Severe acute respiratory syndrome coronavirus 2
(SARS-CoV-2) has resulted in a global pandemic of coronavirus
disease 2019 (COVID-19). Although infection typically starts with
flu-like symptoms (1), patients
can also be asymptomatic and may go on to have a mild to severe
disease course (2).
Mechanistically, COVID-19 is characterized by an important burden
of inflammation mainly in the respiratory system, although it can
also affect other organ systems (3,4).
Previous studies have assessed the association between hemogram
indices and COVID-19 pathogenesis, improving the prognostic
judgement and patient management of this disease (5,6).
Nevertheless, a requirement for safe prevention and treatment
methods for COVID-19 remains. Efforts for this are currently
ongoing, including the development of numerous types of vaccines,
attempts to establish active herd immunity, exploration of the
efficacy of existing drugs and the development of novel
small-molecule drugs targeting viral or host proteins to inhibit
the replication of viruses (7–9). The
majority of patients with SARS-CoV-2 infections typically exhibit
an antibody response 5–15 days after the occurrence of symptoms,
peaking at 21–28 days, before declining (10,11).
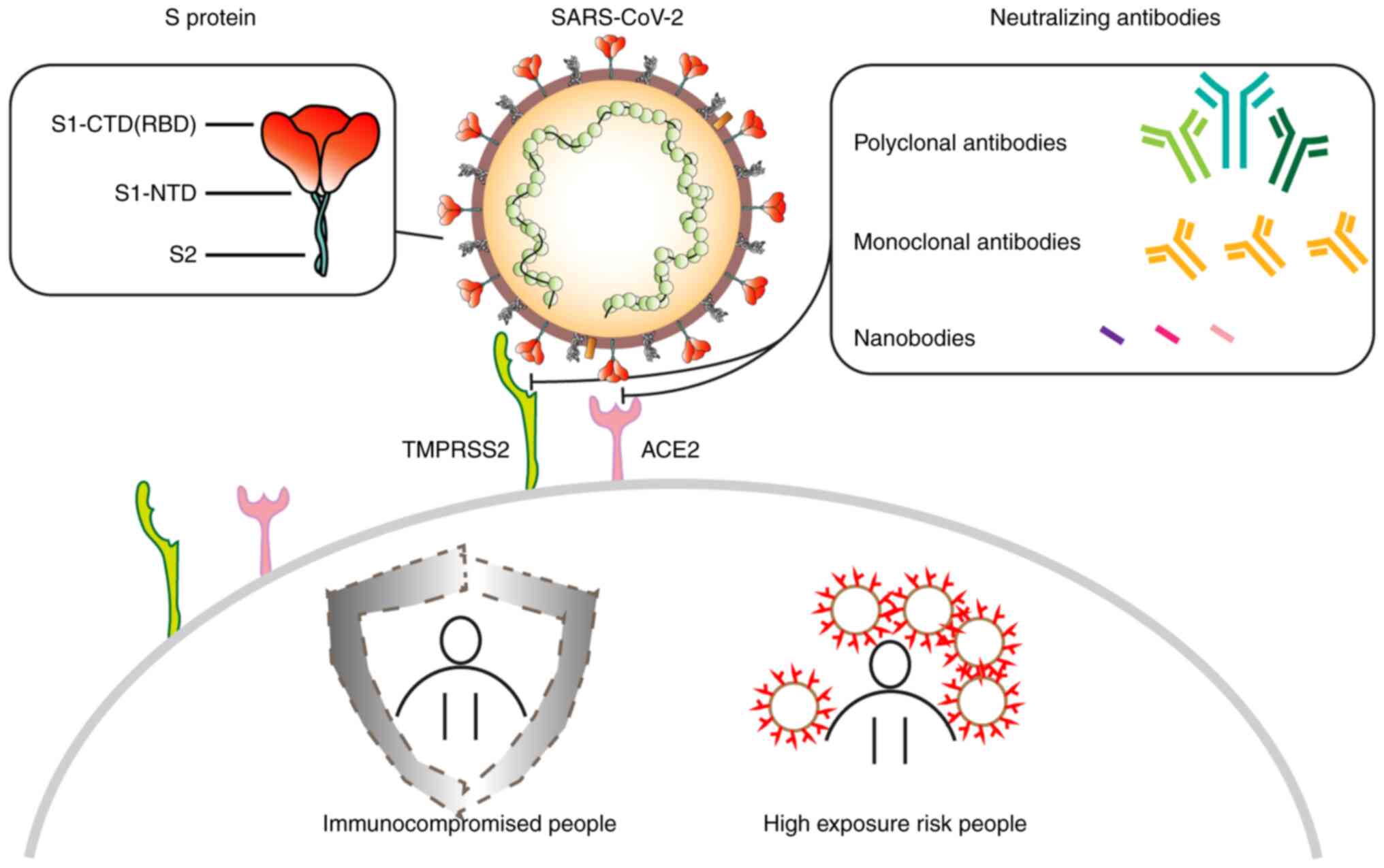
Given the time it takes to develop immunity, this is a dangerous
period for individuals who are at high risk of exposure, who are
immunocompromised, or who are unable to acquire antibodies from
active immunization. Thus, prompt passive immunization is urgently
required (12).
The earliest attempt at passive immunization was
through convalescent plasma infusion, which provides neutralizing
antibodies (NAbs), which can alleviate the inflammatory burden
(13). Convalescent plasma is
plasma donated by individuals previously infected with infectious
diseases, who have produced protective NAbs 14–28 days after
infection (14). Although early
clinical trials on convalescent plasma transfusion reported
promising results, its shortcomings were not negligible. For the
treatment to work effectively, patients should ideally receive an
infusion during the early stages of infection, whilst being
monitored to avoid risks associated with blood transfusions, such
as hyperinflammatory immune reactions and transfusion-associated
circulatory overload (15).
Moreover, the screening and monitoring of plasma donors is labor
and resource-intensive. There is also a lack of a universally
accepted standard for the infusion volume. Recent clinical trials
have reported that convalescent plasma transfusion does not
significantly reduce mortality, as its neutralization capacity
declines after viral mutations (16,17).
However, the total antibody titer following human COVID-19
immunoglobulin intravenous injection was previously reported to be
three times higher compared with that of convalescent plasma,
suggesting this treatment to be more effective. pH4 is manufactured
from already inactivated, filtered and purified convalescent plasma
of patients infected with SARS-CoV-2. It contains high purity and
high titer SARS-CoV-2 NAbs, which have been reported to effectively
neutralize SARS-CoV-2 in vivo. A number of clinical trials
have reported that the intravenous injection of human
immunoglobulin can abridge the duration of positive PCR
confirmation and inflammation, as evidenced by computed tomography
data in patients (18,19). According to the diagnostic and
treatment guidelines in China, intravenous immunoglobulins can be
used in an emergency for patients with rapid severe disease
progression (20). Furthermore,
another study has previously proposed the potential application of
pH4 for patients with SARS-CoV-2 infection with low immunoglobin M
levels (21).
However, one limitation of convalescent plasma and
intravenous immunoglobulin is the requirement to recruit blood
donors with high titers of NAbs against the pathogens of interest
to maintain a stable, sufficient supply. Identification of single B
cells that produce virus-specific neutralizing monoclonal
antibodies (mAbs) from these donors can potentially be used to
circumvent this limitation. The immunoglobulin gene expressed in B
cells can be cloned and expressed to produce high titers of
neutralizing monoclonal antibodies (22). Large-scale preparations of
specifically targeting mAbs that are of high purity with potent
neutralizing activity can render them powerful tools for the
prevention and treatment of infectious diseases (23). For SARS-CoV-2, specific B cells
have been obtained from patients recovered from COVID-19 to produce
large quantities of humanized mAbs through numerous methods,
including mouse hybridoma fusion, single B cell sorting, phage
display and transgenic mice and antibody screening technologies
(24–26). A number of NAbs against SARS-CoV-2
have been developed. According to randomized double-blinded
controlled clinical trials, the Food and Drug Administration has
previously approved the emergency use of certain NAb drugs for the
treatment of SARS-CoV-2 such as bamlanivimab, etesevimab,
casirivmab and imdevimab (27,28).
Furthermore, The State Drug Administration of China has authorized
Amubarvimab and Romlusevimab cocktail therapies, which were
designed for treating adults or young individuals (12–17 years old,
weight ≥40 kg) with mild and common types of SARS-CoV-2 infection
who are at high risk of developing severe type infections (29). This treatment strategy is now
serving a pivotal role in the clinical treatment process in Chinese
hospitals to reduce incidence of serious adverse events (30). Compared with convalescent plasma,
these types of NAbs confer a number of distinct advantages. The
final selection of the most effective candidate antibody can be
evaluated, with the IC50 value typically on nanomolar or
picomolar levels, and the dose can be evaluated more accurately
(31). Furthermore, the
probability of the antibody-dependent enhancement (ADE) phenomenon
associated with this type of NAbs, wherein non-neutralizing
antibodies facilitate the entry or replication of a pathogen,
thereby potentiating infection rather than providing protection, is
considerably lower (32).
New variants continue to emerge, such as the ο
variants, which are notably more transmissible compared with those
of the original strain, and have negatively impacted the health and
normal life of the global population (33). Mutations that produce mutant forms
of the spike (S) protein have resulted in the reduction or
disappearance of the efficacy of vaccines and NAbs, increasing the
demand for the development of next generation vaccines and antibody
drugs (34). Nanobodies (Nbs) are
heavy chain antibodies, which have been previously reported to be
found in camelids and cartilaginous fish, such as sharks (35). They form the smallest known
complete functional structure that can target virus antigens.
Numerous characteristics of Nbs, such as their small sizes, high
specificity, stability, ease of production, potent penetration and
low immunogenicity, render them able to recognize antibody epitopes
that cannot be readily recognized by conventional antibodies
(36). This would in turn increase
the diversity of potential targets and binding ability of
antibodies, providing a broader and more targeted choice for the
research and development (R&D) of next generation antibody
drugs with wide clinical implications.
There are currently four known genera of
coronaviruses, specifically α, β, γ and δ. β coronaviruses can be
classified into subgenera A, B, C and D (37). SARS-CoV-2 is a member of the B
subgenus of β coronaviruses. SARS-CoV-2 is a single-stranded,
positive-sense RNA virus that can directly guide protein synthesis
upon entering the cell to replicate itself, by generating negative
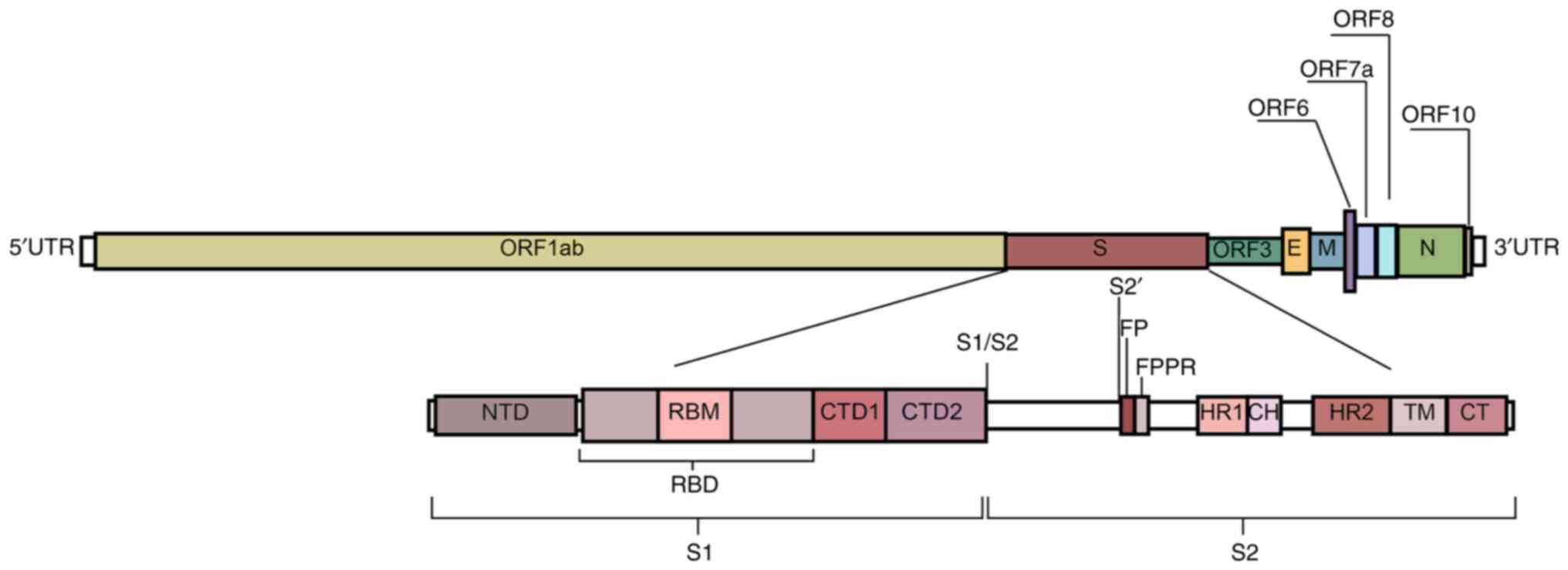
strands using RNA polymerase (38). The RNA sequence of SARS-CoV-2
contains 29,891 nucleotides, encoding 9,860 amino acids with a GC
percentage of ~38%. Furthermore, there is a 5′ cap-like structure
and a 3′ poly-A tail on its genome (Fig. 2) (39). In total, two overlapping open
reading frames (ORFs), namely ORF1a and ORF1b, encode 16
non-structural proteins (40). The
−1 frameshift between ORF1a and ORF1b contributes to the production
of polypeptide 1a and a larger polypeptide 1ab. For genome
amplification, SARS-CoV-2 viruses generate antisense RNAs as
templates for creating the sense genomic RNA (gRNA) and subgenomic
RNA (sgRNA). gRNA together with the structural proteins expressed
contributes to producing the viral offspring. The shorter sgRNA has
the role of expressing the four types of conserved structural
proteins in coronaviruses, namely S protein, membrane protein,
envelope protein and nucleocapsid protein, as well as six auxiliary
proteins (3, 6, 7a, 7b, 8 and 10) (41).
The S protein is a type I fusion protein that
mediates viral entry into targeted cells. It is a major target of
post-infection NAbs and the main focus of numerous therapy and
vaccine studies. The S protein forms a surface-exposed trimer on
the viral particles and consists of a long extracellular region, a
transmembrane region and an intramembrane region (42). On the surfaces of targeted cells,
the S protein binds with angiotensin converting enzyme 2 (ACE2)
before undergoing structural transformations to induce the fusion
of the viral and cell membranes. Each S protein contains 1,273
amino acids, including an N-terminal signal peptide, a
receptor-binding part S1 and a fusion part S2. The S protein has
been previously studied using cryoelectron microscopy (Cryo-EM),
which reported two structural states (43,44).
While in the closed state, three receptor binding domains (RBDs) of
the S protein are locked in the ‘down’ conformation, whereas during
the open state, there is one RBD in the ‘up’ conformation. The RBDs
form the key region necessary for SARS-CoV-2 interactions with
ACE2, where its open state is the prerequisite for virus-cell
membrane fusion (44,45).
The S1 subunit has four domains, namely A, B, C and
D, with domains A and B being responsible for receptor binding. The
structure of domain A consists of a galectin-like β-fold, whereas
domain B has a reverse parallel β-fold structure. There is an
extended loop of domain B at the end of the virion that is
structurally different depending on the species of the
β-coronavirus, known as the hypervariable region (46). Domains C and D on the C-terminus
constitute discrete segments of the primary protein sequence and
are directly linked to the stem core of the S2 subunit to form a
β-fold structure. The entire S1 subunit is connected by a ring
covering the surface of S2 (47).
From the perspective of its linear peptide
structure, S1 consists of the N-terminal domain (NTD), RBD and the
C-terminal domain (CTD). Due to the swing of the S protein on the
viral membrane with a 40 main angle of inclination, the perceived
primary point of interaction with the epithelium is the NTD, which
can be targeted by numerous powerful Nabs (48–50).
The NTD of the SARS-CoV-2 S protein can form a ribbon structure
that is analogous to that of human galectin and is located at
residues 14–305. This mediates weak and reversible interactions
with superficial glycans, such as sialic acid, through low-affinity
hydrogen bonds, is crucial for the virus to attach to and navigate
along the cell surface, a process referred to as ‘viral surfing’.
Subsequently, numerous critical residues of the NTD combine with
sialic acid (51,52), forming a flat surface to strengthen
the primary interaction between SARS-CoV-2 and targeted cells,
consolidating the infection (53).
The RBD is located at residues 336–525 of the S protein, which has
two domains: A central structure consisting of five parallel
β-sheets and an extended loop known as the receptor-binding motif
(RBM) at residues 437–508. This extended loop serves to surround
the edge of the central structure and interacts with ACE2 (31). In the down state, the
receptor-binding motif (RBM) is partially obstructed, limiting its
engagement with ACE2. Upon RBD's transition to the up state, the
RBM becomes accessible to interact with ACE2, facilitating viral
entry (54,55). At residues 528–685 of the S protein
is the CTD, which mainly consists of a β-structure. CTD1 serves to
‘sense’ changes in its neighboring sites, whilst CTD2 is vital for
membrane fusion with the entire rearranged S protein (46,56).
The S2 subunit promotes membrane fusion and binds
the S protein onto the host cell membrane. It is highly conserved
among coronaviruses and contains important regions for promoting
fusion with target cells (45).
Upon RBD's engagement with the receptor, the fusion peptide (FP) is
inserted into the cell membrane, which then triggers the unfolding
of the heptad repeat 1 (HR1) domain and the folding back of the HR2
domain. This sequence of events leads to the domains coming
together, causing the membranes to bend towards each other and
facilitating membrane fusion, thus enabling viral entry (57). In particular, the FP proximal
region in S2 appears to serve a supporting role in clamping the RBD
and stabilizing the closed conformation of the S protein (46).
For all enveloped viruses, membrane fusion is the
critical initial phase for entry into the targeted cell and the
establishment of infection. There is a high dynamic barrier when
the two membranes approach each other, where the free energy
required to overcome the kinetic barrier comes from the
rearrangement of the fusion protein encoded by the virus,
specifically by changing from the basal variable conformational
state to the stable state upon fusion. Subsequently, through two
protein cleavage events, the S protein transforms into a state that
can readily transition into a low-energy state. The first cleavage
occurs at the boundary between S1 and S2, commonly known as the
S1/S2 site. A four-amino acid residue Arg-Arg-Ala-Arg sequence is
present at the S1/S2 site, which is cleaved by the protease furin
(58). The prefusion S protein
trimer fluctuates between closed and open conformations. It has
been previously hypothesized that the near-universal expression of
furin-like protease may have a role in enhancing the cellular and
tissue tropism of SARS-CoV-2, thus enhancing its infectivity and/or
modifying its pathogenicity. The second cleavage event occurs at
the S2′ site. This cleavage site is only accessible after the
initial S1/S2 cleavage and RBD-ACE2 binding. The differential
access route to SARS-CoV-2 results in the S2′ site being cleaved by
distinct proteases. The cleavage of S2′ site occurs on the cell
surface and is mediated by transmembrane protease serine 2
(TMPRSS2). In the absence of TMPRSS2 or when the likelihood of
encountering TMPRSS2 diminishes on the cell surface, the virus-ACE2
complex will be internalized by lectin-mediated endocytosis into
the endolysosome, where the S2′ site is cleaved by cathepsins,
particularly cathepsin L (59). In
both of these access routes, S2′ cleavage releases the structural
constraints on the FP, whilst the dissociation of S1 from S2
results in a drastic change in the conformation of the S2 subunit,
particularly in HR1, driving the FP into the cell membrane. This
forms a fusion pore through which viral RNA is delivered into the
cytosol of the target cell.
Moreover, SARS-CoV-2 can be cleaved by serine
endonuclease protein convertase 1, trypsin and trypsin-like
integral membrane serine peptidase, all of which can readily
recognize and cleave the S1/S2 site. The S protein is cleavable by
a broader range of proteases compared with the SARS-CoV-related
viruses, which is a critical contributor to its ease of entrance
into targeted cells through the ACE2 pathway and infectiousness
(60).
pAbs are prepared by the direct injection of
antigens into animals for immunization, followed by serum
collection and purification (61).
Compared with monoclonal antibodies, they possess a higher affinity
for target antigens as they can target multiple binding sites on a
single antigen. Moreover, due to the inherent diversity of pAbs,
they tend to be more resistant to the polymorphism of target
antigens, retaining activity despite antigen glycosylation or other
post-translational modifications (62). pAb therapeutics are associated with
abbreviated production timelines and reduced manufacturing expenses
relative to monoclonal antibody counterparts (63). However, due to the large divergence
among batches of pAb therapeutics and the rapid development of more
effective vaccines and monoclonal antibody therapy, the popularity
of pAb drugs has gradually waned. At present, to the best of our
knowledge, there have only been a small number of studies on the
use of pAb against COVID-19.
A horse pAb therapy was previously reported to be
safe and potent for treating SARS-CoV-2 (64). The efficiency of this antibody
against RBD was reported to be ~50 times greater than that of
normal convalescent plasma, and the resultant drug INM 005
(COVID-19) derived from this horse pAb has been approved for
SARS-CoV-2 treatment in Argentina (65). Moreover, another previous study
used the RBD region of the S protein to immunize pigs, which then
produced polyclonal NAbs that do not interact with human Fc
receptors, avoiding potential ADE effects (66). A purified polyclonal IgG fraction
drug, XAV-19, was also reported to be able to neutralize the
original Wuhan strain and subsequent variants, including the γ and
δ variants. XAV-19 was also well tolerated in hospitalized patients
with moderate COVID-associated pneumonia who required low-flow
oxygenation, according to results from clinical trials (67,68).
Commonly used monoclonal antibody screening
technologies include traditional murine hybridoma fusion,
transgenic mouse technology, single B cell isolation, cloning and
phage display (69). The
conventional murine hybridoma method yields antibodies derived from
mice, which are susceptible to human anti-mouse antibody responses,
limiting their clinical applicability. Advances in transgenic mouse
technology have enabled the direct production of fully humanized
antibodies without the need for subsequent humanization steps
(70). This method entails
genetically integrating human antibody genes into the mouse genome,
followed by immunization of these transgenic mice to elicit fully
human antibodies. These antibodies mature in vivo,
exhibiting high affinity and specificity, thus establishing this
platform as the preferred and auspicious route for antibody-based
pharmaceutical development (71).
Single B-cell sorting is a technique used to isolate individual B
cells capable of producing specific NAbs from recovered
individuals. The genes encoding the antibody are sequenced to allow
the recombinant expression and purification of the required mAbs.
This method has emerged as a cornerstone for the expedited
development of anti-SARS-CoV-2 NAbs, as it allows for the rapid and
high-throughput generation of human antibodies from peripheral
blood mononuclear cells, while maintaining the native pairing of
heavy and light chains (72).
Phage display is a method that involves cloning the entire
repertoire of genes from the variable regions of human antibodies
and inserting them into phages harboring the coat protein gene.
This results in the display of exogenous genes on the phage surface
as fusion proteins, creating an antibody library. Screening this
phage library with target proteins facilitates the rapid isolation
of antibodies with high affinity for the desired target (73,74).
Phage display technology outperforms the conventional hybridoma
method in terms of speed, efficiency, and simplicity. Furthermore,
it can be coupled with Nb technology to generate novel Nb variants
characterized by reduced molecular weight, enhanced stability and
increased neutralizing capacity (75).
RBD NAbs are considered to be the most abundant and
potent of the SARS-CoV-2 Nabs (76,77).
A prior investigation into the humoral immune response to
SARS-CoV-2 infection showed that antibodies blocking the
interaction between RBD and ACE2 led to a decrease in viral RNA
expression to levels below detection, suggesting a crucial role for
RBD-specific neutralizing antibodies in the response to SARS-CoV-2
infection (78). A number of RBD
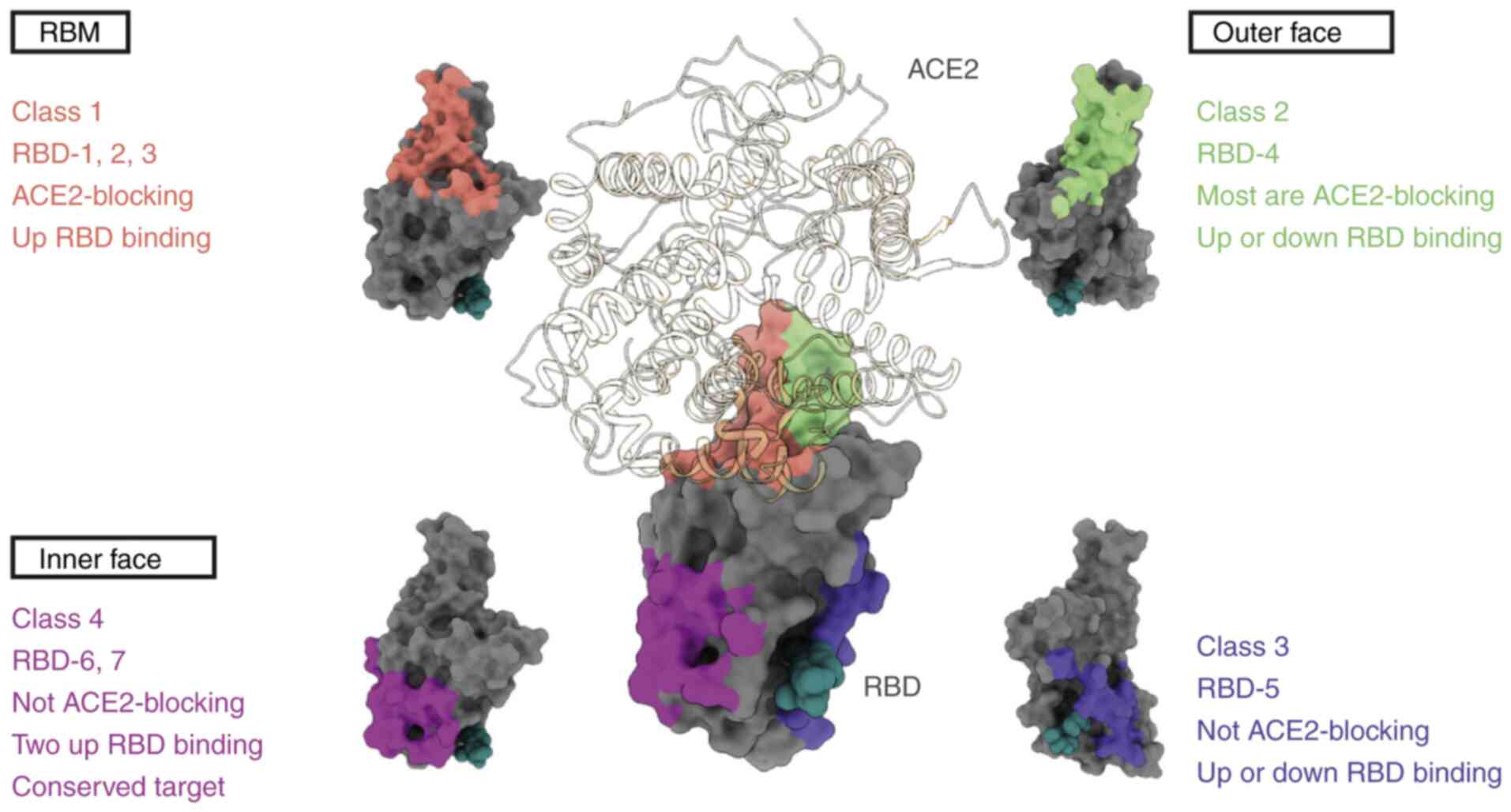
antibody categorization systems have been proposed, with the one
devised by Barnes et al (76) being the most widely accepted. The
approach employs a classification system comprising four
categories, which are delineated by the structural characteristics
and binding sites of the NAbs (Fig.
3). Class 1 RBD NAbs are characterized by their immunoglobulin
heavy chain variable region (IGHV) of 3–53 or 3–66 genetic origin,
short complementarity-determining region (CDR)3 of antibody heavy
chains (CDRH3) of <15 residues in length, RBD ‘up’-state binding
and ACE2 blocking capability. C105 (79), LY-CoV016 (27), B38 (80), CB6 (81) and CT-P59 (31), 1–20, 4–20 (49), 910–30 (82), S2E12 (83) and S2K146 (84) are representative examples of Class
1 RBD NAbs. They predominantly target or overlap with the ACE2
binding site at the RBM, competing with ACE2 for binding at this
site. However, these antibodies do not bind adjacent RBDs (85,86).
Class 2 RBD antibodies are also able to bind to the
ACE2 binding site by recognizing both the ‘up’- and ‘down’-state
RBDs, with added specificity to neighboring RBDs. Unlike the Class
1 IGHV3-53 antibodies, Class 2 antibodies have CDRH3 loops that are
>15 residues. The prominence of Class 2 antibodies in the
RBD-targeting fraction of plasma may be partially attributed to
their binding capacity to both the ‘up’ and ‘down’ states of RBD.
They are typically produced by germline genes, including variable
heavy-chain (VH) 1–2, VH1-69 and VH3-53 (87). C135, C110 (87), C144, C002, C104, C119, C121
(76), DH1041, DH1042 and DH1043
(88) belong to this class of RBD
NAbs. These antibodies not only directly obstruct ACE2 engagement
but can also bridge adjacent ‘down’ state RBDs, locking S proteins
in a ‘closed’ pre-fusion state to inhibit S-ACE2 engagement. Class
1 and Class 2 NAbs do not cross-neutralize viruses due to the
limited conservation of RBM structures in different β-coronavirus
species. By contrast, a number of SARS-CoV-2 NAbs that can identify
conserved RBD epitopes further away from the ACE2 engaging site
have been reported to exhibit considerable cross-neutralizing
ability. Such non-RBM-targeting RBD antibodies are categorized into
structural classes 3 or 4. Class 3 RBD NAbs can identify both the
‘up’ state and ‘down’ state RBDs but do not inhibit ACE2 binding,
whilst Class 4 RBD NAbs do not recognize the ‘down’ RBD
conformation. For example, CR3022 is a weak Class 4 cross-reactive
neutralizing antibody and is one of the most broad-spectrum
coronavirus mAbs identified to date (89,90).
The cryptic site targeted by CR3022 shares 86% similarity among
SARS and SARS-CoV-2 (91).
Furthermore, the Class 3 antibody S309, which was first identified
in a blood sample from a patient with SARS in 2003, can inhibit a
range of associated coronaviruses, including SARS-CoV-2. S309
recognizes a proteoglycan epitope accessible in both the ‘up’ and
‘down’ conformations of RBD but differs in the mode of binding to
RBM on SARS-CoV-2 (25). In
addition, the IgG1κ mAb sotrovimab was developed from the S309
antibody and was previously approved under emergency use
authorization for mild-to-moderate COVID-19 cases in patients aged
≥12 years and weighing >40 kg because of its ability to reduce
the risk of disease progression (92). However, since sotrovimab only
targets a single viral antigenic epitope, resistance can readily
develop in patients (93).
Using high-throughput surface plasmon resonance
analysis and Cryo-EM structure determination, RBD antibodies can be
classified into seven different ‘communities’ ranging from RBD-1 to
RBD-7 according to their binding epitopes, giving rise to another
categorization system (98). This
categorization system offers a more detailed landscape of RBD NAbs,
which can be used to complement the four aforementioned classes.
Possessing non-overlapping epitopes and considerable potency, the
neutralizing effects of RBD-1 to −4 clusters are highly susceptible
to deletions and mutations in emerging SARS-CoV-2 variants. By
contrast, RBD-5 to −7 antibodies are generally less potent, but the
epitopes they target are highly conserved and therefore more
resistant to mutations. Therefore, combining and/or engineering
these antibodies into multivalent formulas can produce
mutagenesis-resistant NAb therapeutic cocktails (98).
Although NAbs generally achieved effective
neutralization against the original 2019 strain of SARS-CoV-2, with
the emergence of variants of concerns (VOCs), the NTD region has
been reported to be highly variable. Antibodies targeting the
supersite of NTD appeared to be especially susceptible to potency
loss, with a large number losing their affinity to α, β and γ VOCs.
However, numerous NTD NAbs, such as 5–7, C1520 and C1565, remain
capable of binding other sites, thereby remaining effective even
for the ο variant BA.1. However, NTD NAbs combined with RBD
antibodies have been proposed to be a more effective therapeutic
cocktail for treating the variants. The combination of FC05 with
H014, HB27 and P17 (106), as
well as the combination of ADI-56479 with ADI-56443 (107), are both examples of combining
NTD-targeting mAbs with RBD-targeting mAbs, and have been reported
to result in lower virus escape compared with either class of NAbs
when used alone. This combination was also proposed to reduce S
protein mutations that lead to neutralization escape.
In a study investigating SARS-CoV-2-related IgG
antibodies, the majority of healthy individuals who had not been
exposed to SARS-CoV-2 exhibited the presence of IgG antibodies
targeting the S2 subunit in their sera (108). Numerous studies have previously
suggested that S2-targeting antibody responses against SARS-CoV-2
are associated with superior outcomes for patients and broader
neutralization, suggesting the crucial protective effect of this
type of antibodies (109–111). S2 is more conserved compared with
S1 and shows 63–98% sequence similarity among the same protein from
seven different human coronaviruses (112). S2 contains numerous conserved
antigenic sites, including the stem helix, the FP and the hinge
region. S2 NAbs tend to inhibit the formation of the six-helix
bundle structures by HR1 or HR2, thereby blocking membrane fusion
and viral entry. Certain S2 antibodies are summarized in Table II. Nevertheless, S2 NAbs appear to
rely on the Fc mechanism for protection in vivo, although
combination with the Fc has been reported to be a promising
strategy in the development of antibody drugs (113).
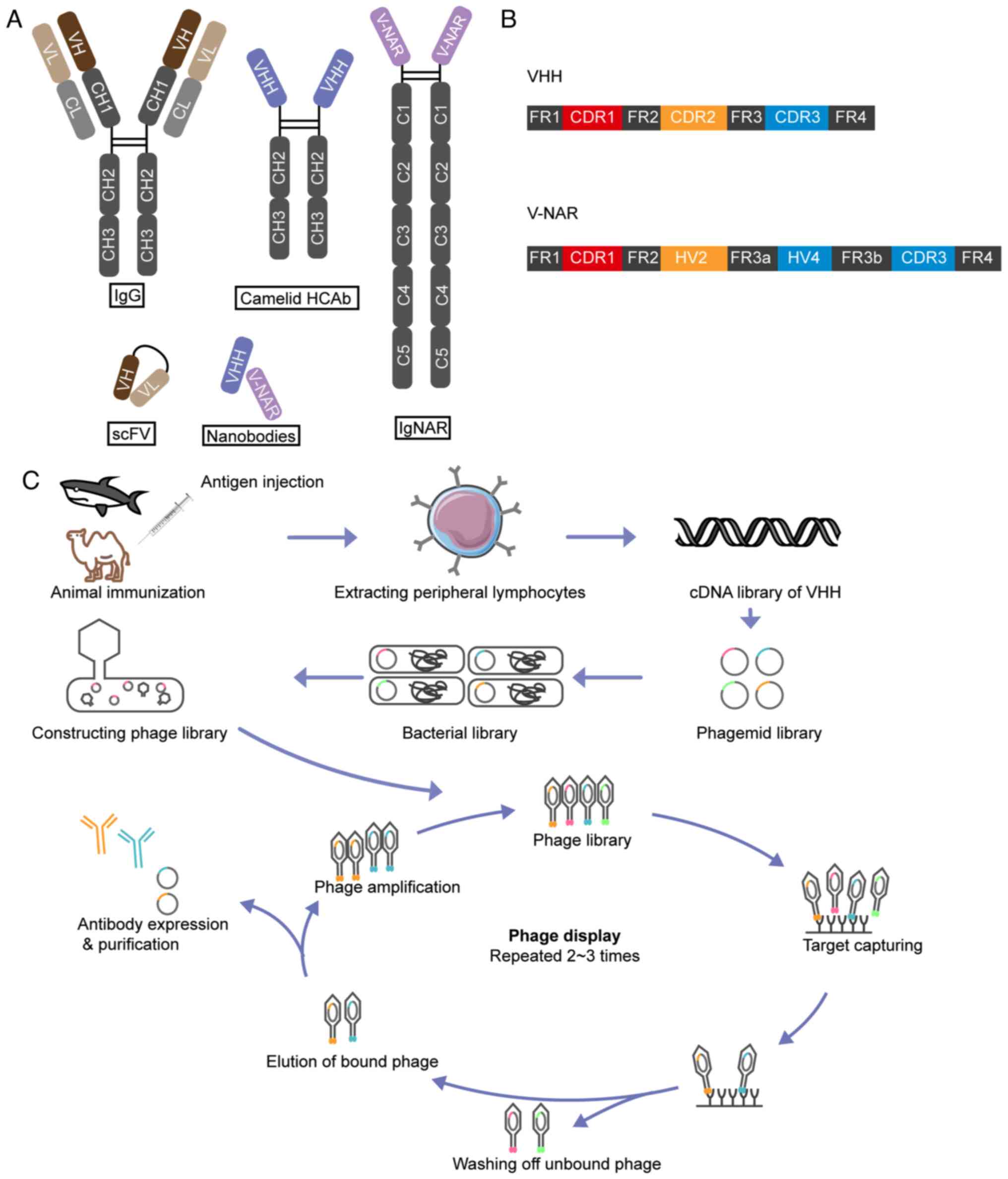
Heavy-chain-only antibodies (HCAbs) in camelids
include two constant structural domains CH2 and CH3, a hinge region
and a variable domain heavy-chain (VHH), and retain complete
antigen binding capability (Fig.
4A) (114). In cartilaginous
fish, their immunoglobulin new antigen receptors (NARs) consist of
a homodimer of five constant domains and a variable domain (V-NAR)
(Fig. 4A) (115). Recombinantly expressed VHH and
V-NAR domains exhibit remarkable structural stability under extreme
temperature and pH conditions, and their antigen-binding
capabilities are comparable to those of HCAbs (116). These fragments represent the
minimal functional units necessary for antigen targeting. Due to
their low molecular weight (<15 kDa), VHH and V-NAR are also
classed as Nbs. The modularity and small size of Nbs allow them to
be readily linked to other molecules, rendering them optimal for
generating bispecific or multispecific antibodies with ideal
affinity or effectiveness (117).
Furthermore, VHH-72 is an Nb that has been previously generated by
immunizing camelids with SARS-CoV and middle East respiratory
syndrome coronavirus RBDs. After the attachment of human IgG Fc to
induce bivalency, it was reported to be able to neutralize the
SARS-CoV-2 pseudovirus in vitro and can be expressed in
transiently transfected ExpiCHO cells (101).
VHHs consist of four conserved sequence regions that
surround three highly variable CDRs, whilst V-NARs possess two CDRs
(CDR1 and CDR3) (118,119) (Fig.
4B). CDR3 is the primary binding domain responsible for 60–80%
of antigen interactions (120).
The SARS-CoV-2 spike protein exhibits an average spacing of 25 nm,
which is not optimal for the 5–10 nm range required for efficient
B-cell response activation. Consequently, this larger spacing
results in inadequate stimulation of B cells and complement
recruitment, leading to a less efficient and transient neutralizing
antibody response (121).
Compared with the poor diversity in CDR loop lengths in
conventional antibodies, VHH and V-NAR possess long protruding CDR3
loops that allow them to access more occluded antigenic epitopes.
Due to their low molecular weight and weak cohesive interactions
between monomers, Nbs can be readily concatenated through genetic
engineering to form multimers. When designed as therapeutic agents,
this property allows for the creation of multivalent and bispecific
antibodies, enhancing their binding capabilities to antigens
(122).
Immunization of animals with specific antigens is
the first step in Nb production (Fig.
4C). Multiple immunizations are typically administered to
stimulate the immune system in the animal into producing antibodies
against specific antigens. After isolating the B lymphocytes from
the blood of immunized animals, molecular biology techniques, such
as reverse transcription PCR, are used to amplify the antibody
genes from the B cells. Specific screening methods, such as phage
display or yeast surface display technology, are then used to
select Nb genes with high affinity from the amplified antibody gene
library, which are subsequently cloned into the expression vectors.
These vectors are transformed into suitable host cells, such as
Escherichia coli or mammalian cells, for recombinant protein
expression. Nbs are then purified from cell culture supernatants or
cell lysates using a range of purification methods, such as
affinity chromatography, ion exchange chromatography and gel
filtration. The purified Nbs undergo numerous bioactivity assays,
cell-based assays and animal experiments, to verify their affinity
and specificity. Depending on the requirements, further
modification and optimization of the Nbs may be performed to
enhance their stability and affinity (123).
Although VHHs are not of human origin, they exhibit
low immunogenicity because of the substantial sequence identity
with the human VH gene family III (124). However, due to the evolutionary
distance between sharks and humans, V-NARs from sharks exhibit
minimal sequence identity with the human VH and variable region
light chain structural domains (~30%). Therefore, humanization of
V-NARs would be desirable prior to clinical application, which at
present is the routine practice. However, in fully humanized VHH,
conserved hydrophilic amino acids in framework region (FR)2 are
changed, leading to unsatisfactory solubility and stability.
Therefore, partial humanization is typically performed.
Traditionally, there are two main approaches for humanization, CDR
grafting and resurfacing. CDR grafting is a process in which the
CDR region is directly grafted from a heterologous antibody to
human FRs (125). Resurfacing is
the replacement of exposed FR residues on the surface of non-human
antibodies with corresponding residues of human antibody FRs,
minimizing the immunogenicity of the antibody in humans (126). Humanized antibodies can also be
produced directly through the immunization of humanized mice.
Moreover, it is theoretically possible to directly extract antibody
gene clones from humans and combine them with Nb. A full Nb library
was constructed by grafting the CDR sequences from a natural
antibody repertoire derived from healthy donor blood onto human
IGHV gene framework regions. A total of 18 unique single-domain
antibodies were isolated from the library, which demonstrated
efficient and specific binding to the SARS-CoV-2 RBD and were
categorized into three competitive groups (127). Among the isolated antibodies,
n3088 and n3130 were found to bind to a concealed epitope of the
SARS-CoV-2 RBD and exhibited significant neutralizing activity.
Due to their stability during long-term storage,
VHHs serve as a viable therapeutic option for future epidemic
responses (128). Their favorable
biophysical attributes facilitate large-scale production via
prokaryotic expression systems in a matter of weeks, which allows
for their swift deployment in an emergency situation (129). A Nb termed Nb6 has been
previously reported to stabilize two adjacent RBDs in the ‘down’
state after binding to S proteins, which may then reorganize the
second and third binding sites of Nb6 to maintain the closed
conformation, causing the RBD to detach from ACE2. Moreover, the
affinity and neutralizing capacity of Nb6 was reported to be
enhanced further after dimerization and trimerization, where the
trimerized version of Nb6 showed picomolar-range neutralization and
femtomolar-range affinity for the SARS-CoV-2 RBD. Efficient
neutralization was even maintained after nebulization,
lyophilization and thermal processing (130).
The majority of mAbs must be generated in mammalian
cells and require intravenous injection. In comparison, Nbs are
produced in bacteria or yeast and can be delivered to the lungs by
inhalation, which can confer significant advantages for SARS-CoV-2
treatment. Possible benefits include low systemic exposure, rapid
onset of action and high concentrations at the lesion site
(131). After immunization of
alpacas with SARS-CoV-2 S protein, Xiang et al (132) identified a large number of Nbs
with high affinity to S protein RBD by using a custom-designed Nb
platform technology. Through further screening, purification and
testing, a number of neutralizing Nbs with exceptional antiviral
ability were identified. Among them, Nb21 was reported to bind to
RBD with picomolar-range affinity and displayed neutralizing
ability against viruses. Upon formation into multivalent
antibodies, their antiviral capabilities are further enhanced. In
particular, Nb21 was reported to be highly thermally stable and
retained the same antiviral capacity after lyophilization and
nebulization. An inhalable nebulizer based on this antibody,
Pittsburgh inhalable Nb-21, was reported to be effective against
severe SARS-CoV-2 infection in hamsters in an in vitro viral
infection assay (133). Nbs, such
as NIH-CoVnb-112, Nb11-59, bn03, 2–3-Fc, Nb22, RBD-1-2G,
pan-Sarbecovirus Nbs, TP17, TP86, R14 and S43 have all also been
demonstrated to exert positive neutralizing effects against
SARS-CoV-2 following respiratory administration (134–141).
ADE has been reported to impact the degranulation of
mast cells, contributing to heart damage or multisystem
inflammatory syndrome (151,152). Due to the complex nature of the
immune system in vivo, whether complement-dependent
cytotoxicity and/or antibody-dependent cell-mediated cytotoxicity
occurs during ADE will likely depend on the balance of virus
removal and infection augmentation. ADE risk has been previously
associated with the concentration of antibodies (153). The balance between neutralizing
and deleterious antibodies in the body favors the neutralization of
SARS-CoV-2 (154). In vivo
experiments prior to clinical trials are necessary for avoiding
ADE. Furthermore, non-neutralizing antibodies have been reported to
be mostly responsible for ADE. Focusing on the RBM or other highly
neutralizing sites may mitigate these drawbacks. Likewise,
engineering antibodies to avoid contact with FcRs or using Nbs
without a Fc region may also reduce these side effects.
Additionally, studies have identified associations between ADE and
specific epitopes on the RBD and NTD of SARS-CoV-2, providing
crucial insight for the development of safe and effective Nabs
(88,155).
Neutralizing antibody development is challenged by
the continuous emergence of mutant variants (156). As an RNA virus, SARS-CoV-2
undergoes constant mutation during replication and under the
pressure of antibody selection. The first prevalent S protein
mutation that received worldwide attention was D614G, a site that
does not come into direct contact with any RBD antibodies, and
therefore had no significant impact on antibody neutralization
activity. By contrast, the ο variant, which was first detected at
the end of 2021, rapidly replaced the δ variant as the major
epidemic strain worldwide due to its high transmissibility. The ο
variants with enhanced resistance to NAbs also challenged
vaccination and infection-induced immunity, rendering therapeutic
mAbs ineffective (157).
Continued mutational evolution of the virus has led to the
emergence of a wide range of variants with greater growth
advantage, which evaded almost all current neutralizing antibody
drugs and vaccinated or convalescent plasma, as represented by the
XBB strain, BQ 1.1 strain and CH 1.1 strain, subvariants of the ο
variant of the SARS-CoV-2 (158).
Breakthrough infections with the BA.2 and BA.5 subvariants of
SARS-CoV-2 have reduced the diversity of NAb binding sites and
increased the proportion of non-NAb clones. This, in turn, has
increased the selective pressure on the humoral immune response,
fostering the convergent evolution of RBD (158,159). Subsequent analysis uncovered that
the ο variant harbors 15 mutation sites within the RBD, which
confer the ability to circumvent antibody neutralization. Among
them, the K417, E484, G446 and S371 mutations mediated the binding
inhibition of Class 1–4 antibodies, respectively. Specifically,
Class 4 antibodies were rendered ineffective against the variants,
leading to a marked reduction in the plasma neutralization capacity
among recovered and vaccinated individuals (84,160).
Antibodies against conserved epitopes are promising
in dealing with the ever-emerging variants. Starr et al
(83) previously identified a
broad-spectrum neutralizing antibody referred to as S2H97, which
binds to a previously unidentified hidden epitope on RBD, causing a
conformational change in the RBD, thereby preventing the binding of
ACE2. This antibody was reported to retain neutralizing activity
against a wide range of strains of Sarbecovirus, including SARS-CoV
and SARS-CoV-2. Antibodies which interact with the same binding
site as S2H97 could not be found in the blood of convalescent
individuals, which is most likely due to the fact that these
epitopes are not easily accessible and therefore cannot
sufficiently trigger an immune response (83).
Through biological computing technologies,
antibodies with superior neutralizing potency can be designed and
produced. An antibody named AI-1028, which is a modified S2H97 with
an improved neutralization capacity compared with S2H97, was
previously reported to be capable of broadly neutralizing
Sarbecovirus, including the emerging ο subvariants XBB, BQ.1.1 and
BA.2.3.20 (161).
It is of strategic importance to predict the
direction of virus evolution to anticipate possible mutant strains
in advance. Cao et al (162) developed a computational model for
predicting the trend of mutational evolution in SARS-CoV-2 using a
high-throughput deep mutation scanning method. Specifically,
mutations in BA.5 and BA.2 that may escape existing herd immunity
were analyzed, identified and validated.
The simultaneous use of multiple antibodies that
target distinct epitopes are known as cocktail therapeutic
regimens, which presents potential for disease treatment and
advancement of novel antibodies. In cases where existing antibody
products exhibit diminished ability to neutralize viral variants,
it would appear logical to modify them using engineering
techniques, such as site mutation, creating bispecific or
multispecific antibodies, bivalent or multivalent constructs
(163,164). Furthermore, the combination of
soluble cytokine receptors or exosomes with antiviral Nbs has
increased effectiveness. c19s130Fc is a bispecific therapeutic that
hinders both the IL-6 signaling pathway and the SARS-CoV-2 RBD. It
is created by fusing a soluble cytokine receptor with an antiviral
Nb, enabling it to block viral entry and dampen the inflammatory
response induced by the virus simultaneously (165). In addition, researchers have
engineered exosomes with a S-protein-targeting Nb and human IFN-β
bound to MFG-E8, creating a dual-action system. The Nbs on the
exosome surface bind to the SARS-CoV-2 S protein, blocking its
entry into host cells, while the encapsulated IFN-β is delivered to
infected cells to trigger antiviral responses and boost the
expression of interferon-stimulated genes (166). In summary, further exploration of
antibody engineering is justified to improve the neutralization
capacity of available antibodies.
Novel NAbs that can target different parts of
SARS-CoV-2 proteins, including the S protein, are constantly being
discovered. It is likely that highly effective novel antibodies
will be screened in the future for treating patients with COVID-19,
especially during the early stages of the disease. Combination
therapy using multiple different NAbs may improve treatment
efficacy and reduce the likelihood of resistance. Furthermore, the
development of long-acting NAbs may reduce the required frequency
of administration and improve patient compliance. This may involve
modifications in the engineering process of the antibody to extend
its half-life in the body. NAbs can be used not only for treatment
but also for pre- or post-exposure prophylaxis, particularly in
high-risk groups, such as healthcare workers and the elderly.
Broad-spectrum neutralizing antibodies, such as the SA55 and SA58
antibody combination, can be administered as a nasal spray to
establish rapid short-acting prophylaxis in the respiratory tract.
These antibodies can also be injected during the initial stages of
infection to provide medium-to-long-term prophylaxis, which is
particularly suitable for the protection of high-risk healthcare
workers, patients with compromised immune systems who cannot be
vaccinated and the elderly (162). Therefore, investigations into
enhanced delivery methods will likely contribute to the advancement
and use of NAbs. Nebulized inhalation and nasal dripping have also
demonstrated promising outcomes in animal models (138–140). Further research into the
mechanism of NAbs to understand how they interact with the
different regions of the virus may be helpful in designing more
effective antibodies and vaccines. Reducing the cost of producing
NAbs by optimizing the production process would make them more
widely available and economically viable. To aid end, research
institutions and companies worldwide should strengthen their
collaboration and share resources and data to accelerate the
R&D of neutralizing antibodies. Moreover, large-scale clinical
trials are warranted to assess the safety and efficacy of
neutralizing antibodies in different populations. Governments and
regulatory agencies should also provide policy support to
accelerate the R&D and approval processes of NAbs, whilst
ensuring product quality and safety.
The present review summarized the current research
progress on antibodies against SARS-CoV-2 by considering the target
sites of antibodies, the SARS-CoV-2 invasion mechanism, as well as
the preparation methods, structural properties, mechanisms of
action and clinical applications of different NAbs. Following the
analysis of a broad body of research, a number of conclusions can
be drawn. Firstly, NAbs serve a role in the prevention and
treatment of SARS-CoV-2. These antibodies are capable of
recognizing and binding to key parts of SARS-CoV-2, preventing
viral invasion into host cells and viral replication. Screening and
evaluation of SARS-CoV-2 NAbs is another required step for
optimization. Through the screening of a large number of candidate
antibodies, antibodies with high neutralizing activity have been
identified, providing a foundation for subsequent research and
applications. Additionally, evaluating these antibodies for
numerous parameters, such as affinity, stability and safety may
ensure their effectiveness and safety in clinical applications.
Furthermore, a series of notable advancements have been made in the
practical clinical application of SARS-CoV-2 NAbs, especially in
the elderly, immunosuppressed or critically ill patients. Moreover,
NAbs can be used as an emergency treatment to reduce viral
replication and disease severity. For individuals at high risk of
exposure to SARS-CoV-2, such as healthcare workers, NAbs can be
used as a preventive treatment. NAbs can also be combined with
vaccines as a ‘passive immunization’ strategy to provide immediate
protection whilst awaiting the establishment of the ‘active
immunization’ response induced by the vaccine. For patients who
continue to experience long-term symptoms after recovery, NAbs may
also help alleviate symptoms and improve quality of life. In
summary, the research and application of SARS-CoV-2 NAbs not only
provide a notable therapeutic tool for understanding the COVID-19
pandemic, but also hold potential preventive and therapeutic value
for future outbreaks. With increased understanding and the
advancement of technology, the clinical application of NAbs will
likely become more widespread, making a greater contribution to
global public health security.
Not applicable.
The present study was supported by the National Natural Science
Foundation of China (grant nos. 81973995 and 82170131) and the
Program for HUST Academic Frontier Youth Team (grant no.
2018QYTD14).
Not applicable.
The present manuscript was written by TZ, DY, LT and
YH. Images were prepared by TZ and DY. All authors read and
approved the final manuscript. Data authentication is not
applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Aktas G: A comprehensive review on
rational and effective treatment strategies against an invisible
enemy; SARS Cov-2 infection. Exp Biomed Res. 3:293–311. 2020.
View Article : Google Scholar
|
|
2
|
Wu Z and McGoogan JM: Characteristics of
and Important Lessons From the Coronavirus Disease 2019 (COVID-19)
Outbreak in China: Summary of a Report of 72 314 Cases From the
Chinese Center for Disease Control and Prevention. JAMA.
323:1239–1242. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aktas G, Balci B, Yilmaz S, Bardak H and
Duman TT: Characteristics of Covid-19 infection with the original
SARS-Cov-2 virus and other variants: A comparative review. J Bionic
Mem. 2:96–112. 2022.
|
|
4
|
Ceasovschih A, Sorodoc V, Shor A, Haliga
RE, Roth L, Lionte C, Onofrei Aursulesei V, Sirbu O, Culis N,
Shapieva A, et al: Distinct features of vascular diseases in
COVID-19. J Inflamm Res. 16:2783–2800. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Khalid A, Ali Jaffar M, Khan T, Abbas Lail
R, Ali S, Aktas G, Waris A, Javaid A, Ijaz N and Muhammad N:
Hematological and biochemical parameters as diagnostic and
prognostic markers in SARS-COV-2 infected patients of Pakistan: A
retrospective comparative analysis. Hematology. 26:529–542. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aktas G: Hematological predictors of novel
Coronavirus infection. Rev Assoc Med Bras (1992). 67 (Suppl
1):S1–S2. 2021. View Article : Google Scholar
|
|
7
|
Fiolet T, Kherabi Y, MacDonald CJ, Ghosn J
and Peiffer-Smadja N: Comparing COVID-19 vaccines for their
characteristics, efficacy and effectiveness against SARS-CoV-2 and
variants of concern: A narrative review. Clin Microbiol Infect.
28:202–221. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng B, Zhao Q, Yang W, Feng P, Xin C,
Ying Y, Yang B, Han B, Zhu J, Zhang M and Li G: Small-molecule
antiviral treatments for COVID-19: A systematic review and network
meta-analysis. Int J Antimicrob Agents. 63:1070962024. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Saul S and Einav S: Old drugs for a new
virus: Repurposed approaches for combating COVID-19. ACS Infect
Dis. 6:2304–2318. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Crawford KHD, Dingens AS, Eguia R, Wolf
CR, Wilcox N, Logue JK, Shuey K, Casto AM, Fiala B, Wrenn S, et al:
Dynamics of neutralizing antibody titers in the months after severe
acute respiratory syndrome coronavirus 2 infection. J Infect Dis.
223:197–205. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Prévost J, Gasser R, Beaudoin-Bussières G,
Richard J, Duerr R, Laumaea A, Anand SP, Goyette G, Benlarbi M,
Ding S, et al: Cross-Sectional Evaluation of Humoral Responses
against SARS-CoV-2 Spike. Cell Rep Med. 1:1001262020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li M, Wang H, Tian L, Pang Z, Yang Q,
Huang T, Fan J, Song L, Tong Y and Fan H: COVID-19 vaccine
development: milestones, lessons and prospects. Signal Transduct
Target Ther. 7:1462022. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wakefield TW, Strieter RM, Wilke CA,
Kadell AM, Wrobleski SK, Burdick MD, Schmidt R, Kunkel SL and
Greenfield LJ: Venous thrombosis-associated inflammation and
attenuation with neutralizing antibodies to cytokines and adhesion
molecules. Arterioscler Thromb Vasc Biol. 15:258–268. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cagdas D: Convalescent plasma and
hyperimmune globulin therapy in COVID-19. Expert Rev Clin Immunol.
17:309–316. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li L, Zhang W, Hu Y, Tong X, Zheng S, Yang
J, Kong Y, Ren L, Wei Q, Mei H, et al: Effect of convalescent
plasma therapy on time to clinical improvement in patients with
severe and life-threatening COVID-19: A Randomized clinical trial.
JAMA. 324:460–470. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang J, Grubbs G, Lee Y, Golding H and
Khurana S: Impact of convalescent plasma therapy on severe acute
respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody profile in
coronavirus disease 2019 (COVID-19) Patients. Clin Infect Dis.
74:327–334. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Ma Y, Xu Y, Liu J, Li X, Chen Y,
Chen Y, Xie J, Xiao L, Xiang Z, et al: Resistance of SARS-CoV-2
Omicron variant to convalescent and CoronaVac vaccine plasma. Emerg
Microbes Infect. 11:424–427. 2022.PubMed/NCBI
|
|
18
|
Cao W, Liu X, Hong K, Ma Z, Zhang Y, Lin
L, Han Y, Xiong Y, Liu Z, Ruan L and Li T: High-Dose intravenous
immunoglobulin in severe coronavirus disease 2019: A multicenter
retrospective study in China. Front Immunol. 12:6278442021.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao W, Liu X, Bai T, Fan H, Hong K, Song
H, Han Y, Lin L, Ruan L and Li T: High-Dose intravenous
immunoglobulin as a therapeutic option for deteriorating patients
with coronavirus disease 2019. Open Forum Infect Dis.
7:ofaa1022020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiang HR, Cheng X, Li Y, Luo WW, Zhang QZ
and Peng WX: Efficacy of IVIG (intravenous immunoglobulin) for
corona virus disease 2019 (COVID-19): A meta-analysis. Int
Immunopharmacol. 96:1077322021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kindgen-Milles D, Feldt T, Jensen BEO,
Dimski T and Brandenburger T: Why the application of IVIG might be
beneficial in patients with COVID-19. Lancet Respir Med.
10:e152022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Breedveld FC: Therapeutic monoclonal
antibodies. Lancet. 355:735–740. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Buss NA, Henderson SJ, McFarlane M,
Shenton JM and de Haan L: Monoclonal antibody therapeutics: History
and future. Curr Opin Pharmacol. 12:615–622. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ren Z, Shen C and Peng J: Status and
developing strategies for neutralizing monoclonal antibody therapy
in the omicron Era of COVID-19. Viruses. 15:12972023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pinto D, Park YJ, Beltramello M, Walls AC,
Tortorici MA, Bianchi S, Jaconi S, Culap K, Zatta F, De Marco A, et
al: Cross-neutralization of SARS-CoV-2 by a human monoclonal
SARS-CoV antibody. Nature. 583:290–295. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hillenbrand M, Esslinger C, Seidenberg J,
Weber M, Zingg A, Townsend C, Eicher B, Rutkauskaite J, Riese P,
Guzman CA, et al: Fast-Track Discovery of SARS-CoV-2-neutralizing
antibodies from human B Cells by direct functional screening.
Viruses. 16:3392024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gottlieb RL, Nirula A, Chen P, Boscia J,
Heller B, Morris J, Huhn G, Cardona J, Mocherla B, Stosor V, et al:
Effect of bamlanivimab as monotherapy or in combination with
etesevimab on viral load in patients with mild to moderate
COVID-19: A Randomized clinical trial. JAMA. 325:632–644. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Weinreich DM, Sivapalasingam S, Norton T,
Ali S, Gao H, Bhore R, Musser BJ, Soo Y, Rofail D, Im J, et al:
REGN-COV2, a neutralizing antibody cocktail, in outpatients with
Covid-19. N Engl J Med. 384:238–251. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ji Y, Zhang Q, Cheng L, Ge J, Wang R, Fang
M, Mucker EM, Chen P, Ma J, Zhang R, et al: Preclinical
characterization of amubarvimab and romlusevimab, a pair of
non-competing neutralizing monoclonal antibody cocktail, against
SARS-CoV-2. Front Immunol. 13:9804352022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Evering TH, Chew KW, Giganti MJ, Moser C,
Pinilla M, Wohl DA, Currier JS, Eron JJ, Javan AC, Bender Ignacio
R, et al: Safety and efficacy of combination SARS-CoV-2
neutralizing monoclonal antibodies amubarvimab plus romlusevimab in
nonhospitalized patients with COVID-19. Ann Intern Med.
176:658–666. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kim C, Ryu DK, Lee J, Kim YI, Seo JM, Kim
YG, Jeong JH, Kim M, Kim JI, Kim P, et al: A therapeutic
neutralizing antibody targeting receptor binding domain of
SARS-CoV-2 spike protein. Nat Commun. 12:2882021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang YT, Allen RD, Kim K, Shafee N,
Gonzalez AJ, Nguyen MN, Valentine KM, Cao X, Lu L, Pai CI, et al:
SARS-CoV-2 monoclonal antibodies with therapeutic potential: Broad
neutralizing activity and No evidence of antibody-dependent
enhancement. Antiviral Res. 195:1051852021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian D, Sun Y, Xu H and Ye Q: The
emergence and epidemic characteristics of the highly mutated
SARS-CoV-2 Omicron variant. J Med Virol. 94:2376–2383. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Guo H, Gao Y, Li T, Li T, Lu Y, Zheng L,
Liu Y, Yang T, Luo F, Song S, et al: Structures of Omicron spike
complexes and implications for neutralizing antibody development.
Cell Rep. 39:1107702022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Muyldermans S: Applications of Nanobodies.
Annu Rev Anim Biosci. 9:401–421. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu J, Xu K, Jung S, Conte A, Lieberman J,
Muecksch F, Lorenzi JCC, Park S, Schmidt F, Wang Z, et al:
Nanobodies from camelid mice and llamas neutralize SARS-CoV-2
variants. Nature. 595:278–282. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Weiss SR and Navas-Martin S: Coronavirus
pathogenesis and the emerging pathogen severe acute respiratory
syndrome coronavirus. Microbiol Mol Biol Rev. 69:635–664. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song
ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al: A new coronavirus
associated with human respiratory disease in China. Nature.
579:265–269. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang H and Rao Z: Structural biology of
SARS-CoV-2 and implications for therapeutic development. Nat Rev
Microbiol. 19:685–700. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen Y, Liu Q and Guo D: Emerging
coronaviruses: Genome structure, replication, and pathogenesis. J
Med Virol. 92:418–423. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kim D, Lee JY, Yang JS, Kim JW, Kim VN and
Chang H: The Architecture of SARS-CoV-2 Transcriptome. Cell.
181:914–921.e10. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hoffmann M, Kleine-Weber H, Schroeder S,
Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu NH,
Nitsche A, et al: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2
and is blocked by a clinically proven protease inhibitor. Cell.
181:271–280.e8. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Huang Y, Yang C, Xu XF, Xu W and Liu SW:
Structural and functional properties of SARS-CoV-2 spike protein:
potential antivirus drug development for COVID-19. Acta Pharmacol
Sin. 41:1141–1149. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Walls AC, Park YJ, Tortorici MA, Wall A,
McGuire AT and Veesler D: Structure, Function, and Antigenicity of
the SARS-CoV-2 Spike Glycoprotein. Cell. 181:281–292.e6. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Q, Zhang Y, Wu L, Niu S, Song C,
Zhang Z, Lu G, Qiao C, Hu Y, Yuen KY, et al: Structural and
Functional Basis of SARS-CoV-2 Entry by Using Human ACE2. Cell.
181:894–904.e9. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cai Y, Zhang J, Xiao T, Peng H, Sterling
SM, Walsh RM Jr, Rawson S, Rits-Volloch S and Chen B: Distinct
conformational states of SARS-CoV-2 spike protein. Science.
369:1586–1592. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Song W, Gui M, Wang X and Xiang Y: Cryo-EM
structure of the SARS coronavirus spike glycoprotein in complex
with its host cell receptor ACE2. PLoS Pathog. 14:e10072362018.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chi X, Yan R, Zhang J, Zhang G, Zhang Y,
Hao M, Zhang Z, Fan P, Dong Y, Yang Y, et al: A neutralizing human
antibody binds to the N-terminal domain of the Spike protein of
SARS-CoV-2. Science. 369:650–655. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu L, Wang P, Nair MS, Yu J, Rapp M, Wang
Q, Luo Y, Chan JF, Sahi V, Figueroa A, et al: Potent neutralizing
antibodies against multiple epitopes on SARS-CoV-2 spike. Nature.
584:450–456. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C,
Zhang J, Weng T, Zhang Z, Wu Z, et al: Molecular Architecture of
the SARS-CoV-2 Virus. Cell. 183:730–738.e13. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fantini J, Di Scala C, Chahinian H and
Yahi N: Structural and molecular modelling studies reveal a new
mechanism of action of chloroquine and hydroxychloroquine against
SARS-CoV-2 infection. Int J Antimicrob Agents. 55:1059602020.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Fantini J, Chahinian H and Yahi N:
Synergistic antiviral effect of hydroxychloroquine and azithromycin
in combination against SARS-CoV-2: What molecular dynamics studies
of virus-host interactions reveal. Int J Antimicrob Agents.
56:1060202020. View Article : Google Scholar
|
|
53
|
Seyran M, Takayama K, Uversky VN, Adadi P,
Mohamed Abd El-Aziz T, Soares AG, Kandimalla R, Tambuwala M, Hassan
SS, Azad GK, et al: The structural basis of accelerated host cell
entry by SARS-CoV-2†. FEBS J. 288:5010–5020. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S,
Zhang Q, Shi X, Wang Q, Zhang L and Wang X: Structure of the
SARS-CoV-2 spike receptor-binding domain bound to the ACE2
receptor. Nature. 581:215–220. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li F, Li W, Farzan M and Harrison SC:
Structure of SARS coronavirus spike receptor-binding domain
complexed with receptor. Science. 309:1864–1868. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang J, Cai Y, Xiao T, Lu J, Peng H,
Sterling SM, Walsh RM Jr, Rits-Volloch S, Zhu H, Woosley AN, et al:
Structural impact on SARS-CoV-2 spike protein by D614G
substitution. Science. 372:525–530. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jackson CB, Farzan M, Chen B and Choe H:
Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol.
23:3–20. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Benton DJ, Wrobel AG, Xu P, Roustan C,
Martin SR, Rosenthal PB, Skehel JJ and Gamblin SJ: Receptor binding
and priming of the spike protein of SARS-CoV-2 for membrane fusion.
Nature. 588:327–330. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bayati A, Kumar R, Francis V and McPherson
PS: SARS-CoV-2 infects cells after viral entry via
clathrin-mediated endocytosis. J Biol Chem. 296:1003062021.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jaimes JA, Millet JK and Whittaker GR:
Proteolytic Cleavage of the SARS-CoV-2 Spike protein and the role
of the novel S1/S2 Site. iScienc. 23:1012122020. View Article : Google Scholar
|
|
61
|
Newcombe C and Newcombe AR: Antibody
production: Polyclonal-derived biotherapeutics. J Chromatogr B
Analyt Technol Biomed Life Sci. 848:2–7. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ascoli CA and Aggeler B: Overlooked
benefits of using polyclonal antibodies. Biotechniques. 65:127–136.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Leenaars M and Hendriksen CF: Critical
steps in the production of polyclonal and monoclonal antibodies:
Evaluation and recommendations. ILAR J. 46:269–279. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Zylberman V, Sanguineti S, Pontoriero AV,
Higa SV, Cerutti ML, Morrone Seijo SM, Pardo R, Muñoz L, Acuña
Intrieri ME, Alzogaray VA, et al: Development of a hyperimmune
equine serum therapy for COVID-19 in Argentina. Medicina (B Aires).
80 (Suppl 3):S1–S6. 2020.
|
|
65
|
Lopardo G, Belloso WH, Nannini E, Colonna
M, Sanguineti S, Zylberman V, Muñoz L, Dobarro M, Lebersztein G,
Farina J, et al: RBD-specific polyclonal F(ab´)2 fragments of
equine antibodies in patients with moderate to severe COVID-19
disease: A randomized, multicenter, double-blind,
placebo-controlled, adaptive phase 2/3 clinical trial.
EClinicalMedicine. 34:1008432021. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Vanhove B, Duvaux O, Rousse J, Royer PJ,
Evanno G, Ciron C, Lheriteau E, Vacher L, Gervois N, Oger R, et al:
High neutralizing potency of swine glyco-humanized polyclonal
antibodies against SARS-CoV-2. Eur J Immunol. 51:1412–1422. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Gaborit B, Dailly E, Vanhove B, Josien R,
Lacombe K, Dubee V, Ferre V, Brouard S, Ader F, Vibet MA, et al:
Pharmacokinetics and Safety of XAV-19, a Swine Glyco-humanized
Polyclonal Anti-SARS-CoV-2 Antibody, for COVID-19-Related Moderate
Pneumonia: A Randomized, Double-Blind, Placebo-Controlled, Phase
IIa Study. Antimicrob Agents Chemother. 65:e01237212021. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Vanhove B, Marot S, So RT, Gaborit B,
Evanno G, Malet I, Lafrogne G, Mevel E, Ciron C, Royer PJ, et al:
XAV-19, a swine glyco-humanized polyclonal antibody against
SARS-CoV-2 spike receptor-binding domain, targets multiple epitopes
and broadly neutralizes variants. Front Immunol. 12:7612502021.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Singh R, Chandley P and Rohatgi S: Recent
advances in the development of monoclonal antibodies and
next-generation antibodies. Immunohorizons. 7:886–897. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Safdari Y, Farajnia S, Asgharzadeh M and
Khalili M: Antibody humanization methods-a review and update.
Biotechnol Genet Eng Rev. 29:175–186. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yu H, Borsotti C, Schickel JN, Zhu S,
Strowig T, Eynon EE, Frleta D, Gurer C, Murphy AJ, Yancopoulos GD,
et al: A novel humanized mouse model with significant improvement
of class-switched, antigen-specific antibody production. Blood.
129:959–969. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Pedrioli A and Oxenius A: Single B cell
technologies for monoclonal antibody discovery. Trends Immunol.
42:1143–1158. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Winter G and Milstein C: Man-made
antibodies. Nature. 349:293–299. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
McCafferty J, Griffiths AD, Winter G and
Chiswell DJ: Phage antibodies: Filamentous phage displaying
antibody variable domains. Nature. 348:552–554. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen F, Liu Z, Kang W, Jiang F, Yang X,
Yin F, Zhou Z and Li Z: Single-domain antibodies against SARS-CoV-2
RBD from a two-stage phage screening of universal and focused
synthetic libraries. BMC Infect Dis. 24:1992024. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Barnes CO, Jette CA, Abernathy ME, Dam KA,
Esswein SR, Gristick HB, Malyutin AG, Sharaf NG, Huey-Tubman KE,
Lee YE, et al: SARS-CoV-2 neutralizing antibody structures inform
therapeutic strategies. Nature. 588:682–687. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Piccoli L, Park YJ, Tortorici MA,
Czudnochowski N, Walls AC, Beltramello M, Silacci-Fregni C, Pinto
D, Rosen LE, Bowen JE, et al: Mapping neutralizing and
immunodominant sites on the SARS-CoV-2 spike receptor-binding
domain by structure-guided high-resolution serology. Cell.
183:1024–1042.e21. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Röltgen K, Powell AE, Wirz OF, Stevens BA,
Hogan CA, Najeeb J, Hunter M, Wang H, Sahoo MK, Huang C, et al:
Defining the features and duration of antibody responses to
SARS-CoV-2 infection associated with disease severity and outcome.
Sci Immunol. 5:eabe02402020. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Barnes CO, West AP Jr, Huey-Tubman KE,
Hoffmann MAG, Sharaf NG, Hoffman PR, Koranda N, Gristick HB,
Gaebler C, Muecksch F, et al: Structures of Human Antibodies Bound
to SARS-CoV-2 spike reveal common epitopes and recurrent features
of antibodies. Cell. 182:828–842.e16. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Wu Y, Wang F, Shen C, Peng W, Li D, Zhao
C, Li Z, Li S, Bi Y, Yang Y, et al: A noncompeting pair of human
neutralizing antibodies block COVID-19 virus binding to its
receptor ACE2. Science. 368:1274–1278. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Shi R, Shan C, Duan X, Chen Z, Liu P, Song
J, Song T, Bi X, Han C, Wu L, et al: A human neutralizing antibody
targets the receptor-binding site of SARS-CoV-2. Nature.
584:120–124. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Banach BB, Cerutti G, Fahad AS, Shen CH,
Oliveira De Souza M, Katsamba PS, Tsybovsky Y, Wang P, Nair MS,
Huang Y, et al: Paired heavy- and light-chain signatures contribute
to potent SARS-CoV-2 neutralization in public antibody responses.
Cell Rep. 37:1097712021. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Starr TN, Czudnochowski N, Liu Z, Zatta F,
Park YJ, Addetia A, Pinto D, Beltramello M, Hernandez P, Greaney
AJ, et al: SARS-CoV-2 RBD antibodies that maximize breadth and
resistance to escape. Nature. 597:97–102. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Cameroni E, Bowen JE, Rosen LE, Saliba C,
Zepeda SK, Culap K, Pinto D, VanBlargan LA, De Marco A, di Iulio J,
et al: Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron
antigenic shift. Nature. 602:664–670. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Brouwer PJM, Caniels TG, van der Straten
K, Snitselaar JL, Aldon Y, Bangaru S, Torres JL, Okba NMA,
Claireaux M, Kerster G, et al: Potent neutralizing antibodies from
COVID-19 patients define multiple targets of vulnerability.
Science. 369:643–650. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Kim SI, Noh J, Kim S, Choi Y, Yoo DK, Lee
Y, Lee H, Jung J, Kang CK, Song KH, et al: Stereotypic neutralizing
VH antibodies against SARS-CoV-2 spike protein receptor binding
domain in patients with COVID-19 and healthy individuals. Sci
Transl Med. 13:eabd69902021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Greaney AJ, Starr TN, Barnes CO, Weisblum
Y, Schmidt F, Caskey M, Gaebler C, Cho A, Agudelo M, Finkin S, et
al: Mapping mutations to the SARS-CoV-2 RBD that escape binding by
different classes of antibodies. Nat Commun. 12:41962021.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Li D, Edwards RJ, Manne K, Martinez DR,
Schäfer A, Alam SM, Wiehe K, Lu X, Parks R, Sutherland LL, et al:
In vitro and in vivo functions of SARS-CoV-2 infection-enhancing
and neutralizing antibodies. Cell. 184:4203–4219.e32. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tian X, Li C, Huang A, Xia S, Lu S, Shi Z,
Lu L, Jiang S, Yang Z, Wu Y and Ying T: Potent binding of 2019
novel coronavirus spike protein by a SARS coronavirus-specific
human monoclonal antibody. Emerg Microbes Infect. 9:382–385. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
ter Meulen J, van den Brink EN, Poon LL,
Marissen WE, Leung CS, Cox F, Cheung CY, Bakker AQ, Bogaards JA,
van Deventer E, et al: Human monoclonal antibody combination
against SARS coronavirus: synergy and coverage of escape mutants.
PLoS Med. 3:e2372006. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yuan M, Wu NC, Zhu X, Lee CD, So RTY, Lv
H, Mok CKP and Wilson IA: A highly conserved cryptic epitope in the
receptor binding domains of SARS-CoV-2 and SARS-CoV. Science.
368:630–633. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Gupta A, Gonzalez-Rojas Y, Juarez E,
Crespo Casal M, Moya J, Falci DR, Sarkis E, Solis J, Zheng H, Scott
N, et al: Early Treatment for Covid-19 with SARS-CoV-2 Neutralizing
Antibody Sotrovimab. N Engl J Med. 385:1941–1950. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Rockett R, Basile K, Maddocks S, Fong W,
Agius JE, Johnson-Mackinnon J, Arnott A, Chandra S, Gall M, Draper
J, et al: Resistance Mutations in SARS-CoV-2 delta variant after
sotrovimab use. N Engl J Med. 386:1477–1479. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Martinez DR, Schaefer A, Gobeil S, Li D,
De la Cruz G, Parks R, Lu X, Barr M, Manne K, Mansouri K, et al: A
broadly neutralizing antibody protects against SARS-CoV,
pre-emergent bat CoVs, and SARS-CoV-2 variants in mice. bioRxiv
(Preprint). doi: 10.1101/2021.04.27.441655.
|
|
95
|
Wec AZ, Wrapp D, Herbert AS, Maurer DP,
Haslwanter D, Sakharkar M, Jangra RK, Dieterle ME, Lilov A, Huang
D, et al: Broad neutralization of SARS-related viruses by human
monoclonal antibodies. Science. 369:731–736. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Rappazzo CG, Tse LV, Kaku CI, Wrapp D,
Sakharkar M, Huang D, Deveau LM, Yockachonis TJ, Herbert AS,
Battles MB, et al: Broad and potent activity against SARS-like
viruses by an engineered human monoclonal antibody. Science.
371:823–829. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Li D, Sempowski GD, Saunders KO, Acharya P
and Haynes BF: SARS-CoV-2 Neutralizing Antibodies for COVID-19
Prevention and Treatment. Annu Rev Med. 73:1–16. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hastie KM, Li H, Bedinger D, Schendel SL,
Dennison SM, Li K, Rayaprolu V, Yu X, Mann C, Zandonatti M, et al:
Defining variant-resistant epitopes targeted by SARS-CoV-2
antibodies: A global consortium study. Science. 374:472–478. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
99
|
McCallum M, De Marco A, Lempp FA,
Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z,
Zatta F, et al: N-terminal domain antigenic mapping reveals a site
of vulnerability for SARS-CoV-2. Cell. 184:2332–2347.e16. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Chi XY, Yan RH, Zhang J, Zhang G, Zhang Y,
Hao M, Zhang Z, Fan P, Dong Y, Yang Y, et al: A neutralizing human
antibody binds to the N-terminal domain of the Spike protein of
SARS-CoV-2. Science. 369:650–655. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wrapp D, De Vlieger D, Corbett KS, Torres
GM, Wang N, Van Breedam W, Roose K, van Schie L; VIB-CMB COVID-19
Response Team; Hoffmann M, ; et al: Structural basis for potent
neutralization of betacoronaviruses by single-domain camelid
antibodies. Cell. 181:1436–1441. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Hoffmann M, Kleine-Weber H and Pöhlmann S:
A Multibasic Cleavage Site in the Spike Protein of SARS-CoV-2 is
essential for infection of human lung cells. Mol Cell.
78:779–784.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Cerutti G, Guo Y, Zhou T, Gorman J, Lee M,
Rapp M, Reddem ER, Yu J, Bahna F, Bimela J, et al: Potent
SARS-CoV-2 neutralizing antibodies directed against spike
N-terminal domain target a single supersite. Cell Host Microbe.
29:819–833.e7. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Nielsen SCA, Yang F, Jackson KJL, Hoh RA,
Röltgen K, Jean GH, Stevens BA, Lee JY, Rustagi A, Rogers AJ, et
al: Human B Cell Clonal Expansion and Convergent Antibody Responses
to SARS-CoV-2. Cell Host Microbe. 28:516–525.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Boyd SD, Gaëta BA, Jackson KJ, Fire AZ,
Marshall EL, Merker JD, Maniar JM, Zhang LN, Sahaf B, Jones CD, et
al: Individual variation in the germline Ig gene repertoire
inferred from variable region gene rearrangements. J Immunol.
184:6986–6992. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wang N, Sun Y, Feng R, Wang Y, Guo Y,
Zhang L, Deng YQ, Wang L, Cui Z, Cao L, et al: Structure-based
development of human antibody cocktails against SARS-CoV-2. Cell
Res. 31:101–103. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Haslwanter D, Dieterle ME, Wec AZ, O'Brien
CM, Sakharkar M, Florez C, Tong K, Rappazzo CG, Lasso G, Vergnolle
O, et al: A Combination of Receptor-Binding Domain and N-Terminal
Domain Neutralizing Antibodies Limits the Generation of SARS-CoV-2
Spike Neutralization-Escape Mutants. mBio. 12:e02473212021.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Nguyen-Contant P, Embong AK, Kanagaiah P,
Chaves FA, Yang H, Branche AR, Topham DJ and Sangster MY: S
Protein-Reactive IgG and Memory B Cell Production after Human
SARS-CoV-2 Infection Includes Broad Reactivity to the S2 Subunit.
mBio. 11:e01991–20. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Guo L, Wang Y, Kang L, Hu Y, Wang L, Zhong
J, Chen H, Ren L, Gu X, Wang G, et al: Cross-reactive antibody
against human coronavirus OC43 spike protein correlates with
disease severity in COVID-19 patients: A retrospective study. Emerg
Microbes Infect. 10:664–676. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Zohar T, Loos C, Fischinger S, Atyeo C,
Wang C, Slein MD, Burke J, Yu J, Feldman J, Hauser BM, et al:
Compromised humoral functional evolution tracks with SARS-CoV-2
Mortality. Cell. 183:1508–1519.e12. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Ma X, Zou F, Yu F, Li R, Yuan Y, Zhang Y,
Zhang X, Deng J, Chen T, Song Z, et al: Nanoparticle vaccines based
on the receptor binding Domain (RBD) and Heptad Repeat (HR) of
SARS-CoV-2 elicit robust protective immune responses. Immunity.
53:1315–1330.e9. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Silva RP, Huang Y, Nguyen AW, Hsieh CL,
Olaluwoye OS, Kaoud TS, Wilen RE, Qerqez AN, Park JG, Khalil AM, et
al: Identification of a conserved S2 epitope present on spike
proteins from all highly pathogenic coronaviruses. Elife.
12:e837102023. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Hsieh CL, Werner AP, Leist SR, Stevens LJ,
Falconer E, Goldsmith JA, Chou CW, Abiona OM, West A, Westendorf K,
et al: Stabilized coronavirus spike stem elicits a broadly
protective antibody. Cell Rep. 37:1099292021. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Hamers-Casterman C, Atarhouch T,
Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N and
Hamers R: Naturally occurring antibodies devoid of light chains.
Nature. 363:446–448. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Tanaka Y, Nishikawa M, Kamisaki K, Hachiya
S, Nakamura M, Kuwazuru T, Tanimura S, Soyano K and Takeda K:
Marine-derived microbes and molecules for drug discovery. Inflamm
Regen. 42:182022. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Muyldermans S: Nanobodies: Natural
single-domain antibodies. Annu Rev Biochem. 82:775–797. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Jovčevska I and Muyldermans S: The
therapeutic potential of nanobodies. BioDrugs. 34:11–26. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Muyldermans S, Baral TN, Retamozzo VC, De
Baetselier P, De Genst E, Kinne J, Leonhardt H, Magez S, Nguyen VK,
Revets H, et al: Camelid immunoglobulins and nanobody technology.
Vet Immunol Immunopathol. 128:178–183. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Zielonka S, Empting M, Grzeschik J,
Könning D, Barelle CJ and Kolmar H: Structural insights and
biomedical potential of IgNAR scaffolds from sharks. MAbs. 7:15–25.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Transue TR, De Genst E, Ghahroudi MA, Wyns
L and Muyldermans S: Camel single-domain antibody inhibits enzyme
by mimicking carbohydrate substrate. Proteins. 32:515–522. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Bachmann MF, Mohsen MO, Zha L, Vogel M and
Speiser DE: SARS-CoV-2 structural features may explain limited
neutralizing-antibody responses. NPJ Vaccines. 6:22021. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Steeland S, Vandenbroucke RE and Libert C:
Nanobodies as therapeutics: Big opportunities for small antibodies.
Drug Discov Today. 21:1076–1113. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Holliger P and Hudson PJ: Engineered
antibody fragments and the rise of single domains. Nat Biotechnol.
23:1126–1136. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Vu KB, Ghahroudi MA, Wyns L and
Muyldermans S: Comparison of llama VH sequences from conventional
and heavy chain antibodies. Mol Immunol. 34:1121–1131. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zhang YF, Sun Y, Hong J and Ho M:
Humanization of the Shark VNAR Single Domain Antibody Using CDR
Grafting. Curr Protoc. 3:e6302023. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Almagro JC and Fransson J: Humanization of
antibodies. Front Biosci. 13:1619–1633. 2008.PubMed/NCBI
|
|
127
|
Wu Y, Li C, Xia S, Tian X, Kong Y, Wang Z,
Gu C, Zhang R, Tu C, Xie Y, et al: Identification of Human
Single-Domain Antibodies against SARS-CoV-2. Cell Host Microbe.
27:891–898.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hassanzadeh-Ghassabeh G, Devoogdt N, De
Pauw P, Vincke C and Muyldermans S: Nanobodies and their potential
applications. Nanomedicine (Lond). 8:1013–1026. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Harmsen MM and De Haard HJ: Properties,
production, and applications of camelid single-domain antibody
fragments. Appl Microbiol Biotechnol. 77:13–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Schoof M, Faust B, Saunders RA, Sangwan S,
Rezelj V, Hoppe N, Boone M, Billesbølle CB, Puchades C, Azumaya CM,
et al: An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by
stabilizing inactive Spike. Science. 370:1473–1479. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Van Heeke G, Allosery K, De Brabandere V,
De Smedt T, Detalle L and de Fougerolles A: Nanobodies®
as inhaled biotherapeutics for lung diseases. Pharmacol Ther.
169:47–56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Xiang Y, Nambulli S, Xiao Z, Liu H, Sang
Z, Duprex WP, Schneidman-Duhovny D, Zhang C and Shi Y: Versatile
and multivalent nanobodies efficiently neutralize SARS-CoV-2.
Science. 370:1479–1484. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Nambulli S, Xiang Y, Tilston-Lunel NL,
Rennick LJ, Sang Z, Klimstra WB, Reed DS, Crossland NA, Shi Y and
Duprex WP: Inhalable Nanobody (PiN-21) prevents and treats
SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. Sci
Adv. 7:eabh03192021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Gai J, Ma L, Li G, Zhu M, Qiao P, Li X,
Zhang H, Zhang Y, Chen Y, Ji W, et al: A potent neutralizing
nanobody against SARS-CoV-2 with inhaled delivery potential.
MedComm (2020). 2:101–113. 2021.PubMed/NCBI
|
|
135
|
Li C, Zhan W, Yang Z, Tu C, Hu G, Zhang X,
Song W, Du S, Zhu Y, Huang K, et al: Broad neutralization of
SARS-CoV-2 variants by an inhalable bispecific single-domain
antibody. Cell. 185:1389–1401.e18. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Ma H, Zhang X, Zeng W, Zhou J, Chi X, Chen
S, Zheng P, Wang M, Wu Y, Zhao D, et al: A bispecific nanobody
dimer broadly neutralizes SARS-CoV-1 & 2 variants of concern
and offers substantial protection against Omicron via low-dose
intranasal administration. Cell Discov. 8:1322022. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Wu X, Wang Y, Cheng L, Ni F, Zhu L, Ma S,
Huang B, Ji M, Hu H, Li Y, et al: Short-Term Instantaneous
Prophylaxis and Efficient Treatment Against SARS-CoV-2 in hACE2
Mice Conferred by an Intranasal Nanobody (Nb22). Front Immunol.
13:8654012022. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Xiang Y, Huang W, Liu H, Sang Z, Nambulli
S, Tubiana J, Williams KL Jr, Duprex WP, Schneidman-Duhovny D,
Wilson IA, et al: Superimmunity by pan-sarbecovirus nanobodies.
Cell Rep. 39:1110042022. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Nagata K, Utsumi D, Asaka MN, Maeda R,
Shirakawa K, Kazuma Y, Nomura R, Horisawa Y, Yanagida Y, Kawai Y,
et al: Intratracheal trimerized nanobody cocktail administration
suppresses weight loss and prolongs survival of SARS-CoV-2 infected
mice. Commun Med (Lond). 2:1522022. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Maeda R, Fujita J, Konishi Y, Kazuma Y,
Yamazaki H, Anzai I, Watanabe T, Yamaguchi K, Kasai K, Nagata K, et
al: A panel of nanobodies recognizing conserved hidden clefts of
all SARS-CoV-2 spike variants including Omicron. Commun Biol.
5:6692022. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Liu H, Wu L, Liu B, Xu K, Lei W, Deng J,
Rong X, Du P, Wang L, Wang D, et al: Two pan-SARS-CoV-2 nanobodies
and their multivalent derivatives effectively prevent Omicron
infections in mice. Cell Rep Med. 4:1009182023. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Bournazos S, Gupta A and Ravetch JV: The
role of IgG Fc receptors in antibody-dependent enhancement. Nat Rev
Immunol. 20:633–643. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Wang TT, Sewatanon J, Memoli MJ, Wrammert
J, Bournazos S, Bhaumik SK, Pinsky BA, Chokephaibulkit K, Onlamoon
N, Pattanapanyasat K, et al: IgG antibodies to dengue enhanced for
FcγRIIIA binding determine disease severity. Science. 355:395–398.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Iwasaki A and Yang Y: The potential danger
of suboptimal antibody responses in COVID-19. Nat Rev Immunol.
20:339–341. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Ubol S and Halstead SB: How innate immune
mechanisms contribute to antibody-enhanced viral infections. Clin
Vaccine Immunol. 17:1829–1835. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Haynes BF, Corey L, Fernandes P, Gilbert
PB, Hotez PJ, Rao S, Santos MR, Schuitemaker H, Watson M and Arvin
A: Prospects for a safe COVID-19 vaccine. Sci Transl Med.
12:eabe09482020. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Yang Y and Xu F: Evolving understanding of
antibody-dependent enhancement (ADE) of SARS-CoV-2. Front Immunol.
13:10082852022. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Wang S, Peng Y, Wang R, Jiao S, Wang M,
Huang W, Shan C, Jiang W, Li Z, Gu C, et al: Characterization of
neutralizing antibody with prophylactic and therapeutic efficacy
against SARS-CoV-2 in rhesus monkeys. Nat Commun. 11:57522020.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Liu Y, Soh WT, Kishikawa JI, Hirose M,
Nakayama EE, Li S, Sasai M, Suzuki T, Tada A, Arakawa A, et al: An
infectivity-enhancing site on the SARS-CoV-2 spike protein targeted
by antibodies. Cell. 184:3452–3466.e18. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Wang Z, Deng T, Zhang Y, Niu W, Nie Q,
Yang S, Liu P, Pei P, Chen L, Li H and Cao B: ACE2 can act as the
secondary receptor in the FcγR-dependent ADE of SARS-CoV-2
infection. iScience. 25:1037202022. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Tkaczyk C, Okayama Y, Woolhiser MR,
Hagaman DD, Gilfillan AM and Metcalfe DD: Activation of human mast
cells through the high affinity IgG receptor. Mol Immunol.
38:1289–1293. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Darrell DO, Gherlone N, Fremont-Smith P,
Tisdall P and Fremont-Smith M: Kawasaki Disease, Multisystem
Inflammatory Syndrome in Children: Antibody-Induced Mast Cell
Activation Hypothesis. J Pediatrics & Pediatr Med. 4:1–7. 2020.
View Article : Google Scholar
|
|
153
|
Ricke DO: Two Different Antibody-Dependent
Enhancement (ADE) Risks for SARS-CoV-2 Antibodies. Front Immunol.
12:6400932021. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Yahi N, Chahinian H and Fantini J:
Infection-enhancing anti-SARS-CoV-2 antibodies recognize both the
original Wuhan/D614G strain and Delta variants. A potential risk
for mass vaccination? J Infect. 83:607–635. 2021.PubMed/NCBI
|
|
155
|
Zhou Y, Liu Z, Li S, Xu W, Zhang Q, Silva
IT, Li C, Wu Y, Jiang Q, Liu Z, et al: Enhancement versus
neutralization by SARS-CoV-2 antibodies from a convalescent donor
associates with distinct epitopes on the RBD. Cell Rep.
34:1086992021. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Hachmann NP, Miller J, Collier AY, Ventura
JD, Yu J, Rowe M, Bondzie EA, Powers O, Surve N, Hall K and Barouch
DH: Neutralization Escape by SARS-CoV-2 Omicron Subvariants
BA.2.12.1, BA.4, and BA.5. N Engl J Med. 387:86–88. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Jiang XL, Zhu KL, Wang XJ, Wang GL, Li YK,
He XJ, Sun WK, Huang PX, Zhang JZ, Gao HX, et al: Omicron BQ.1 and
BQ.1.1 escape neutralisation by omicron subvariant breakthrough
infection. Lancet Infect Dis. 23:28–30. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Cao Y, Jian F, Wang J, Yu Y, Song W,
Yisimayi A, Wang J, An R, Chen X, Zhang N, et al: Imprinted
SARS-CoV-2 humoral immunity induces convergent Omicron RBD
evolution. Nature. 614:521–529. 2023.PubMed/NCBI
|
|
159
|
Röltgen K, Nielsen SCA, Silva O, Younes
SF, Zaslavsky M, Costales C, Yang F, Wirz OF, Solis D, Hoh RA, et
al: Immune imprinting, breadth of variant recognition, and germinal
center response in human SARS-CoV-2 infection and vaccination.
Cell. 185:1025–1040.e14. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Liu L, Iketani S, Guo Y, Chan JF, Wang M,
Liu L, Luo Y, Chu H, Huang Y, Nair MS, et al: Striking antibody
evasion manifested by the Omicron variant of SARS-CoV-2. Nature.
602:676–681. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Yang X, Duan H, Liu X, Zhang X, Pan S,
Zhang F, Gao P, Liu B, Yang J, Chi X and Yang W: Broad sarbecovirus
neutralizing antibodies obtained by computational design and
synthetic library screening. J Virol. 97:e00610232023. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Cao Y, Jian F, Zhang Z, Yisimayi A, Hao X,
Bao L, Yuan F, Yu Y, Du S, Wang J, et al: Rational identification
of potent and broad sarbecovirus-neutralizing antibody cocktails
from SARS convalescents. Cell Rep. 41:1118452022. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Ma H, Zhang X, Zheng P, Dube PH, Zeng W,
Chen S, Cheng Q, Yang Y, Wu Y, Zhou J, et al: Hetero-bivalent
nanobodies provide broad-spectrum protection against SARS-CoV-2
variants of concern including Omicron. Cell Res. 32:831–842. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Mendon N, Ganie RA, Kesarwani S, Dileep D,
Sasi S, Lama P, Chandra A and Sirajuddin M: Nanobody derived using
a peptide epitope from the spike protein receptor-binding motif
inhibits entry of SARS-CoV-2 variants. J Biol Chem. 299:1027322023.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Ettich J, Werner J, Weitz HT, Mueller E,
Schwarzer R, Lang PA, Scheller J and Moll JM: A Hybrid Soluble
gp130/Spike-Nanobody Fusion Protein Simultaneously Blocks
Interleukin-6 trans-Signaling and Cellular Infection with
SARS-CoV-2. J Virol. 96:e01622212022. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Lyu X, Imai S, Yamano T and Hanayama R:
Preventing SARS-CoV-2 Infection Using Anti-spike Nanobody-IFN-β
Conjugated Exosomes. Pharm Res. 40:927–935. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Gruell H, Vanshylla K, Weber T, Barnes CO,
Kreer C and Klein F: Antibody-mediated neutralization of
SARS-CoV-2. Immunity. 55:925–944. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Suryadevara N, Shrihari S, Gilchuk P,
VanBlargan LA, Binshtein E, Zost SJ, Nargi RS, Sutton RE, Winkler
ES, Chen EC, et al: Neutralizing and protective human monoclonal
antibodies recognizing the N-terminal domain of the SARS-CoV-2
spike protein. Cell. 184:2316–2331.e15. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Voss WN, Hou YJ, Johnson NV, Delidakis G,
Kim JE, Javanmardi K, Horton AP, Bartzoka F, Paresi CJ, Tanno Y, et
al: Prevalent, protective, and convergent IgG recognition of
SARS-CoV-2 non-RBD spike epitopes. Science. 372:1108–1112. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Cerutti G, Guo Y, Wang P, Nair MS, Huang
Y, Yu J, Liu L, Katsamba PS, Bahna F, Reddem ER, et al:
Neutralizing antibody 5–7 defines a distinct site of vulnerability
in SARS-CoV-2 spike N-terminal domain. Cell Rep. 37:1099282021.
View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Graham C, Seow J, Huettner I, Khan H,
Kouphou N, Acors S, Winstone H, Pickering S, Galao RP, Dupont L, et
al: Neutralization potency of monoclonal antibodies recognizing
dominant and subdominant epitopes on SARS-CoV-2 Spike is impacted
by the B.1.1.7 variant. Immunity. 54:1276–1289.e6. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Wang Z, Muecksch F, Cho A, Gaebler C,
Hoffmann HH, Ramos V, Zong S, Cipolla M, Johnson B, Schmidt F, et
al: Analysis of memory B cells identifies conserved neutralizing
epitopes on the N-terminal domain of variant SARS-Cov-2 spike
proteins. Immunity. 55:998–1012.e8. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Pinto D, Sauer MM, Czudnochowski N, Low
JS, Tortorici MA, Housley MP, Noack J, Walls AC, Bowen JE, Guarino
B, et al: Broad betacoronavirus neutralization by a stem
helix-specific human antibody. Science. 373:1109–1116. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Zhou P, Yuan M, Song G, Beutler N,
Shaabani N, Huang D, He WT, Zhu X, Callaghan S, Yong P, et al: A
human antibody reveals a conserved site on beta-coronavirus spike
proteins and confers protection against SARS-CoV-2 infection. Sci
Transl Med. 14:eabi92152022. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Shi W, Wang L, Zhou T, Sastry M, Yang ES,
Zhang Y, Chen M, Chen X, Choe M, Creanga A, et al: Vaccine-elicited
murine antibody WS6 neutralizes diverse beta-coronaviruses by
recognizing a helical stem supersite of vulnerability. Structure.
30:1233–1244.e7. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
176
|
Li W, Chen Y, Prévost J, Ullah I, Lu M,
Gong SY, Tauzin A, Gasser R, Vézina D, Anand SP and Goyette G:
Structural basis and mode of action for two broadly neutralizing
antibodies against SARS-CoV-2 emerging variants of concern. Cell
Rep. 38:1102102022. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Dacon C, Tucker C, Peng L, Lee CD, Lin TH,
Yuan M, Cong Y, Wang L, Purser L, Williams JK, et al: Broadly
neutralizing antibodies target the coronavirus fusion peptide.
Science. 377:728–735. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Low JS, Jerak J, Tortorici MA, McCallum M,
Pinto D, Cassotta A, Foglierini M, Mele F, Abdelnabi R, Weynand B,
et al: ACE2-binding exposes the SARS-CoV-2 fusion peptide to
broadly neutralizing coronavirus antibodies. Science. 377:735–742.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Sun X, Yi C, Zhu Y, Ding L, Xia S, Chen X,
Liu M, Gu C, Lu X, Fu Y, et al: Neutralization mechanism of a human
antibody with pan-coronavirus reactivity including SARS-CoV-2. Nat
Microbiol. 7:1063–1074. 2022. View Article : Google Scholar : PubMed/NCBI
|