309A-CNU-155; Daejeon, South Korea). All mice were bred and
maintained in a controlled environment (free access to food and
water, 12-h light/dark cycle, 50–60% humidity, 22°C ambient
temperature). A total of 18 male C57BL/6 mice, 10 to 12 weeks old,
were supplied by Narabiotech (Seoul, South Korea). A total of five
12-week-old male PHF20-overexpressing C57BL/6J mice that we had
previously generated and maintained were used (17). At the end of the study, mice were
euthanized by CO2 gas inhalation at a CO2
filling rate of 30% of the chamber volume/min. Before cardiac
perfusion of all the mice, the mice were anesthetized with Avertin
(200 mg/kg) and perfused first with PBS and then with 4%
paraformaldehyde. To ensure sacrifice, euthanasia was performed
using CO2 at the aforementioned rate before specimen
collection. The gastrocnemius, tibialis and soleus muscles of the
mice were collected and used in the experiments. All mice
experiments were conducted at animal facilities in accordance with
institutional guidelines.
Cell-free (cf)RNA isolation from
plasma
Human plasma stored at −80°C was rapidly thawed at
room temperature, centrifuged at 4°C for 10 min at 2,000 × g, and
the supernatant was transferred to a sterile 1.5-ml tube.
Subsequently, 15 µl concentrated cfRNA was extracted using the
Quick-cfRNA Serum & Plasma Kit (cat. no. 1059; Zymo Research
Corp.) according to the manufacturer's instructions, and was
immediately stored at −80°C.
RNA extraction from skeletal
muscle
Blood effects were minimized through cardiac
perfusion before muscle collection. The gastrocnemius, soleus and
TA muscles located on the hind limb of the mice were collected, and
any fat tissue was carefully removed without further injuring the
muscle tissues. For cfRNA isolation, fresh skeletal muscle tissues
were collected from mice and immediately placed in a 3.5-cm dish
containing Dulbecco's Modified Eagle's Medium (cat. no. LM001-06;
Welgene, Inc.) supplemented with 10% exosome-depleted FBS, 4 mM
L-glutamine and 5.5 mM D-glucose. The tissues were incubated in a
humidified incubator at 37°C and 5% CO2 for 24 h,
allowing them to release muscle-derived cfRNA under conditions
resembling the natural environment within the body. Subsequently,
15 µl concentrated cfRNA was extracted using the Quick-cfRNA Serum
& Plasma Kit, according to the manufacturer's instructions. In
addition, total RNA was extracted from the gastrocnemius and TA
muscles using TRIzol according to the manufacturer's protocol.
Reverse transcription-quantitative PCR
(RT-qPCR)
cDNA synthesis was performed using SuperScript™ II
Reverse Transcriptase (cat. no. 18064022; Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. For
cDNA synthesis, 300 ng human plasma-derived cfRNA, 500 ng mouse
gastrocnemius and tibialis muscle-derived cfRNA, 300 ng mouse
soleus muscle-derived cfRNA, and 1 µg mouse gastrocnemius and
tibialis anterior (TA) muscle-derived total RNA samples were used.
cDNA was diluted 1:50 for qPCR on a Bio-Rad CFX96 Real-Time PCR
Detection System (cat. no. 3600037; Bio-Rad Laboratories, Inc.)
using GoTaq® qPCR Master Mix (cat. no. A6001; Promega
Corporation). The expression levels of each mature mRNA and
precursor of hsa-miRNA and mmu-miRNA were normalized to the
expression levels of Gapdh for mature mRNAs, to U6
for hsa-miRNA and to small nucleolar RNA (snoRNA)202
for mmu-miRNAs. The qPCR thermal cycling conditions were as
follows: Initial heat activation at 95°C for 2 min, followed by 40
cycles at 95°C for 15 sec and 60°C for 1 min. The Cq value was
defined using Bio-Rad CFX Maestro 2.8 software (Bio-Rad
Laboratories, Inc.). The relative RNA expression levels were
quantified using the 2−ΔΔCq method (ΔCq=Cq Target
gene-Cq Normalization gene) (18).
The primer sequences are listed in Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene symbol | Primer sequence
(5′-3′) |
|---|
| hsa-miR-12136 | F:
CCATGGGGTTGGCTTGAAAC |
| precursor | R:
CAAAAAAGGAAGGAATCGAACCCC |
| hsa-miR-1291 | F:
TGTACTGTGGCTGTTGGTTTCA |
| precursor | R:
CAGGAAGACAGTCCTTTAGGCCTC |
| hsa-miR-3651 | F:
GATTCGATGGGCCATAGCA |
| precursor | R:
TGAGGAGAAGCAGCCTCC |
| hsa-miR-206 | F:
TTCCCGAGGCCACATGCTTC |
| precursor | R:
CCATAGCAAAGTAATCCATATGGGG |
| hsa-miR-133b | F:
CCTCAGAAGAAAGATGCCC |
| precursor | R:
TCTCCAAGGACTGGGCAT |
| hsa-miR-664a | F:
GAACATTGAAACTGGCTAGG |
| precursor | R:
TTTTTCATTTTGTAGGCTGG |
| human
U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| mmu-miR-206 | F:
CCAGGCCACATGCTT |
| precursor | R:
TTCCATAGTGCTGAGATATC |
| mmu-miR-1291 | F:
AGAATCAAGGGATGGGAGGTTACC |
| precursor | R:
GAAGACAGTTCTCTAGGCGTCTGC |
| mmu-miR-6516 | F:
AACCTCTTCCCTGGGGTTAG |
| precursor | R:
CCACCAAACTGCTGCTAGG |
| mmu-miR-23a | F:
GATTTGATGCCAGTCACA |
| precursor | R:
GGGTCAGTTGGAAATCC |
| mmu-miR-664 | F:
TGACTGGATAGAAAACATTATTC |
| precursor | R:
CTTTCATGTGTAGGCTGG |
| mouse | F:
GCTGTACTGACTTGATGAAAGTAC |
| snoRNA202 | R:
CATCAGATGGAAAAGGGTTCAA |
| mouse
Phf20 | F:
CATTGACTACGAAGAAGGGAG |
|
| R:
CTTCTCTAAAGGGCGCAGATA |
| mouse
Trim63 | F:
GCTGGTGGAAAACATCATTGACAT |
|
| R:
CATCGGGTGGCTGCCTTT |
| mouse
Cdkn1b | F:
TCAAACGTGAGAGTGTCTAACGG |
|
| R:
AGGGGCTTATGATTCTGAAAGTCG |
| mouse
Usp25 | F:
CAGAAGCACCAGCAGACATTT |
|
| R:
TGGCATTCTTTGCAGTGAGGA |
| mouse
Pax7 | F:
GTGCCCTCAGTGAGTTCGATTAGC |
|
| R:
CCACATCTGAGCCCTCATCCA |
| mouse
Myod1 | F:
CCACTCCGGGACATAGACTTG |
|
| R:
AAAAGCGCAGGTCTGGTGAG |
| mouse
Gapdh | F:
GACCCCTTCATTGACCTC |
|
| R:
GCCATCCACAGTCTTCTG |
Cell culture
Human skeletal muscle cells obtained at passage two
(cat. no. CC-2561; Lonza Group, Ltd.) were cultured in SkBM Basal
Medium (cat. no. CC-3161; Lonza Group, Ltd.) containing SkGM
SingleQuots Supplements and Growth Factors (cat. no. CC-4139; Lonza
Group, Ltd.). Exosome-depleted FBS (10%; cat. no. A2720801; Gibco;
Thermo Fisher Scientific, Inc.) was added to the cell medium. The
cells were cultured at 37°C in a humidified incubator containing 5%
CO2. Cells were subcultured at 80% confluence. Human
skeletal muscle cells were differentiated when they reached 60%
confluence for 5 days using Human Skeletal Muscle Cell
Differentiation Medium (cat. no. 151D-250; Cell Applications,
Inc.). The medium was changed every day during differentiation. The
morphology of differentiated muscle cells was confirmed using an
optical microscope. Cells were used for experiments for up to 5
passages. Mycoplasma negativity was confirmed before all
cell experiments.
Western blot analysis
As described previously (19), after the completion of experiments
for western blot analysis, cells and muscles from mice were placed
on ice and proteins were extracted using PRO-PREP™ Protein
Extraction Solution (cat. no. 17081; Intron Biotechnology, Inc.).
The lysates were then centrifuged for 30 min, 20,000 × g at 4°C.
The extracted proteins was quantified using the Bradford method and
25 µg protein was loaded per lane. Quantified proteins were
separated by SDS-PAGE on 10.0% gels and were transferred to PVDF
blotting membranes (cat. no. 10600023; Cytiva). Subsequently, the
transferred membranes were blocked in 1X Tris-buffered saline [140
mM NaCl, 2.7 mM KCl and 250 mM Tris-HCl (pH 7.4)] containing 5%
skimmed milk and 0.2% Tween-20 for 1 h at room temperature. Blocked
membranes were then incubated with primary antibodies (dilution
ratio, 1:1,000) overnight at 4°C, followed by incubation with
secondary antibodies (dilution ratio, 1:10,000) for 1 h at room
temperature. The following primary antibodies were used: Anti-MYOD
(cat. no. sc-377460; Santa Cruz Biotechnology, Inc.), anti-PHF20
(cat. no. 3934S; Cell Signaling Technology, Inc.), anti-muscle
RING-finger protein-1 (MuRF1; cat. no. sc-398608; Santa Cruz
Biotechnology, Inc.), anti-tumor susceptibility gene 101 (TSG101;
cat. no. sc-7964; Santa Cruz Biotechnology, Inc.), anti-calnexin
(cat. no. 2679; Cell Signaling Technology, Inc.) and anti-GAPDH
(cat. no. A19056; ABclonal Biotech Co., Ltd.). The following
secondary antibodies were used: HRP-conjugated anti-rabbit (cat.
no. 7074V; Cell Signaling Technology, Inc.) and HRP-conjugated
anti-mouse (cat. no. 31430; Invitrogen; Thermo Fisher Scientific,
Inc.). Protein expression was visualized using ProNA ECL (cat. no.
TLP-112.1; TransLab, Co., Ltd.) according to the manufacturer's
instructions. All protein expression levels were normalized to the
expression levels of the loading control GAPDH. ImageJ software
(version 1.49; National Institutes of Health) was used for
semi-quantification of blotting.
Exosome isolation from conditioned
media
Two 10-cm plates were used for exosome isolation.
ExoQuick-TC™ (cat. no. EXOTC50A-1; System Biosciences, LLC) was
used to isolate exosomes from the growth media, cultured for 24 h
after media change, of pre-differentiation and post-differentiation
skeletal muscle cells. Concentrated media (20 ml) were centrifuged
at 3,000 × g for 15 min at room temperature to remove cells and
cell debris. Subsequently, 4 ml ExoQuick-TC was added to the
supernatant. After inverting at least five times, the samples were
incubated at 4°C overnight (≥12 h), then centrifuged at 1,500 × g
for 30 min at room temperature. The supernatant was discarded and
the pellet was resuspended in TRIzol® (cat. no.
15596026; Invitrogen; Thermo Fisher Scientific, Inc.) for precursor
miRNA profile analysis. After resuspending the exosomes in TRIzol,
they were immediately stored at −80°C before starting precursor
miRNA profiling analysis.
Precursor miRNA profile analysis
The miRNeasy® Serum/Plasma Kit (cat. no.
217184; Qiazen, Inc.) was used to prepare RNA samples. The RNA
isolated from each sample was used to construct sequencing
libraries with the SMARTer smRNA-Seq Kit for Illumina (Takara Bio,
Inc.), following the manufacturer's protocol. Briefly, input RNA
was first polyadenylated in order to provide a priming sequence for
an oligo-(dT) primer. cDNA synthesis was primed by the 3′ smRNA dT
Primer, which incorporated an adapter sequence at the 5′ end of
each first-strand cDNA molecule. When the MMLV-derived PrimeScript™
Reverse Transcriptase (cat. no. 2680A; Takara Bio Inc.) reached the
5′ end of each RNA template, it added non-templated nucleotides,
which are bound by the SMART smRNA Oligo-enhanced with locked
nucleic acid (LNA) technology for greater sensitivity. In the
template-switching step, PrimeScript RT used the SMART smRNA Oligo
as a template for the addition of a second adapter sequence to the
3′ end of each first-strand cDNA molecule. In the next step,
full-length Illumina adapters (including index sequences for sample
multiplexing) were added during PCR amplification. The Forward PCR
Primer bound to the sequence added by the SMART smRNA Oligo, while
the Reverse PCR Primer bound to the sequence added by the 3′ smRNA
dT Primer. Resulting library cDNA molecules included sequences
required for clustering on an Illumina flow cell. The libraries
were validated by checking the size, purity and concentration on
the Agilent Bioanalyzer. The libraries were pooled in equimolar
amounts, and sequenced on an Illumina HiSeq 2500 instrument. Image
decomposition and quality values calculation were performed using
the modules of the Illumina pipeline All exosomal precursors of
miRNA sequencing procedures were performed by Macrogen, Inc.
Sequence alignment and detection of known and novel precursors of
miRNA were performed using the miRDeep2 software algorithm (Ver.
2.0.0.8; Max Delbrück Center). In the read alignment to precursor
result of the Quantifier module result of miRDeep2, the read
aligned to each precursor was selected. Among the selected reads,
reads that were assigned to mature miRNA were excluded, and reads
with a length of >50% of the precursor were selected. Reads per
million (RPM) was calculated using the total sum of reads for each
sample obtained from the selected results. All exosomal precursors
of miRNA sequencing procedures were performed by Macrogen, Inc.
From the analysis results of precursor miRNA sequencing, as a
cutoff point, abundant precursor miRNAs >700 RPM were selected
as biomarker candidates.
Grip strength test
To measure hindlimb grip strength, mice at 12 weeks
old were placed on a grip strength meter, while their tail and nape
were held and pulled gently. Each mouse was allowed to perform the
test five times and was given a 5-min break between each test.
Values for grip strength were recorded and normalized by dividing
by whole body weight.
RNA correlation analysis
To evaluate the relationship between the expression
of genes in skeletal muscle, Gene Expression Profiling Interactive
Analysis 2 (GEPIA2) (http://GEPIA2.cancer-pku.cn/index.html) (20) was used, and a Spearman correlation
analysis was performed. Only human skeletal muscle tissue databases
(n=396) from GTEx (21) were used
for the analysis.
Histological analysis of muscle
tissue
Gastrocnemius and tibialis muscle tissues from mice
were fixed with 4% paraformaldehyde for 16 h at 4°C and 4-µm
paraffin-embedded sections were generated. The paraffin-embedded
sections were stained with hematoxylin (for 5 min at room
temperature) and eosin (for 1 min at room temperature) according to
standard protocols. Light microscopy was used for histological
observation. Measurements of cross-sectional area (CSA) were
performed using ImageJ software (version 1.49; National Institutes
of Health). For each CSA, six different views were randomly
selected for measurement.
Immobilization model
The right hind limbs of 13 10-week-old male C57BL/6
mice supplied by Narabiotech (Seoul, South Korea) were used. As
described in a previous study, surgical tape was wrapped around
starting from the distal aspect of the foot to the ankle area.
Subsequently, Velcro was wrapped around the hindlimb, starting at
the distal end of the foot. Velcro was replaced if adverse effects
(e.g., skin injury and edema) or loose Velcro was observed. The
forepaw and left hindlimb were free, so that the mice had free
access to food and water.
Bioinformatic analysis
Using Human TargetScan (Ver 8.0; http://www.targetscan.org/vert_80/) and mouse
TargetScan (Ver 8.0; http://www.targetscan.org/mmu_80/), target mRNAs for
microRNAs were scrutinized.
Muscle injury and administration of
miRNA
To transfer miR-6516 to TA muscles of
immobilization-injured mice, mimic-miR-6516 (cat. no. SMM-003; 20
nM; Bioneer Corporation) was mixed with RNAiMAX (cat. no. 13778030,
Invitrogen; Thermo Fisher Scientific, Inc.), as described
previously (22). For the control
group, mimic-miR-control (cat. no. SMC-2003; Bioneer Corporation)
was used and a mixture was prepared in the same manner as the
mimic-miR-6516 mixture. The resulting mixture (50 µl) was injected
on the 5th and 10th day during the 14-day immobilization period,
and the Velcro was replaced after injection.
Statistical analysis
Data are expressed as the mean and standard error of
the mean (SEM) from at least three separate experiments. GraphPad
Prism (version 8.1.1; Dotmatics) was used for statistical analysis.
Quantitative data are presented as the mean ± SEM unless indicated
otherwise. Comparisons between two groups were evaluated using
unpaired Student's t-test or Mann-Whitney U test, depending on the
results of the Shapiro-Wilk test. For multiple comparisons, one-way
ANOVA was performed, followed by Dunnett's post hoc test. P<0.05
was considered to indicate a statistically significant
difference.
Results
Circulating precursors of miR-206 and
miR-664a increase on development of sarcopenia-related disease
It has previously been shown that miRNA expression
levels are not always proportional to those of precursor miRNAs
(14), suggesting that the changes
in miRNA levels observed as muscular atrophy develops may differ
from the changes in precursor miRNA levels. To profile the skeletal
muscle-derived miRNA precursors secreted into the plasma (possible
biomarkers of muscle atrophy), the exosomal miRNA precursors
(cfRNAs) of human muscle cells were first sequenced. Muscle cell
differentiation was confirmed by the gross changes in fused cell
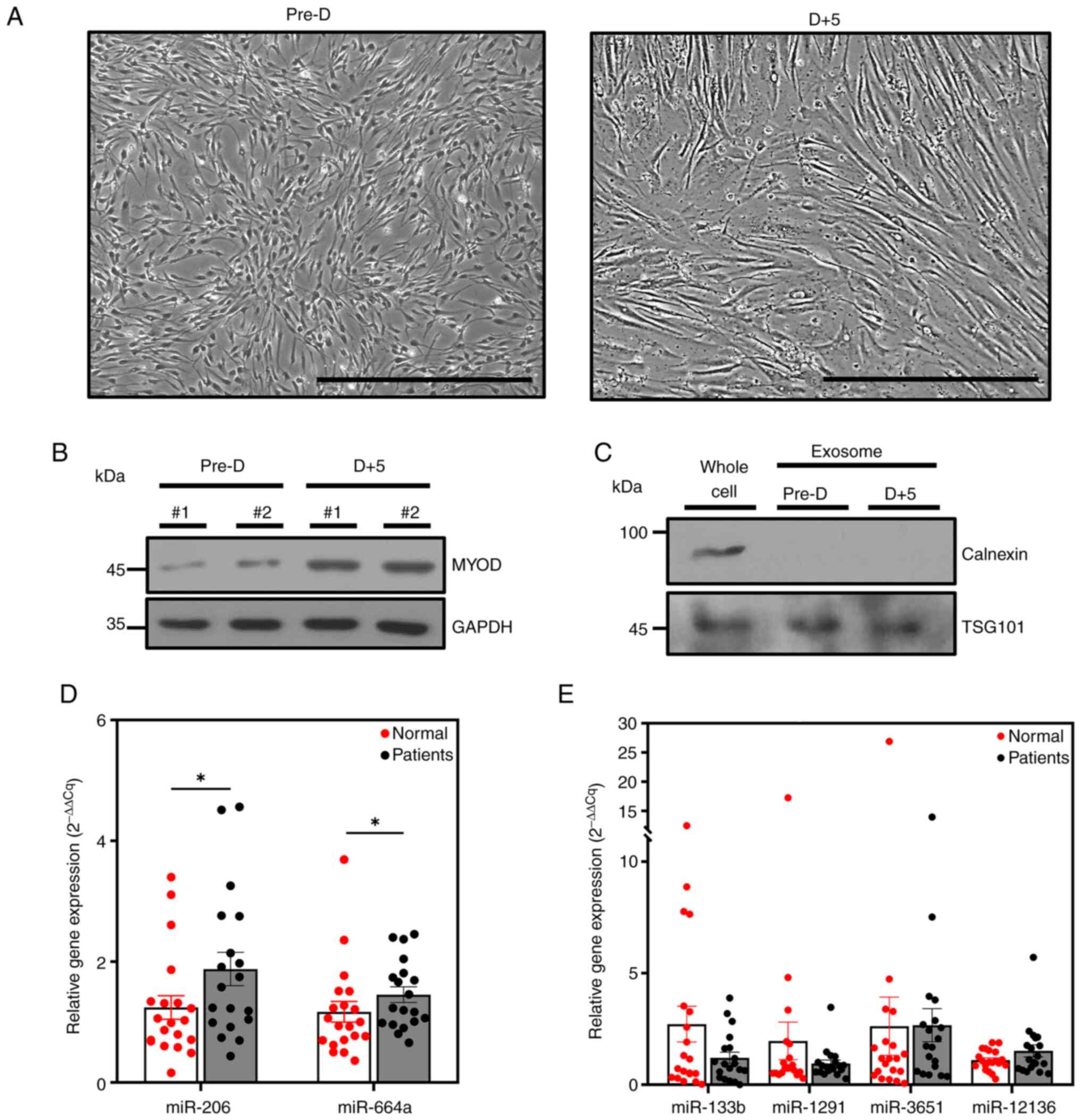
morphology and increased expression of MYOD (Fig. 1A and B). Exosomes were extracted
using ExoQuick, and western blotting detected the positive exosomal
marker TSG101 but not the negative marker calnexin (Fig. 1C). Exosomal precursor miRNA
sequencing results confirmed that precursor miRNAs were abundant in
exosomes derived from pre- and post-differentiated muscle cells
(Table II). Of such precursor
miRNAs, any that overlapped with other genes or were difficult to
analyze via qPCR were excluded. The precursor of miR-206, known to
be a muscle-only miRNA (23), and
the precursor of miR-133b, which is downregulated in the plasma of
patients with sarcopenia (24),
were analyzed in the present study. Previous studies have shown
that some snoRNAs also function as precursors of miRNAs (25–28).
Precursors of miRNAs that overlapped with snoRNAs that were not
identified as producing miRNAs were excluded from the analysis,
even if they did not overlap in sequence with a specific mRNA.
Another study reported that miR-1291 and miR-3651 are derived from
snoRA2C and snoRD84, respectively, and these snoRNAs are very
similar in terms of their sequences to the precursors of the
corresponding miRNAs (29). For
this reason, the precursors of miR-1291 and miR-3651 were not
excluded from the analysis, despite their significant sequence
overlap with snoRA2C and snoRD84. In addition, the precursor of
miR-4449 was excluded from the analysis, as primer design and
RT-qPCR were difficult to perform given the 86% G+C content. In
summary, in human plasma-derived cfRNAs, the precursors of miR-206,
miR-12136, miR-1291, miR-664a, miR-3651 and miR-133b were selected
for analysis. Given the low number of patients diagnosed with
sarcopenia, patients with diseases likely to cause sarcopenia as a
complication were included. Previous studies have revealed that the
incidence of sarcopenia is high in patients with diabetes and
chronic kidney disease (30,31).
Therefore, the biomarker candidates were quantitatively analyzed in
the plasma of patients with either of these diseases, and of normal
people aged 20–30 years. Of the biomarker candidates, the
expression levels of precursors of miR-206 and miR-664a were
significantly increased in the patient group with diabetes and
chronic kidney disease (Fig. 1D).
By contrast, there were no significant differences in the
expression levels of the precursors of miR-133b, miR-1291, miR-3651
and miR-12136 between patients and control individuals (Fig. 1E). These results suggested that
increased expression of the precursors of miR-206 and miR-664a in
circulating plasma cfRNAs may be associated with sarcopenia, which
is characterized by muscle atrophy.
 | Table II.Abundant pre-miRNAs in exosomes
derived from human skeletal muscle cells. |
Table II.
Abundant pre-miRNAs in exosomes
derived from human skeletal muscle cells.
|
| Reads per
million |
|---|
|
|
|
|---|
| Precursor
miRNA |
Pre-differentiated | Differentiated |
|---|
| miR-12136 |
6.7×105 |
1.18×105 |
| miR-1291 |
6.72×103 |
7.97×102 |
| miR-23a |
8.52×102 |
5.31×102 |
| miR-3651 |
2.7×103 |
8.4×104 |
| miR-4449 |
4.83×103 |
1.75×104 |
| miR-6516 |
2.79×103 |
1.38×104 |
| miR-664a |
1.04×103 | - |
| miR-664b |
7.1×102 | - |
| let-7a-3 |
1.89×102 |
7.97×102 |
| miR-10394 |
9.47×101 |
7.97×102 |
PHF20 overexpression induces muscular
atrophy in vivo
The patients analyzed in the present study were
expected to develop muscle atrophy as a complication of diabetes or
chronic kidney disease; however, atrophy was not definitively
confirmed. Therefore, the present study analyzed muscle-derived
cfRNAs in a mouse model of muscular atrophy. Induction of atrophy
was confirmed in mice overexpressing PHF20. In a previous study,
PHF20 exerted a negative effect on muscle differentiation via
positive regulation of YY1 (17).
Subsequently, correlations between PHF20 levels and those of gene
products associated with muscle differentiation, namely MYH2, TNNT1
and TNNT3, were assessed (32).
The relationships between the levels of the muscle atrophy markers
MuRF1 (TRIM63) and atrogin-1 (FBXO32), and their
upstream transcription factor FOXO3a (33,34),
were also investigated. These studies confirmed the negative
association between the levels of PHF20 and those of muscle
differentiation genes, and the positive association between the
levels of PHF20 and muscle atrophy genes. The correlation pattern
between PHF20 and each marker gene was similar to the correlation
pattern between YY1 and each marker gene (Fig. 2A). The correlation of genes was
analyzed using the dataset from GTEx. These findings indicated the
possibility that PHF20 overexpression may inhibit muscle
differentiation and facilitate muscle atrophy. A previous study
confirmed that overexpression of PHF20 inhibits muscle
differentiation and triggers defects in muscle morphology in
vivo (17). This suggests that
overexpression of PHF20 may induce muscle atrophy; additional
experiments were thus performed in the present study to assess
this. In PHF20 TG mice, the expression levels of both PHF20 and the
muscle atrophy marker MuRF1 were increased (Fig. 2B), as were the mRNA expression
levels of Phf20 and Trim63, which encode PHF20 and
MuRF1 (Fig. 2C). The grip strength
test was used to evaluate the muscle strength of PHF20 TG mice
(35), and the strength was
significantly lower than that of wild-type mice (Fig. 2D). Additionally, the muscle CSA was
significantly reduced in PHF20 TG mice compared with that in
wild-type mice (Fig. 2E). These
results provided clear evidence that overexpression of PHF20
induced muscular atrophy in vivo.
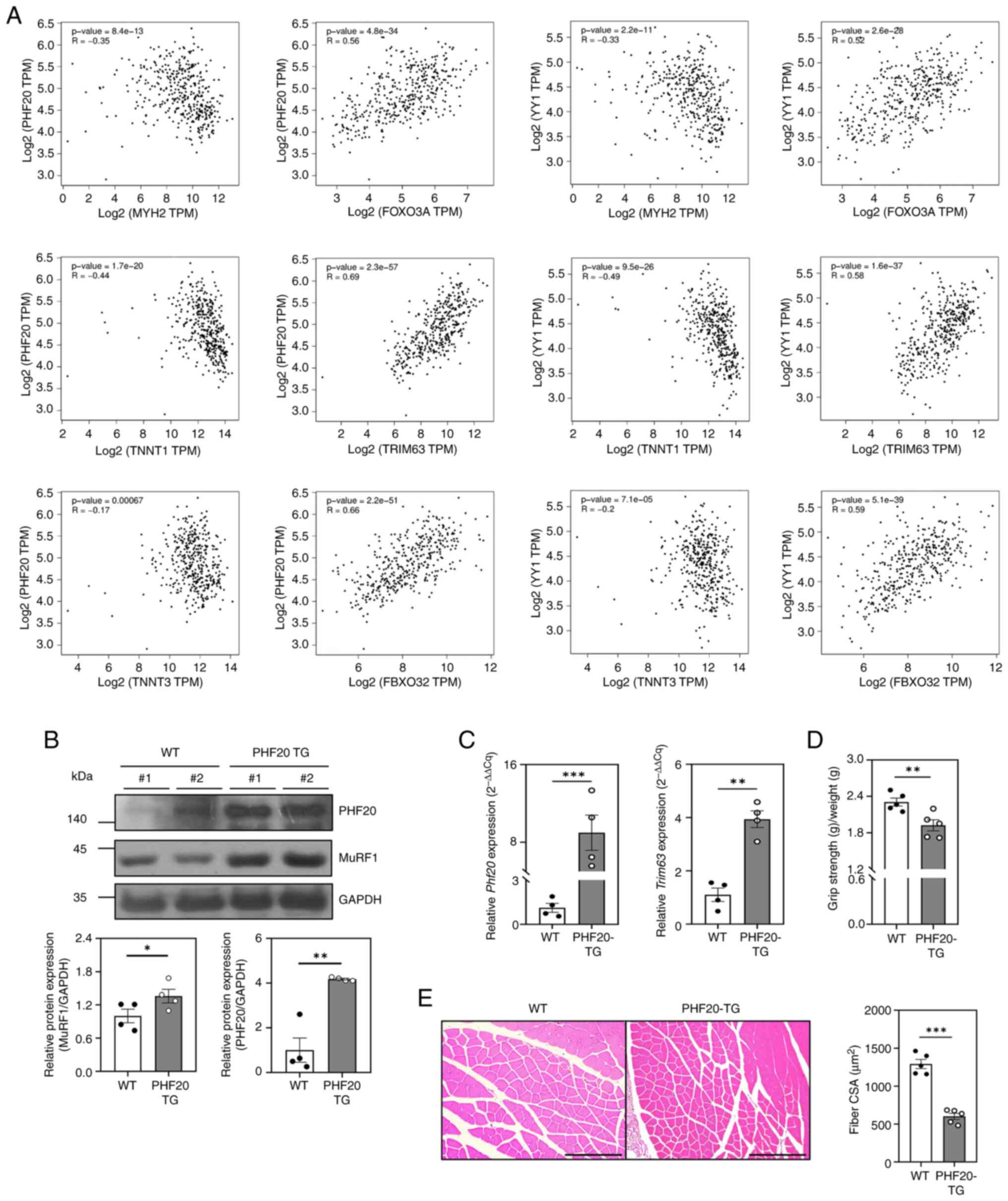 | Figure 2.Effect of PHF20 overexpression on
muscles in vivo. (A) Correlation of gene pairs in skeletal
muscle samples (n=396) present in the GTEx database. Correlation
analysis of PHF20 and YY1 with muscle differentiation marker genes
MYH2, TNNT1 and TNNT3, and muscle atrophy marker genes FOXO3A,
TRIM63 and FBXO32. (B) Lysates were extracted from gastrocnemius
muscle tissues from WT (n=4) and PHF20-TG mice (n=4). Changes in
protein expression levels were analyzed by western blotting with
the corresponding antibodies. (C) Total RNA was extracted from
gastrocnemius muscle tissues from 12-week-old male WT (n=4) and
PHF20-TG mice (n=4). Expression levels of Phf20 and
Trim63 were analyzed by reverse transcription-quantitative
PCR. Expression levels were normalized to Gapdh. (D)
Hindlimb grip strength test of 12-week-old male WT (n=5) and PHF20
TG (n=5) mice. (E) Gastrocnemius muscle cross sections of
12-week-old male WT (n=5) and PHF20-TG (n=5) mice stained with
hematoxylin and eosin. Each CSA was measured with ImageJ software,
and six different views were randomly selected for CSA measurement.
Scale bars, 200 µm. Data are presented as the mean ± SEM.
*P<0.05, **P<0.01, ***P<0.001. CSA, cross-sectional area;
MuRF1, muscle RING-finger protein-1; PHF20, PHD finger protein 20;
TG, transgenic; WT, wild-type. |
Muscle immobilization induces muscle
atrophy and secretion of atrophied muscle-derived cfRNAs
Muscle atrophy attributable to muscle disuse is a
common clinical problem and one of the major causes of muscle
atrophy (36,37). Therefore, to identify candidate
biomarkers of muscle atrophy not only in the PHF20-overexpressing
mouse model but also in a mouse model of muscle atrophy induced by
muscle disuse (immobilization), muscle atrophy was induced in
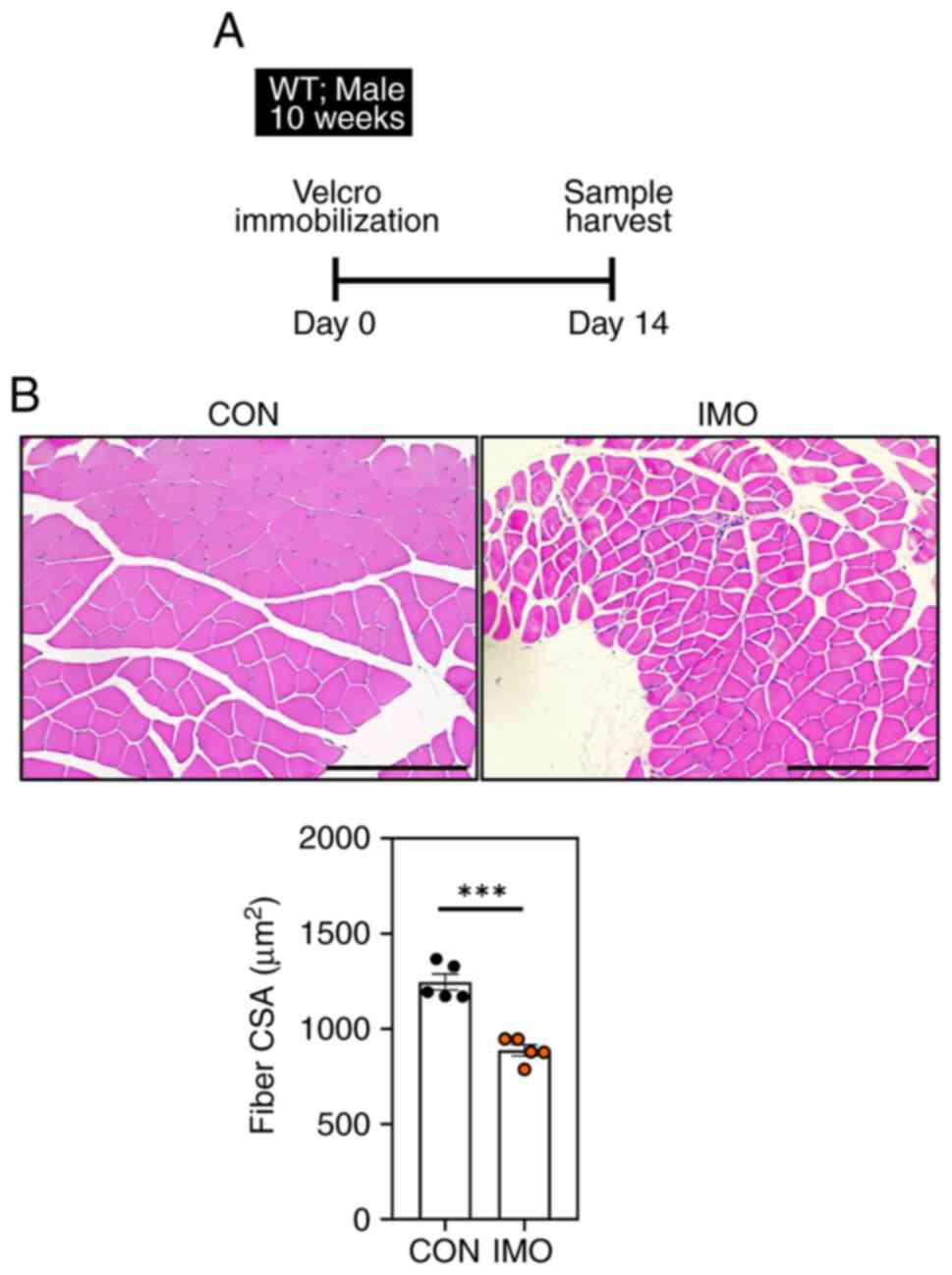
wild-type mice via Velcro immobilization. In previous studies, it
was confirmed that muscle atrophy was induced when the hind limb
was immobilized using Velcro for 2 weeks (38,39).
Therefore, the right hind limbs of 10-week-old male wild-type mice
were immobilized for 2 weeks to induce muscle atrophy (Fig. 3A). Notably, the muscle CSA level of
the hind leg that was fixed with Velcro was decreased compared with
that in non-treated wild type mice (Fig. 3B). Skeletal muscle atrophy causes
biochemical and physiological changes in the muscles, leading to
alterations in gene expression (40). These characteristics suggest that
atrophied muscles in PHF20-overexpressing mice and Velcro-fixed
mice may synthesize cfRNAs, the levels of which may differ from
those of non-fixed wild-type mice; therefore, further analysis of
muscle atrophy biomarker candidates was performed to identify
changes in expression levels.
Muscle atrophy affects the expression
levels of multiple miRNA precursors in skeletal muscle
As aforementioned, to obtain cfRNAs from atrophied
muscles, the right hind limbs of five 10-week-old male wild-type
mice were fixed with Velcro for 2 weeks to induce muscle atrophy;
subsequently, the gastrocnemius, soleus and TA muscles were
dissected, and cfRNAs were extracted from each muscle. Similarly,
the gastrocnemius, soleus and TA muscles of 12-week-old male PHF20
TG mice were prepared, and cfRNAs were extracted from each muscle.
For the same reasons presented when selecting precursor miRNAs to
be analyzed in human plasma, the precursors of miR-206, miR-6516,
miR-1291, miR-23a and miR-664 were selected for mouse analysis. In
mice, miR-664 is not divided into miR-664a and miR-664b; therefore,
the precursor of miR-664 was included in the analysis. In the
immobilized mice, the expression levels of the precursor of miR-206
were increased in cfRNAs derived from the gastrocnemius, soleus and
TA muscles compared with wild type mice; also the expression levels
of the precursor of miR-6516 were decreased (Fig. 4A-C). In addition, the expression
levels of the precursors of miR-1291 and miR-23a were decreased in
the TA muscle-derived cfRNAs from immobilized-mice compared with
those from wild-type mice (Fig.
4B). In the PHF20 TG mice, the expression levels of the
precursor of miR-206 were increased in cfRNAs derived from the TA
muscles compared with those from wild-type mice (Fig. 4B); also the levels of the precursor
of miR-6516 decreased (Fig. 4B).
In addition, the expression levels of the precursors of miR-1291
and miR-23a were decreased in gastrocnemius muscle-derived cfRNAs
from PHF20 TG mice compared with those in wild-type mice (Fig. 4A). The expression levels of the
precursors of miR-1291, miR-23a and miR-664 were also decreased in
the TA muscles of PHF20 TG mice compared with those in wild-type
mice (Fig. 4B). It has previously
been reported that miR-206 is a skeletal muscle-specific miRNA
(23). Thus, as a mature miRNA is
cleaved from a precursor of that miRNA, the precursor of miR-206 is
also muscle-specific. Taken together, these experiments indicated
that the expression levels of the precursor of miR-206 may increase
in mouse muscle-derived cfRNAs during muscle atrophy and in plasma
cfRNAs obtained from patients at risk of sarcopenia.
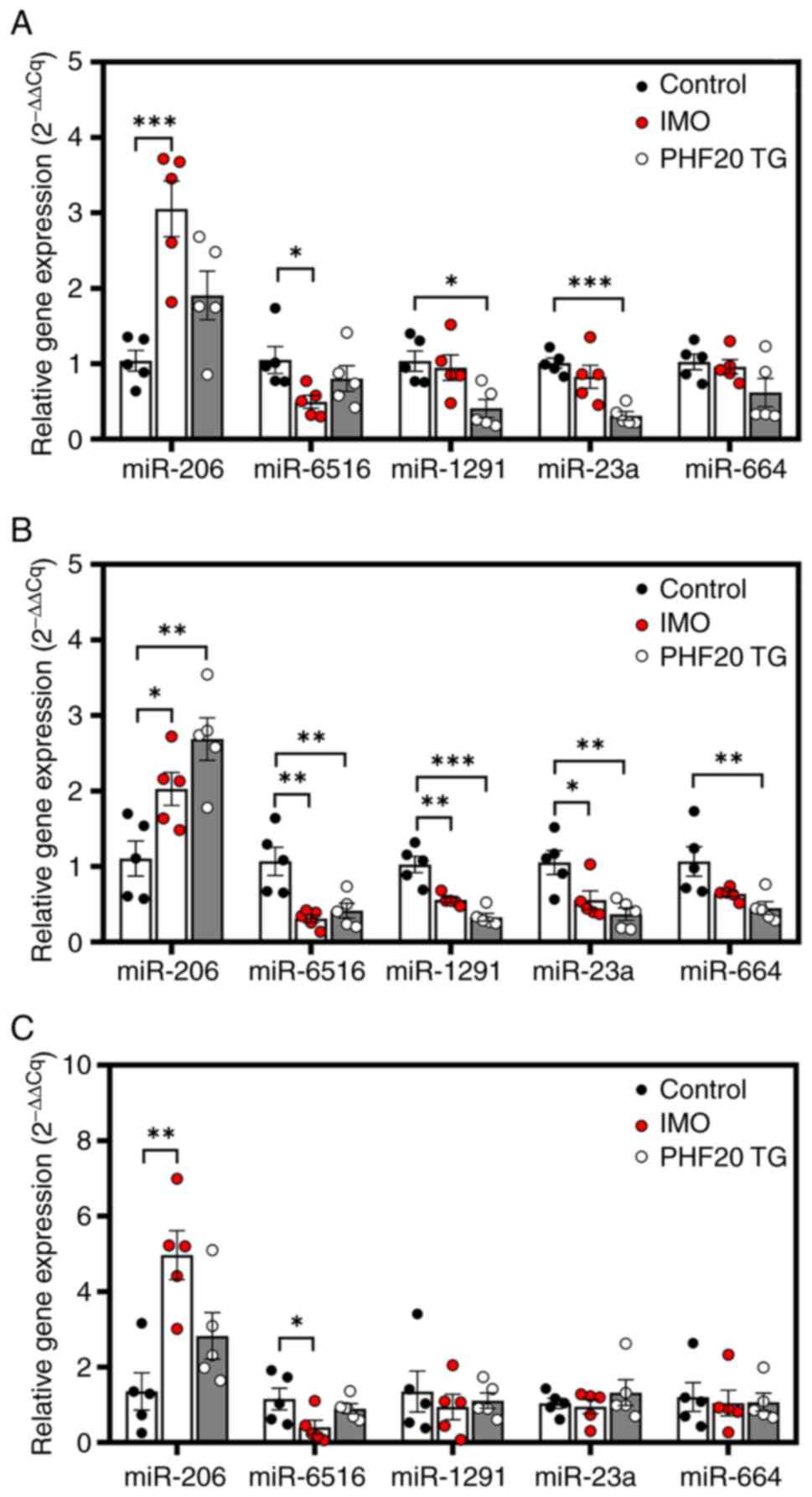 | Figure 4.Expression of miRNA precursors in
mice with IMO-induced muscle atrophy or in PHF20 TG mice with
muscle atrophy. Expression levels of miRNA precursors in cell-free
RNA samples derived from (A) gastrocnemius, (B) TA and (C) soleus
muscles in WT (control), IMO and PHF20-TG mice (n=5/group).
Expression levels were normalized to small nucleolar RNA 202
expression levels. Data are presented as the mean ± SEM.
*P<0.05, **P<0.01, ***P<0.001. IMO, immobilization;
miR/miRNA, microRNA; PHF20, PHD finger protein 20; TA, tibialis
anterior; TG, transgenic; WT, wild-type. |
miR-6516 inhibits
immobilization-induced muscle atrophy by regulating Cdkn1b
Notably, a decrease in the expression levels of the
miR-6516 precursor was observed in all muscle-derived cfRNAs of
Velcro-immobilized mice with muscle disuse atrophy. Based on these
results, additional experiments were performed to determine whether
miR-6516 administration in muscles affected muscle disuse-induced
atrophy. During the 14-day period of muscle atrophy induced via
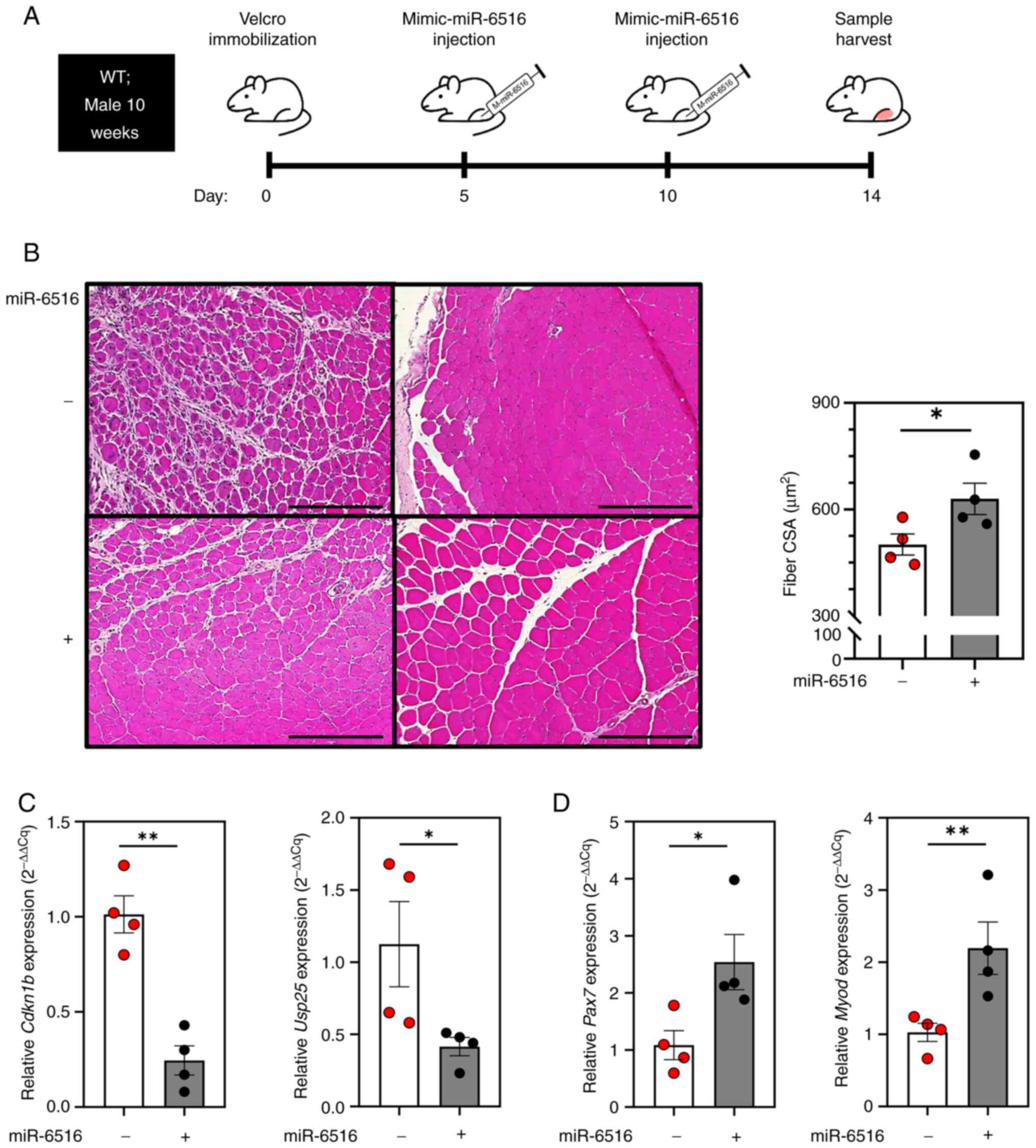
Velcro immobilization of the right hind limb, a mimic-miR-6516 was
injected into the TA muscle on days 5 and 10, and the mice were
sacrificed on day 14 (Fig. 5A). It
was confirmed that the muscle CSA of the mice administered
mimic-miR-6516 was significantly greater than that of mice
administered the mimic-miR-control, indicating that miR-6516
inhibits the progression of muscle atrophy caused by muscle disuse
(Fig. 5B). The genes targeted by
miR-6516 when inhibiting muscle atrophy were subsequently
investigated. Using TargetScan human (Ver. 8.0) and TargetScan
mouse (Ver. 8.0), genes that were predicted to be common targets of
miR-6516-3p or −5p in humans and mice were identified, according to
a cumulative weight context score of ≤-0.4. Additionally, since
muscle atrophy was suppressed upon injection of mimic-miR-6516,
only genes positively correlated with the muscle atrophy markers
MuRF1 (TRIM63), Atrogin1 (FBXO32) and FOXO3a
(FOXO3a) were selected as possible target genes. Only
CDKN1B (p27kip1) and ubiquitin specific peptidase
25 (USP25; USP25) satisfied the aforementioned conditions,
and they were predicted to be target genes of miR-6516 in the
context of inhibition of muscle atrophy (Table III). The mimic-miR-6516 was
injected to determine whether this affected the expression levels
of Usp25 and Cdkn1b. In mice injected with
mimic-miR-6516, the expression levels of Cdkn1b and
Usp25 were significantly reduced (Fig. 5C). A previous study showed that
p27kip1 levels negatively regulate skeletal muscle
satellite cell proliferation and muscle regeneration (41). Additionally, a decrease in muscle
fiber diameter has been confirmed in mice overexpressing
Cdkn1b (42). As expected,
the expression of early muscle regeneration markers Pax7 and
Myod1 was increased in muscles administered mimic-miR-6516
(Fig. 5D). However, additional
investigations are required because no association between
suppression of muscle atrophy and reduction of USP25
expression was conclusively shown (data not shown). Taken together,
these data suggested that injection of mimic-miR-6516 during
immobilization inhibits muscle rest-induced muscle atrophy by
suppressing expression of Cdkn1b and promoting skeletal
muscle satellite cell proliferation and muscle regeneration
(Fig. 6).
 | Table III.CWCS of miR-6516 predicted target
genes. |
Table III.
CWCS of miR-6516 predicted target
genes.
|
| hsa-miR-6516 | mmu-miR-6516 |
|
|---|
|
|
|
|
|
|---|
| Predicted target
gene symbol | −3p | −5p | −3p | −5p | Correlation with
muscle atrophy genes |
|---|
| NPPC | −0.57 | - | −0.49 | - | Negative |
| CDKN1B | −0.47 | - | −0.40 | - | Positive |
| PRND | - | −0.48 | - | −0.53 | Negative |
| USP25 | - | −0.46 | - | −0.80 | Positive |
Discussion
In the present study, precursor miRNAs from skeletal
muscle cell-derived exosomes were sequenced in the search for
biomarker candidates. Among the precursor miRNAs, those that were
abundant were selected as possible candidates. In detail, precursor
miRNAs with sequences that did not overlap with any mRNAs in humans
and mice were selected. Candidate biomarkers were compared between
plasma-derived cfRNAs from patients at a risk of sarcopenia and
normal subjects. In addition, mouse muscle atrophy was confirmed
after muscle immobilization injury and in PHF20 TG mice, and
muscle-derived cfRNAs were compared with wild-type cfRNAs. Notably,
both plasma-derived cfRNAs from patients and muscle-derived cfRNAs
from mice with muscle atrophy due to disuse exhibited increased
expression levels of the precursor of miR-206. Additionally, the
expression levels of the precursor of miRNA-6516 were decreased in
muscle-derived cfRNAs from mice subjected to muscle immobilization
injury. When mimic-miR-6516 was injected into the TA muscle of such
mice, the muscle immobilization-induced muscle atrophy caused by
muscle disuse was suppressed via inhibition of Cdkn1b. Taken
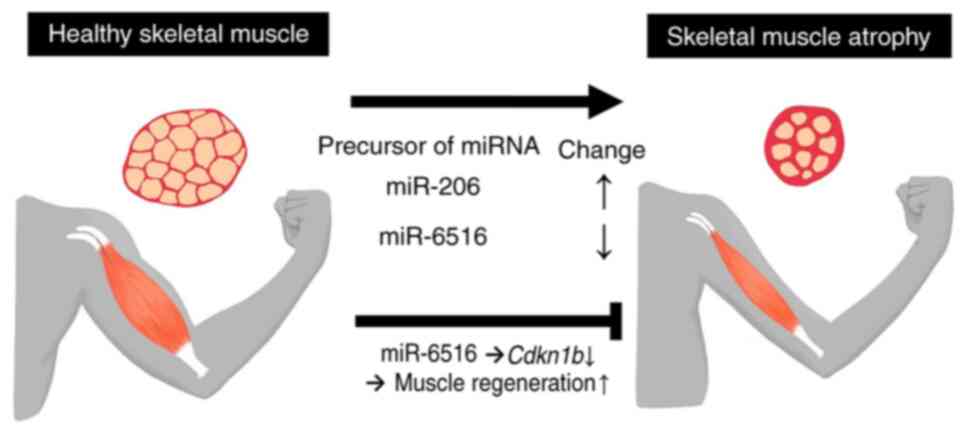
together, these findings indicated that the precursor of miR-206
was increased in plasma and muscle-derived cfRNAs during muscle
atrophy, and that miR-6516 inhibited muscle rest-induced muscle
atrophy, indicating the potential use of these miRNA precursors for
diagnostic and therapeutic purposes.
In previous studies, miR-1, miR-133a, miR-133b and
miR-206 have been reported to be myomiRs, i.e., miRNAs that are
highly expressed specifically in muscle (43–46).
In particular, miR-206 exhibits muscle-specific expression
characteristics (43,45,47),
and miR-133b is downregulated in the plasma of patients with
sarcopenia (24). A previous study
showed that the expression levels of mature miRNAs and their
precursors are not positively correlated (14). However, studies on the precursors
of miRNAs associated with muscle atrophy are lacking; therefore,
miRNA precursors associated with muscle atrophy were targeted in
the present study. Cell-derived miRNA precursors were investigated
in the exosomes of human muscle cells, which also contained the
precursors of muscle-specific miR-206 and sarcopenia-related
miR-133b. It has previously been reported that increased
p27kip1 expression in skeletal muscle inhibits muscle
satellite cell capacity and proliferation, thereby compromising
muscle regeneration (41). The
results of the present study confirmed that the precursor of
miR-6516 was reduced when muscle immobilization triggered atrophy,
and that treatment of muscles with miR-6516 suppressed atrophy by
inhibiting the expression of Cdkn1b. In addition, the
precursor of miR-206 was confirmed to be upregulated in skeletal
muscle-derived cfRNAs from mice with muscle atrophy due to disuse
and in plasma-derived cfRNAs from patients at risk of sarcopenia.
The present study thus suggested that the precursor of miR-206 may
be considered a novel biomarker of muscle atrophy and that miR-6516
could inhibit muscle atrophy by reducing the expression of
Cdkn1b in skeletal muscle. Expression of Usp25,
another target mRNA of miR-6516, was also reduced by administration
of mimic-miR-6516. However, to the best of our knowledge, how
reduced Usp25 expression might prevent muscle atrophy has
not yet been investigated and further research is necessary.
Muscle wastes for a variety of reasons, including
metabolic problems and muscle disuse (48–51),
and it is regenerated from muscle progenitor cells (8,52,53).
Overexpression of PHF20 in muscle inhibits the differentiation of
muscle cells and thus suppresses muscle regeneration, disrupting
the balance between muscle wasting and regeneration (17). In addition, muscle immobilization
injury increases muscle wasting because of muscle disuse, also
disrupting the balance between muscle wasting and regeneration
(51,54–56).
The present results revealed that the precursor of miR-206 was
increased in all muscles of immobilization-injured mice and in the
TA muscle of PHF20-overexpressing mice, whereas miR-6516 was
generally decreased only in the muscles of the
immobilization-injured group. These findings suggested that the
change in the expression levels of the precursor of miR-206 may be
in response to muscle loss, and that the change in the expression
levels of the precursor of miR-6516 may be in response to the
muscle atrophy caused by muscle disuse. The expression levels of
the precursors miR-206 and miR-6516 were decreased in muscle
atrophy-derived cfRNA. Although miR-206 is a well-known
muscle-derived miRNA, there is a lack of research on the
association between miR-6516 and skeletal muscle. Similarly, there
is a lack of research on the association between miR-206 and
miR-6516. Notably, the expression levels of the miR-206 and
miR-6516 precursors were both decreased during skeletal muscle
atrophy, indicating a significant relationship between the
expression levels of precursors and muscle mass maintenance, which
warrants further studies.
Previous studies have reported that miR-664a-5p is
increased in circulating exosomes from patients with obesity and
type 2 diabetes (57). The present
study confirmed that the expression levels of the precursor of
miR-664a were increased in the plasma of patients at risk of
sarcopenia, suggesting that this may be caused by diabetes, which
was common in the patient group.
The present study also revealed that the precursors
of miR-1291, miR-23a and miR-664 were downregulated in the TA
muscles of mice with confirmed muscle atrophy. The TA muscle is
composed principally of fast-twitch muscles (58,59),
suggesting that the precursors of miR-1291, miR-23a and miR-664
function as biomarkers of fast-twitch muscle atrophy. However, as
these three miRNA precursors are produced in tissues other than
skeletal muscle, fast-twitch muscle atrophy cannot be controlled
only by the levels of skeletal muscle-derived miRNA precursors.
It may be beneficial to measure mature miRNAs in
relation to precursor miRNAs in human plasma; however, due to the
limited amount of plasma samples available, only quantitative
analysis of miRNA precursors could be performed in the present
study. The mature form of general precursor miRNA, which did not
exhibit significant differences between the normal and patient
groups, requires further research as it may be discovered as a new
biomarker. Furthermore, due to the limited amount of muscle-derived
cfRNA samples, the present study was unable to conduct quantitative
analysis on targets other than miRNA precursors. It may also be
advantageous to measure not only miRNA precursors, but also mature
miRNAs and the proteins/mRNAs involved in their production. In
addition, the relationship between miR-6516 and Cdkn1b
expression needs to be confirmed. However, for reasons, such as
those aforementioned, the effect of mimic-miR-6516 administration
was confirmed, but it was not confirmed whether the expression
levels of Cdkn1b were dependent on the expression levels of
miR-6516; therefore, further research on this is necessary.
In conclusion, the present study provides evidence
that the expression of the muscle-derived miR-206 precursor may be
increased during muscle atrophy, supporting a role for this
molecule as a muscle atrophy biomarker. Additionally, the present
study provides evidence that miR-6516 administration to muscles
could inhibit muscle rest-induced atrophy by suppressing
Cdkn1b. Taken together, these results indicated that the
miR-206 precursor may serve as a biomarker of muscle atrophy, and
miR-6516 may serve as a candidate therapeutic target to inhibit
muscle atrophy.
Acknowledgements
The biospecimens and data used for the present study
were provided by the Biobank of Korea-Chungbuk National University
Hospital (CBNUH), a member of the Korea Biobank Network. All
materials derived from the National Biobank of Korea-CBNUH were
obtained (with written informed consent) in accordance with
institutional review board-approved protocols.
Funding
This work was financially supported by National Research
Foundation of Korea (NRF) grants funded by the Korea Government
(grant nos. NRF-2021R1A2C1008492 and NRF-2020R1F1A1049801) and by
the Technology Development Program (grant no. S3198556) funded by
the Ministry of SMEs and Startups (Korea).
Availability of data and materials
The data generated in the present study may be found
in the NCBI Sequence Read Archive under accession number
PRJNA1088925 or at the following URL: https://www.ncbi.nlm.nih.gov/sra/PRJNA1088925. The
data generated in the present study may be requested from the
corresponding author.
Authors' contributions
WJ, UJ, SG, HN, BL, SHK, SR, JiP and JoP contributed
to conception and design. WJ, UJ, SG, HN, QH, SL and JoP acquired,
analyzed and interpreted the data. WJ, UJ, SG and HN participated
in the data acquisition and analysis. SHK and JoP contributed the
funding and performed the final revision of the manuscript. WJ and
JoP confirm the authenticity of all the raw data. All authors
agreed to be accountable for all aspects of the work, and all
authors read and approved the final version of the manuscript.
Ethics approval and consent to
participate
Experiments using human derivatives, materials,
methods, ethical considerations and reasons for exemption from
research subject consent were reviewed by the institutional review
board of Chungnam National University Hospital, and exemption from
review was approved on the condition that human derivatives were
provided from a biobank (approval no. CNUH 2022-11-087; Daejeon,
South Korea). The biospecimens and data used for this study were
provided by the Biobank of Korea-CBNUH, a member of the Korea
Biobank Network. Also, the Institutional Animal Care and Use
Committee of Chungnam National University approved all animal
management and experiment protocols (approval nos. 202305A-CNU-083
and 202309A-CNU-155). All mice experiments were conducted in animal
facilities in accordance with institutional guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Larsson L, Degens H, Li M, Salviati L, Lee
YI, Thompson W, Kirkland JL and Sandri M: Sarcopenia: Aging-Related
loss of muscle mass and function. Physiol Rev. 99:427–511. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chianca V, Albano D, Messina C, Gitto S,
Ruffo G, Guarino S, Del Grande F and Sconfienza LM: Sarcopenia:
Imaging assessment and clinical application. Abdom Radiol (NY).
47:3205–3216. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Geladari E, Alexopoulos T, Kontogianni MD,
Vasilieva L, Mani I and Alexopoulou A: Mechanisms of sarcopenia in
liver cirrhosis and the role of myokines. Ann Gastroenterol.
36:392–404. 2023.PubMed/NCBI
|
|
4
|
Xu J, Wan CS, Ktoris K, Reijnierse EM and
Maier AB: Sarcopenia is associated with mortality in adults: A
systematic review and meta-analysis. Gerontology. 68:361–376. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wahlen BM, Mekkodathil A, Al-Thani H and
El-Menyar A: Impact of sarcopenia in trauma and surgical patient
population: A literature review. Asian J Surg. 43:647–653. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mirzai S, Eck BL, Chen PH, Estep JD and
Tang WHW: Current approach to the diagnosis of sarcopenia in heart
failure: A narrative review on the role of clinical and imaging
assessments. Circ Heart Fail. 15:e0093222022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chekulaeva M and Filipowicz W: Mechanisms
of miRNA-mediated post-transcriptional regulation in animal cells.
Curr Opin Cell Biol. 21:452–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brzeszczynska J, Brzeszczynski F, Hamilton
DF, McGregor R and Simpson AHRW: Role of microRNA in muscle
regeneration and diseases related to muscle dysfunction in atrophy,
cachexia, osteoporosis, and osteoarthritis. Bone Joint Res.
9:798–807. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yanai K, Kaneko S, Ishii H, Aomatsu A, Ito
K, Hirai K, Ookawara S, Ishibashi K and Morishita Y: MicroRNAs in
Sarcopenia: A systematic review. Front Med (Lausanne). 7:1802020.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lee J and Kang H: Role of MicroRNAs and
Long Non-Coding RNAs in Sarcopenia. Cells. 11:1872022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gan L and Denecke B: Profiling
Pre-MicroRNA and Mature MicroRNA expressions using a single
microarray and avoiding separate sample preparation. Microarrays
(Basel). 2:24–33. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan B, Yu J and Liu X: Upregulation of
miR-886 indicates poor prognosis and promotes tumour progression of
prostate cancer. Andrologia. 54:e142962022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee K, Kunkeaw N, Jeon SH, Lee I, Johnson
BH, Kang GY, Bang JY, Park HS, Leelayuwat C and Lee YS: Precursor
miR-886, a novel noncoding RNA repressed in cancer, associates with
PKR and modulates its activity. RNA. 17:1076–1089. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee H, Hong Y, Kong G, Lee DH, Kim M, Tran
Q, Cho H, Kim C, Park S, Kim SH, et al: Yin Yang 1 is required for
PHD finger protein 20-mediated myogenic differentiation in vitro
and in vivo. Cell Death Differ. 27:3321–3336. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vo TT, Tran Q, Hong Y, Lee H, Cho H, Kim
M, Park S, Kim C, Bayarmunkh C, Boldbaatar D, et al: AXL is
required for hypoxia-mediated hypoxia-inducible factor-1 alpha
function in glioblastoma. Toxicol Res. 39:669–679. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tang Z, Kang B, Li C, Chen T and Zhang Z:
GEPIA2: An enhanced web server for large-scale expression profiling
and interactive analysis. Nucleic Acids Res. 47:W556–W560. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
GTEx Consortium: Human genomics. The
Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene
regulation in humans. Science. 348:648–660. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lee KP, Shin YJ, Panda AC, Abdelmohsen K,
Kim JY, Lee SM, Bahn YJ, Choi JY, Kwon ES, Baek SJ, et al: miR-431
promotes differentiation and regeneration of old skeletal muscle by
targeting Smad4. Genes Dev. 29:1605–1617. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Salant GM, Tat KL, Goodrich JA and Kugel
JF: miR-206 knockout shows it is critical for myogenesis and
directly regulates newly identified target mRNAs. RNA Biol.
17:956–965. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Iannone F, Montesanto A, Cione E, Crocco
P, Caroleo MC, Dato S, Rose G and Passarino G: Expression Patterns
of Muscle-Specific miR-133b and miR-206 correlate with nutritional
status and Sarcopenia. Nutrients. 12:2972020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Scott MS, Avolio F, Ono M, Lamond AI and
Barton GJ: Human miRNA precursors with box H/ACA snoRNA features.
PLoS Comput Biol. 5:e10005072009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Scott MS, Ono M, Yamada K, Endo A, Barton
GJ and Lamond AI: Human box C/D snoRNA processing conservation
across multiple cell types. Nucleic Acids Res. 40:3676–3688. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Scott MS and Ono M: From snoRNA to miRNA:
Dual function regulatory non-coding RNAs. Biochimie. 93:1987–1992.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rother S and Meister G: Small RNAs derived
from longer non-coding RNAs. Biochimie. 93:1905–1915. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Coley AB, DeMeis JD, Chaudhary NY and
Borchert GM: Small nucleolar derived RNAs as regulators of human
cancer. Biomedicines. 10:18192022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Purnamasari D, Tetrasiwi EN, Kartiko GJ,
Astrella C, Husam K and Laksmi PW: Sarcopenia and chronic
complications of type 2 diabetes mellitus. Rev Diabet Stud.
18:157–165. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sabatino A, Cuppari L, Stenvinkel P,
Lindholm B and Avesani CM: Sarcopenia in chronic kidney disease:
What have we learned so far? J Nephrol. 34:1347–1372. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Owens J, Moreira K and Bain G:
Characterization of primary human skeletal muscle cells from
multiple commercial sources. In Vitro Cell Dev Biol Anim.
49:695–705. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Harding CP and Vargis E: Muscle atrophy
marker expression differs between rotary cell culture system and
animal studies. Biomed Res Int. 2019:20428082019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kang SH, Lee HA, Kim M, Lee E, Sohn UD and
Kim I: Forkhead box O3 plays a role in skeletal muscle atrophy
through expression of E3 ubiquitin ligases MuRF-1 and atrogin-1 in
Cushing's syndrome. Am J Physiol Endocrinol Metab. 312:E495–E507.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chan J, Lu YC, Yao MM and Kosik RO:
Correlation between hand grip strength and regional muscle mass in
older Asian adults: An observational study. BMC Geriatr.
22:2062022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bodine SC: Disuse-induced muscle wasting.
Int J Biochem Cell Biol. 45:2200–2208. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Nunes EA, Stokes T, McKendry J, Currier BS
and Phillips SM: Disuse-induced skeletal muscle atrophy in disease
and nondisease states in humans: Mechanisms, prevention, and
recovery strategies. Am J Physiol Cell Physiol. 322:C1068–C1084.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Urso ML, Scrimgeour AG, Chen YW, Thompson
PD and Clarkson PM: Analysis of human skeletal muscle after 48 h
immobilization reveals alterations in mRNA and protein for
extracellular matrix components. J Appl Physiol (1985).
101:1136–1148. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Aihara M, Hirose N, Katsuta W, Saito F and
Maruyama Hagiwara H: A new model of skeletal muscle atrophy induced
by immobilization using a hook-and-loop fastener in mice. J Phys
Ther Sci. 29:1779–1783. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shen Y, Zhang R, Xu L, Wan Q, Zhu J, Gu J,
Huang Z, Ma W, Shen M, Ding F and Sun H: Microarray analysis of
gene expression provides new insights into denervation-induced
skeletal muscle atrophy. Front Physiol. 10:12982019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Spangenburg EE, Chakravarthy MV and Booth
FW: p27Kip1: A key regulator of skeletal muscle satellite cell
proliferation. Clin Orthop Relat Res (403 Suppl). S221–S227. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pruitt SC, Freeland A, Rusiniak ME, Kunnev
D and Cady GK: Cdkn1b overexpression in adult mice alters the
balance between genome and tissue ageing. Nat Commun. 4:26262013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Toklowicz M, Zbikowska A, Janusz P,
Kotwicki T, Andrusiewicz M and Kotwicka M: MicroRNA expression
profile analysis in human skeletal muscle tissue: Selection of
critical reference. Biomed Pharmacother. 162:1146822023. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mytidou C, Koutsoulidou A, Katsioloudi A,
Prokopi M, Kapnisis K, Michailidou K, Anayiotos A and Phylactou LA:
Muscle-derived exosomes encapsulate myomiRs and are involved in
local skeletal muscle tissue communication. FASEB J. 35:e212792021.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ma G, Wang Y, Li Y, Cui L, Zhao Y, Zhao B
and Li K: MiR-206, a key modulator of skeletal muscle development
and disease. Int J Biol Sci. 11:345–352. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Giagnorio E, Malacarne C, Mantegazza R,
Bonanno S and Marcuzzo S: MyomiRs and their multifaceted regulatory
roles in muscle homeostasis and amyotrophic lateral sclerosis. J
Cell Sci. 134:jcs2583492021. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhelankin AV, Iulmetova LN, Ahmetov II,
Generozov EV and Sharova EI: Diversity and Differential Expression
of MicroRNAs in the human skeletal muscle with distinct fiber type
composition. Life (Basel). 13:6592023.PubMed/NCBI
|
|
48
|
Powers SK, Lynch GS, Murphy KT, Reid MB
and Zijdewind I: Disease-Induced skeletal muscle atrophy and
fatigue. Med Sci Sports Exerc. 48:2307–2319. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Sartori R, Romanello V and Sandri M:
Mechanisms of muscle atrophy and hypertrophy: Implications in
health and disease. Nat Commun. 12:3302021. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Marusic U, Narici M, Simunic B, Pisot R
and Ritzmann R: Nonuniform loss of muscle strength and atrophy
during bed rest: A systematic review. J Appl Physiol (1985).
131:194–206. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gao Y, Arfat Y, Wang H and Goswami N:
Muscle atrophy induced by mechanical unloading: Mechanisms and
potential countermeasures. Front Physiol. 9:2352018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hosoyama T, Van Dyke J and Suzuki M:
Applications of skeletal muscle progenitor cells for neuromuscular
diseases. Am J Stem Cells. 1:253–263. 2012.PubMed/NCBI
|
|
53
|
Pang KT, Loo LSW, Chia S, Ong FYT, Yu H
and Walsh I: Insight into muscle stem cell regeneration and
mechanobiology. Stem Cell Res Ther. 14:1292023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Ji LL and Yeo D: Mitochondrial
dysregulation and muscle disuse atrophy. F1000Res. 8:F1000 Faculty
Rev. 16212019. View Article : Google Scholar
|
|
55
|
Manas-Garcia L, Penedo-Vazquez A,
Lopez-Postigo A, Deschrevel J, Duran X and Barreiro E: Prolonged
immobilization exacerbates the loss of muscle mass and function
induced by cancer-associated cachexia through enhanced proteolysis
in mice. Int J Mol Sci. 21:81672020. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Thompson JM, West DWD, Doering TM, Budiono
BP, Lessard SJ, Koch LG, Britton SL, Byrne NM, Brown MA, Ashton KJ
and Coffey VG: Effect of short-term hindlimb immobilization on
skeletal muscle atrophy and the transcriptome in a low compared
with high responder to endurance training model. PLoS One.
17:e02617232022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kim H, Bae YU, Lee H, Kim H, Jeon JS, Noh
H, Han DC, Byun DW, Kim SH, Park HK, et al: Effect of diabetes on
exosomal miRNA profile in patients with obesity. BMJ Open Diabetes
Res Care. 8:e0014032020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hata J, Nakashima D, Tsuji O, Fujiyoshi K,
Yasutake K, Sera Y, Komaki Y, Hikishima K, Nagura T, Matsumoto M,
et al: Noninvasive technique to evaluate the muscle fiber
characteristics using q-space imaging. PLoS One. 14:e02148052019.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang C, Yue F and Kuang S: Muscle
histology characterization using H&E staining and muscle fiber
type classification using immunofluorescence staining. Bio Protoc.
7:e22792017. View Article : Google Scholar : PubMed/NCBI
|