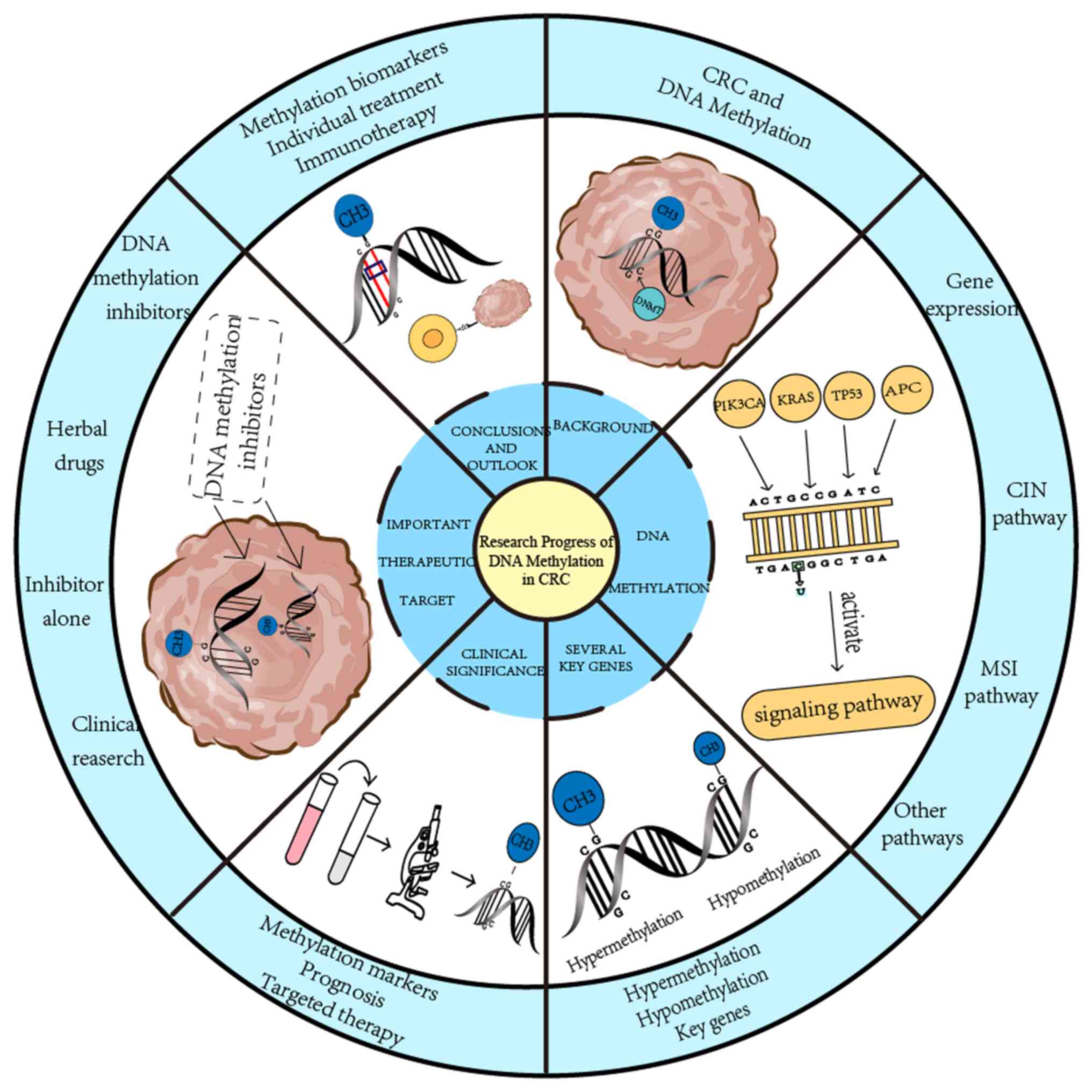
Introduction
Colorectal cancer (CRC) is one of the common
malignant tumors of the digestive tract, and its incidence is
second only to that of gastric and esophageal cancers. CRC ranks
fifth in men and sixth in women as a major cause of tumor-related
deaths in China. In the past 20 years, the morbidity of CRC has
been on the rise, and the onset age has been increasing. In the
western developed countries, CRC is the second most common
malignancy after lung cancer. The incidence rate varies by up to 60
times in different countries. The sites most prone are the rectum
and the junction between the rectum and sigmoid colon, accounting
for 60% of the cancerous region. The incidence of CRC is related to
various factors, such as eating habits, genetics and colitis. The
disease is sporadic in most cases and usually indicates the joint
effect of genes and environment, as only 20–25% of patients
demonstrated a family history. Only 5–6% of CRC cases are
attributed to genetic conditions and are referred to as the CRC
genetic syndrome (1). In recent
years, the morbidity of colon cancer in western developed countries
has demonstrated a declining trend, especially in the 50–74 age
group, which is closely associated with precancer prevention and
treatment (2). One of the reasons
for the high incidence rate of colon cancer in China is the lack of
effective early detection methods. When clinically diagnosed, most
nodules present as advanced colon cancer. CRC is a serious health
risk, ≥1,000,000 new cases are diagnosed every year globally. This
disease occurs as a result of a multi-step process that causes the
accumulation of genetic and epigenetic changes in colonic mucosal
cells, which mainly affect onco-, tumor suppressor, and DNA repair
genes, all of which take part in the key pathways of CRC initiation
and progression (3,4). Among them, p53, the representative
tumor suppressor gene, remains under study, and a number of
scientists are still discussing the therapeutic strategy of
targeting p53 (4). Epigenetic
processes-DNA methylation, histone tail modification and chromatin
remodeling, as well as the mechanisms mediated by noncoding RNA
molecules-are used to describe the mechanisms that can modify the
expression levels of selected genes without necessarily changing
their DNA sequence. Epigenetic modifications are usually
environmentally induced and are tissue-specific phenomena. They can
have similar effects on pathogenic mutations or functional
polymorphisms because they can silence, increase, or reduce the
expression of selected genes in different tissues. This is
particularly relevant to cancer-related genes, such as tumor
suppressor or DNA repair genes (5,6).
With the progress of epigenetics, the association
between DNA methylation and tumors has become a research hotspot. A
series of studies have revealed that there are some specific
abnormal methylations in the process of tumorigenesis, which can be
used as a molecular index for tumor diagnosis. Genome-wide
methylation analysis has now characterized novel genes in patients
with synchronous CRC, which has the clinical potential to improve
the diagnosis and management of patients with CRC (7).
The present study review focused on how DNA
methylation affects key genes, which in turn affect CRC
development, and describes its clinical applications, including the
detection of CRC with methylation biomarkers, the prognostic
analysis of methylated CRC cells, and the relationship between DNA
methylation and targeted CRC therapy. Finally, it was highlighted
that DNA methylation is one of the important therapeutic targets
for CRC, and that its inhibitors provide new ideas for the clinic
therapy of CRC. In summary, compared with other reviews, this
review introduces the relationship between DNA methylation and CRC
more comprehensively, from mechanism and key gene targets to
clinical research, and provides support for the treatment of CRC
with DNA methylation as a breakthrough (Fig. 1).
DNA methylation
Introduction
DNA methylation is one of the different ways of gene
regulation in mammals, and it has been a research hot spot in
epigenetics in recent years. DNA methylation is a chemical
modification of DNA that can regulate genetic expression without
changing the DNA sequence. In a broad sense, DNA methylation refers
to the chemical modification process wherein a methyl group is
added to a specific base of the DNA sequence via covalent bonding,
with S-adenosylmethionine (SAM) as the methyl donor, under the
catalysis of DNA methyltransferase (DNMT). Although there are
numerous variations in methylation modification, the bases of the
modified sites are usually adenine n-6, cytosine n-4, guanine N-7
and cytosine C-5. DNA methylation involved in general research
mainly refers to the methylation process of the fifth carbon atom
of cytosine in cytosine guanine dinucleotide (CpG) dinucleotide,
and the final product of this process is 5-methyl-cytosine.
After fertilization in mammals, entire methylation
of DNA molecules inherited from their parents is cleared by
specific enzymes. After the embryo is implanted into the uterus, a
new round of methylation begins, and the genome of the fertilized
egg is methylated again under the action of DNA methylase. The
stability of DNA methylation is maintained by DNMT1.
In mammals, DNMT1 and DNMT3 are the most essential
DNA methyltransferases. DNMT1 is involved in the maintenance of DNA
methylation and is related to the extension of DNA methylation.
DNMT3 includes DNMT3a and DNMT3b, which are involved in de
novo methylation. Other roles and applications of DNMT family
remain under investigation (8,9).
DNA demethylation is regulated by internal fragments
of genes and their binding factors. There are two hypotheses that
can explain the molecular mechanism underlying DNA demethylation.
One hypothesis associated with DNA semi retention replication is
passive demethylation. If DNA methylation does not occur after
semi-retention replication, the DNA would be in a semi-methylated
state. If the semi-retention replication of semi-methylated DNA
occurs again and the DNA methylation activity is still inhibited,
it indicates that 50% of the cells are in a semi-methylated state.
The second hypothesis is independent of the semi-retention
replication and is an active process, that is to say, DNA
demethylation is catalyzed by DNA demethylase. DNA demethylation is
the removal of methylated bases under the action of DNA
glycosidase, which is equivalent to the repair of damaged DNA by
glycosidases and base free nucleases.
DNA methylation affects gene
expression in tumors
Numerous genes, especially the promoter region of
the housekeeping gene, usually have some regions rich in di-nuclear
glycoside ‘CG’, called ‘CpG islands’. They are usually found in the
promoter and exon regions of genes. Some regions are rich in CpG
dinucleotides, with a length of 300–3,000 base pairs. Methylation
of specific CpG dinucleotides in the promoter region is involved in
transcription regulation. Scientific studies have revealed that DNA
methylation modification demonstrates a stronger inertia in
vitro (10).
Several studies revealed that the methylation status
of tumor cells in the promoter regions of tumor suppressor genes
and repair genes is enhanced, resulting in the inhibition of the
expression of corresponding tumor suppressor gene (11,12).
Moreover, abnormal methylation is common in chronic inflammation
(13), which in turn contributes
directly to tumors and cancers to a large extent.
In 2015, a study on the CRC subtype alliance
summarized four common molecular subtypes (CMS) of CRC: CMS1, CMS2,
CMS3 and CMS4. Among them, CMS1 is highly mutated, and all
microsatellite-unstable CRCs are CMS1 (14). These kinds of tumors have high
methylation status and BRAF mutation rates and a poor prognosis
with the other types. However, the other CMSs demonstrate more
chromosomal instability (CIN) in CRC typing. Although CMS3
demonstrates moderate hypermethylation levels, the KRAS gene is
highly mutated. CMS3 CRC has a high mortality rate once it recurs.
In the present study, the classic chromosome instability (CIN) and
microsatellite instability (MSI) pathways have been used and other
pathways for the pathogenesis of CRC as examples to elucidate the
development process of CRC.
CIN pathway
Studies have revealed that mutations of the tumor
suppressor genes APC regulator of Wnt signaling pathway (APC) and
TP53, and activation mutations of KRAS and PIK3CA are related
events in CIN pathway-induced tumors. Interestingly, the earliest
event in colorectal tumors appears to be mutation inactivation of
APC (15), which causes the Wnt
signaling pathway to be activated, a feature common to almost all
tumors (16).
After the mutation of APC, the mutation of KRAS also
occurs (17): When normal, the
KRAS image molecular switch can control the pathway that regulates
cell proliferation; however, when abnormal, it causes continuous
cell proliferation and prevents self-destruction. The KRAS image
molecular switch participates in intracellular signal transduction.
When the KRAS gene is mutated, it is permanently activated and
cannot produce normal RAS protein, leading to intracellular signal
transduction error, rampant cell proliferation and canceration
(18).
Most sporadic CRC follow the CIN pathway, whereas
85% of sporadic CRC have cyclooxygenase 2 (COX-2) expression
(19). This suggests that COX-2 is
also an important factor in inducing tumors. During the development
of colorectal tumors, COX-2 may convert free arachidonic acid into
prostaglandins and regulate the proliferation of CRC cells; and it
can be used as an alternative approach for the treatment of CRC
(20).
MSI pathway
Regionalized hypermethylation is a common feature of
CRC with MSI phenotype, including CpG island methylation phenotype
(CIMP). In a study by Samowitz et al (21), APC mutations were observed in both
MSI and CIMP high cancers, and there was a significant inverse
correlation trend between APC mutations and CIMP. However, the
mutation frequency of APC and TP53 that was aforementioned is lower
in the MSI pathway than in the CIN pathway (22).
Carcinogenesis of the MSI pathway includes the
mutation of TGF-βR2. The TGF-βR2 gene encodes a protein that
inhibits the proliferation of colonic epithelial cells. The mutated
expression product of the gene no longer sends a signal to prevent
proliferation, which leads to abnormal proliferation of colonic
epithelial cells. Other mutated genes lead to cell cycle arrest
(CASP5 and FAS) and abnormal DNA repair (MBD4, BLM and CHK1)
(23–27).
Other pathways
The serrated pathway is an alternative multistep
mechanism of carcinogenesis. It has been revealed that serrated
colorectal lesions rarely harbor truncated APC mutations, most are
BRAF mutations, and KRAS mutations remain rare. Regarding the
serrated pathway, there was evidence of Fusobacterium
nucleatum in 56% of CRCS, which was associated with CIMP-H
status and large tumors (28).
In addition, IBD-CRC may represent a distinct
tumorigenic pathway. Typical epithelial tumor subtypes associated
with Wnt signaling are completely absent in IBD-CRCs, and different
mechanisms of Wnt pathway dysregulation bias IBD-CRC toward
mesenchymal tumor subtypes (29)
(Table I).
 | Table I.DNA methylation effects gene
expression in tumors. |
Table I.
DNA methylation effects gene
expression in tumors.
| Genes | Roles | Methylation
sites |
|---|
| APC | APC affects cell
growth and division, and also plays an important role in DNA
repair, gene silencing and cell differentiation. | 15 CpG sites within
the APC 1A promoter |
| TP53 | TP53 plays an
important role in cell cycle regulation, DNA damage repair,
apoptosis and anti-aging. | C-terminal,
K305 |
| KRAS | KRAS controls the
pathway that regulates cell growth. | G13D, G12C, G12V,
G12S, G12D |
| MTHFR | MTHFR is involved
in folate metabolism. 5,10-methylenetetrahydrofolate is converted
to 5-methylenetetrahydrofolate with biological functions. It is
essential for maintaining the normal methylation status in the
human body. | C677T, A1298C |
| ADHFE1 | ADHFE1 is
responsible for the oxidation of 4-hydroxybutyrate. Energy
metabolism and detoxification processes may be involved. | 405K |
DNA methylation changes in several key genes
in CRC
DNA methylation refers to the addition of methyl
groups to the bases of DNA molecules by SAM under the action of
DNMT. The covalent bond between cytosine 5 carbon atom and a base
in CpG is the most common mode of modification. DNA methylation
modification demonstrates a stronger inertia in vitro than
in living systems. For example, sodium bisulfite can transform
unmethylated cytosine into uracil, but cannot change the cytosine
in methylated CpG; while in vivo, it leads to a reduction in
gene expression. Therefore, hypermethylation status indicates
inactivation/inhibition/silencing of gene expression (30); however, the hypomethylation state
indicates the activation of gene expression. Early studies revealed
that tumor cells are extensively hypomethylated at the genome-wide
level, resulting in the activation of proto-oncogenes and an
increase in genomic instability (31). It was recently discovered that
tumor cells in the promoter regions of tumor suppressor and repair
genes are hypermethylated, which causes the inhibition of the
expression of corresponding tumor suppressor genes; additionally,
it was revealed that the hypermethylated tumor cells mainly occur
in CpG islands in the promoter region (32), although the corresponding cells are
mostly in the non-methylated state.
Hypermethylation
Hypermethylation of the tumor suppressor gene CpG
island is one of the mechanisms underlying CRC formation. So far,
it has been revealed that numerous cancerous genes experience
hypermethylation, and only a few protective mechanisms, such as
active transcription, active demethylation, replication timing, and
prevention of the acquisition of local chromatin structure of
deoxyribonucleic acid methyltransferase, can prevent
hypermethylation of CpG islands.
Recently, hypermethylation of ADHFE1, CNN1 and NR3C1
has been revealed to play important roles in signal transduction,
cell cycle regulation, angiogenesis, as well as CRC (33–35).
Suzuki et al (36) reported
that the incidence of hypermethylation and downregulation of the
SFRP gene (a negative regulator of Wnt signaling pathway) in normal
colonic mucosae of patients with CRC is higher than in the mucosae
that of patients without CRC.
A meta-analysis revealed a relationship between
ITGA4 promoter methylation status and malignancy, that is, IITGA4
hypermethylation was more frequent in tumor samples than in
non-tumor samples. ITGA4 methylation analysis enables a reliable
method for screening CRC in tissue samples. This has important
implications for the detection of early CRC (37).
Hypermethylation of the promoter may contribute to
the epigenetic silencing of ADAMTS14 in CRC. ADAMTS14 protein
expression was higher in the anterior part of the invasive tumor
than in the tumor center or other regions of the tumor. Its high
expression is associated with poor prognosis in patients with CRC,
suggesting that ADAMTS14 may be a promising indicator for
evaluating the prognosis of CRC (38).
Hypomethylation
Hypomethylation, including CIN and MSI, is
considered to promote tumorigenesis by activating proto-oncogenes,
such as CMYC and HRAS, or causing genomic instability.
There are two common low functional polymorphic
variants of methylenetetrahydrofolate reductase (MTHFR): T variant
at nucleotide 677 (MTHFR C677T) and C variant at nucleotide 1298
(MTHFR A1298C). The first variant, C677T, is related to a reduced
risk of CRC, and the cancer risk associated with MTHFR polymorphism
may be regulated by folate intake. With sufficient folate intake,
individuals carrying the variant MTHFR genotype may have a reduced
risk of cancer. When folate intake is low, DNA methylation and DNA
synthesis/repair in polymorphic individuals may be impaired,
resulting in an increased risk of cancer. In animal studies, folate
deficiency has been revealed to lead to exon-specific
hypomethylation of p53 gene and increased DNA methyltransferase
activity (39). However, moderate
folate deficiency did not cause DNA methylation (40). It was discovered that MTHFR C677T
polymorphism affected DNA methylation status through interaction
with folate status. Another study examined the relationship between
plasma folate status and colorectal adenoma, by assessing the
effect of modification on the polymorphism of its gene (C677T).
Compared with subjects with normal CC folate metabolism or
diminished CT folate metabolism, subjects with poor TT folate
metabolism had a low risk of developing colorectal adenoma at
higher plasma folate levels (adjusted odds ratio, 0.58; 95%
confidence interval, 0.21 to 1.61) and increased risk when their
folate levels were low (adjusted odds ratio, 2.13; 95% confidence
interval, 0.82 to 5.54) (41).
This indicates that the risk of CRC is reduced when folate intake
is low or metabolism is abnormal, that is, when the degree of DNA
methylation is low. This also provides a breakthrough for the
clinical treatment of CRC by controlling folate intake or using DNA
methylation inhibitors.
The present study revealed upregulation and
hypomethylation of HER3 gene expression in CRC cases. High
expression and hypomethylation of HER3 may play an important role
in the occurrence and development of CRC. CpG hypomethylation may
be associated with the early stages of tumorigenesis. The discovery
of this biomarker provides a powerful approach to improve current
diagnostic and therapeutic measures (42).
Key genes
KRAS is a mouse sarcoma virus oncogene. There are
three genes in the RAS gene family associated with human tumors:
HRAS, KRAS and NRAS, located on chromosomes 11, 12, and 1,
respectively. KRAS encodes a 21-kDa RAS protein, also known as the
p21 gene, and has the greatest impact on human cancer. It is like a
molecular switch: When normal, it can control the pathways
regulating cell proliferation, and when abnormalities occur, it
leads to continuous cell proliferation and prevents cell
self-destruction. It is involved in intracellular signal
transmission. When KRAS is mutated, the gene is permanently
activated and cannot produce normal RAS protein, leading to an
intracellular signal transduction disorder, that is, the inability
to control cell proliferation and thus, cancer (43).
CRC is characterized by a series of mutation events
involving APC, KRAS and TP53. KRAS is the most significant
oncogenic mutation in CRC, occurring in 30–40% of patients with CRC
(44). KRAS cooperates with the
Wnt/β-catenin pathway to promote CRC and confers resistance to
anti-EGFR antibodies (45).
The SLC25A22 process is required for the survival of
CRC cells expressing activated KRAS, which are then rapidly
incorporated into the tricarboxylic cycle (glutamine hydrolysis).
In the absence of glutamine, cells can proliferate simply via
incubation with succinate. The cells with mutated KRAS have a lower
ratio of α-ketoglutarate to succinic acid, leading to reduced
hypermethylation of 5-hydroxymethyl cytosine (a marker of DNA
demethylation) and CpG sites. Numerous hypermethylated genes are
located in the pro-cadherin gene cluster in the Wnt signaling
pathway and chromosome 5q31. In CRC cells without KRAS mutations or
with KRAS mutations and SLC25A22 knockout, the pro-cadherin gene
expressed is not methylated at these sites (46).
In summary, in CRC cells expressing activated KRAS,
SLC25A22 promotes the accumulation of succinic acid, resulting in
increased DNA methylation, activation of Wnt signaling for
increased expression of β-catenin, LGR5, proliferation, stem cell
properties and resistance to 5-fluorouracil. Strategies that block
this pathway may be used to treat CRC in the future (Fig. 2).
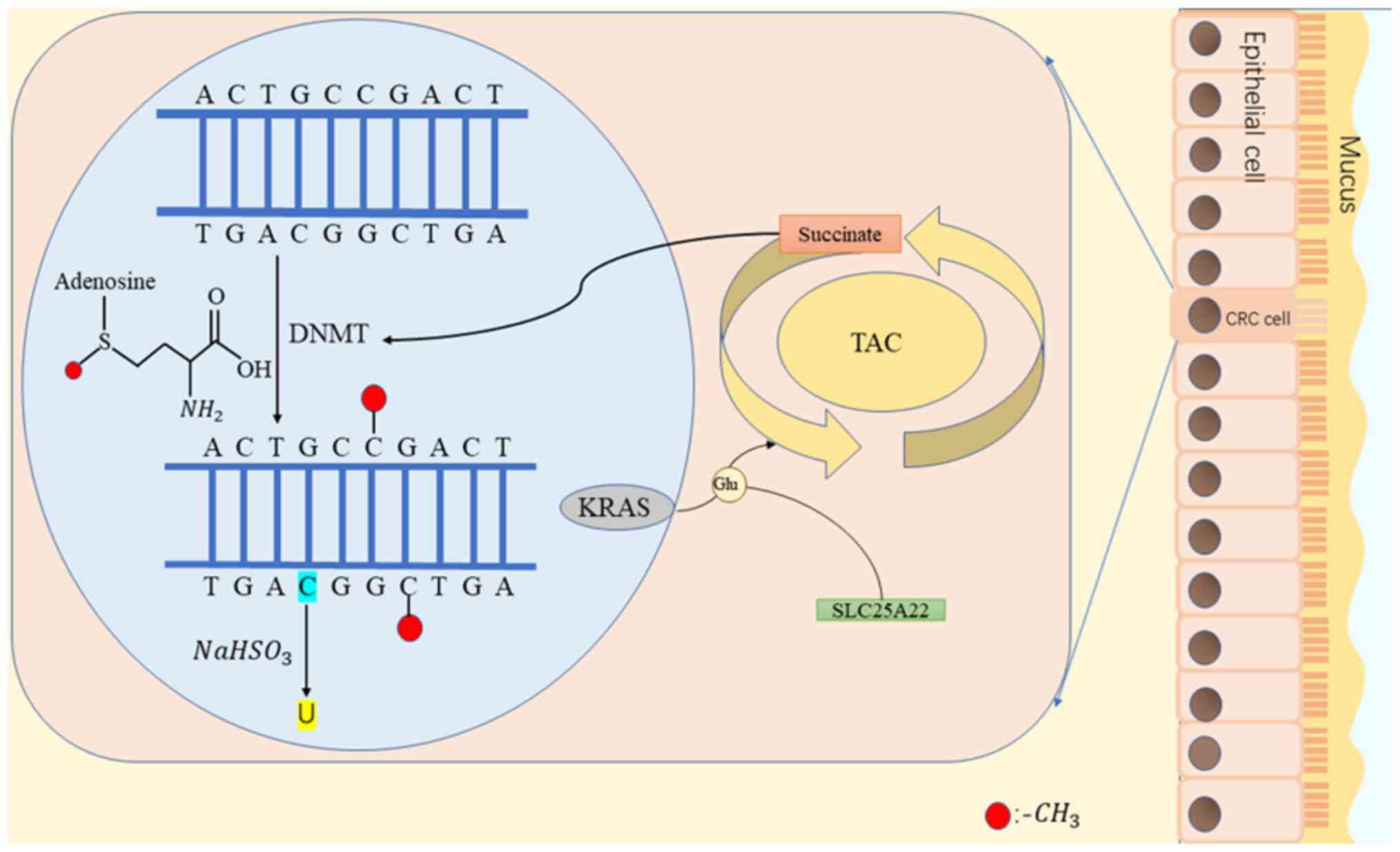 | Figure 2.DNA methylation means that under the
action of DNMT, S-adenosylmethionine adds methyl groups to the
bases of DNA molecules. For example, sodium bisulfite can convert
unmethylated cytosine into uracil, but cannot change the cytosine
in methylated CpG (30). KRAS is
the most important carcinogenic mutation in CRC. In CRC cells
expressing activated KRAS, these KRAS need SLC25A22 to participate
and be rapidly incorporated into the TAC. At the same time,
SLC25A22 promotes the accumulation of succinic acid, resulting in
increased DNA methylation. In CRC cells without KRAS
mutation or KRAS mutation and SLC25A22 knockout, the
expressed pre-adhesin gene was not methylated at these sites. DNMT,
DNA methyltransferase; CRC, colorectal cancer; SLC25A22, solute
carrier family 25 member 22; TAC, tricarboxylic acid cycle; Glu,
glutamine. |
Clinical significance of DNA methylation in
CRC
Using methylation markers to diagnose
CRC
DNA methylation is a chemical modification that can
alter genetic properties without altering the DNA sequence. The
incidence of DNA methylation increases with aging, indicating that
methylation may be related to aging and carcinogenesis (47). In the precursors of CRC (colorectal
polyps) studies, similar abnormal methylation accumulation has been
discovered in DNA sequences of patients, indicating that
methylation biomarker detection in CRC could be a major trend in
the future. Furthermore, it is considered that it is also possible
to trace the origin of tumors through specific methylation of
different types of tumors, which is helpful in determining the
medication and other treatment strategies for patients.
Interestingly, similar methylation also occurs in the colonic
mucosae of healthy people, which means that the use of methylation
biomarkers may predict the possibility of colon cancer (48). As aforementioned, it is considered
by the authors that some of these genes, such as BRAF and KRAS, may
provide a key breakthrough in methylation bioassay research.
Furthermore, MSI and CIN are two important types of
CRC (49). MSI cancer is caused by
the mutation of DNA mismatch repair (MMR) gene and the high
methylation of promoter region. MMR is a protein that is produced
by correcting the wrong base pair during the process of DNA
replication. Therefore, detecting MMR status can predict the
incidence of CRC to a certain extent.
In the study by Hinoue et al (50), CRC cases were divided into
CIMP-high, CIMP-low, and non-CIMP types. Among them, cancer-related
genes in the CIMP-high subgroup demonstrated hypermethylation and
the BRAFV600 gene was mutated, while MLHL demonstrated
hypermethylation. The CIMP-low subgroup is rich in KRAS mutation,
which is characterized by DNA hypermethylation of CIMP-h-related
marker subgroup.
Primarily, the study of Jensen et al
(51) emphasized the potential
utility of using the sensitivity and specificity of DNA methylation
markers in blood samples to detect early tumors. Trimethoxy is a
minimally invasive method for detecting early CRC, derived from the
evaluation of three tumor specific DNA methylation markers in the
blood. According to a study (52),
the sensitivity of CRC cells to trimethoxy reached 78%, and the
specificity was close to 100%. This method undoubtedly lights the
way for the early prediction of CRC. Recently, according to Li
et al (52), SEPT9, SDC2
and ALX4 methylation status can cover multiple molecular pathways
of tumor formation, and combined detection will be hopeful to
further improve the sensitivity of CRC detection.
Furthermore, a new combination of plasma DNA
methylation-based biomarkers has now been developed and validated
using clinical samples from multiple medical centers. It is
expected to provide an alternative and cost-effective strategy for
early detection of targeted gastrointestinal cancers (53).
Effect of DNA methylation on
prognosis
Since the high status of MSI has been detected in
numerous types of CRC, some scholars have proposed MSI status as a
major marker of prognostic analysis (54). However, a meta-analysis has found
that MSI-H is not a robust prognostic marker in stage I and stage
IV CRC without immunotherapy (55). Interestingly, Popat et al
(56) proposed MSI status as a
prospective condition for patient management. Consequently, their
research revealed that patients with MSI administered with
fluorouracil exhibited better survival rates; however, explaining
why MSI patients had an improved prognosis and the mechanism of Fu
was a great challenge for later researchers. A previous study also
posited that gene mutations such as those in the tumor suppressor
gene TP53 and KRAS are rare in MSI tumors, and that these gene
mutations could be associated with poor prognoses (57).
SMAD4 is a tumor suppressor and a component of the
transforming growth factor (TGF)-β signaling pathway, which is
associated with cell proliferation, differentiation, migration, and
apoptosis. Methylation has been reported to lead to activation of
the DPC4/SMAD4 gene, which plays an important role in the
development of CRC. The study also pointed out that DPC4/SMAD4 is
an important factor in prognostic analysis (58).
SDS2 methylation can be used as a potential
biomarker to evaluate preoperative and postoperative fecal DNA in
patients with CRC and can be used to determine whether methylated
SDC2 in stool DNA returns to normal after surgical resection of CRC
(59).
Relationship between DNA methylation
and targeted therapy
CRC is characterized by hypermethylation and overall
hypomethylation of gene promoters. However, DNA methylation is a
double-edged sword. It cannot only affect the development of tumor
but is also a favorable treatment site. In the present study,
several well-studied methylation targeted therapy sites were
summarized.
Cyclin-dependent kinase inhibitor
p16INK4a
Methylation of p16INK4a can be detected in ~30% of
patients with CRC (60), and the
mutation of p16INK4a leads to BRAF mutation, which has been proved
to be a direct cause of tumorigenesis in the intestinal tract of
mice (61). Therefore, p16INK4a
can inhibit tumorigenesis, and targeted demethylation could
potentially be a novel treatment method.
RASSF1
Hypermethylation of the RASSF1 promoter can be
detected in 80% of CRCs. Interestingly, it has been revealed that
knockout of the RASSF1 gene in mice increases the risk of CRC,
which suggests that RASSF1 may be a tumor suppressor gene (62).
Cell division cycle 7 (Cdc7)
Studies have identified somatic mutations of Cdc7 in
CRC, and clinical studies are ongoing. These findings emphasize the
potential of Cdc7 in targeted therapy (63).
DNA methylation is an important therapeutic
target for CRC
Preclinical study: Effects of DNA
methylation inhibitors on colorectal tumor cells
At the time of the formation of colon cancer,
hypermethylation occurs at multiple gene sites on the DNA,
resulting in the inactivation of important genes such as tumor
suppressors. Therefore, DNA methyltransferase inhibitors can
inhibit methylation at specific sites, correct wrong methylation
modifications, and directly alter gene expression.
DNA methylation is a physiological process that
regulates gene expression. The formation and maintenance of DNA
methylation are realized under the action of DNMT. DNMT has three
families, namely DNMT1, whose activity is accelerated during DNA
semi-retention replication, and DNMT2 and DNMT3, which include
DNMT3a, DNMT3b and DNMT3l. The human body possesses DNMT1, DNMT3a
and DNMT3b.
DNMT3a and DNMT3b play important roles in catalytic
de novo methylation during embryonic development. It has
also been suggested that DNMT3a and DNMT3b may correct the errors
left by DNMT1, as they are involved in maintaining DNA methylation
patterns. The three DNMTs maintain DNA methylation and exhibit low
expression in normal tissues and high expression in tumor tissues
(64). Gradually, selecting DNMT
as the target has become a new direction for drug research and
development.
Herbal drugs
In recent years, Traditional Chinese Medicine (TCM)
therapy has become increasingly popular. Since ancient times, there
have been TCM compounds used to treat CRC, such as Wumei Wan and
Sini Tang (65,66). Among them, numerous natural
compounds have been revealed to treat CRC by acting on their
methylated genes. Ginseng is one such case. It enhances apoptosis
by regulating apoptosis-related genes in CRC cells and
down-regulates the expression of DNMTs and reduces the global
methylation level in CRC cells (67). Studies that have revealed promising
results indicate the role of vitamins as DNA methylation modifiers,
but studies with improved study designs are necessary (68).
A clinical study has revealed that 0.5–1 g of
resveratrol administered orally daily produces anticancer effects
in the human GI tract while being well tolerated in patients with
cancer. It actually inhibits cancer spread by regulating epigenetic
changes in tumor cells, through DNA methylation (69).
Curcumin from turmeric induces demethylation of
specific CpG sites in CRC cells, but it does not induce global DNA
methylation changes. Curcumin-induced methylation changes occur in
a gene- and cell line-specific manner, and have a direct impact on
the transcription of various genes involved in important biological
processes (70). A number of
experiments have revealed that some natural compounds can treat CRC
through DNA methylation, proving that DNA methylation can be used
as a target to treat CRC.
Inhibitor alone
There are two classes of DNA methyltransferase
inhibitors: Nucleoside and non-nucleoside, wherein the former
includes 5-azacytosine and its derivatives, and the latter includes
polyphenol epigallocatechin-3-gallate and rgl08 (71).
Although a variety of compounds can inhibit DNA
methylation in mammalian cells, such as genistein (GE) and curcumin
(72,73), the only widely tested DNA methylase
inhibitors are ZCyd (5-azacytidine), DZCyd
(5,6-dihydro-5-azacytidine, also known as DHAC) and ZdCyd
(decitabine). These three compounds can inhibit DNA methylation
only when incorporated into DNA. As aforementioned, ZdCyt is a more
effective DNA methylation inhibitor than ZCyd because it binds only
with DNA. Additionally, ZCyd is synthesized as a more stable
analogue of ZCyd (74), which is
at least one order of magnitude weaker than ZdCyt in blocking
methylation in vivo (75).
This is attributed to the limited incorporation of ZdCyt in DNA
because it is inefficiently phosphorylated by cytidine kinase.
Data, however, revealed that CRC can be treated with ZCyd.
ZNF671 has been discovered to be an important cancer
inhibitor in a variety of tumors. High methylation level of ZNF671
gene promoter region was discovered in CRC, which was negatively
correlated with ZN671 expression. It functions as a tumor
suppressor in CRC through inactivation of Notch signaling. This
suggests that ZNF671 can be used as a candidate target for the
treatment of CRC (76).
Clinical research
Changes in DNA methylation may even be an important
mediator in the transformation of metastatic cancer: Dramatic and
reproducible changes in methylation have been described in several
important longitudinal studies. Such studies have been completed
for CRC (77), prostate (78) and breast cancers (79). E-cadherin (CDH1) is widely
considered to be the target of DNA methylation for breast cancer
metastasis (80). It is worth
noting that DNA methylation of the metastasis suppressor gene is
observed in circulating tumor cells from various tumor types
(81). This increases the
possibility that DNA methylation may also be a useful prognostic
marker for metastasis. It is noteworthy that a pan-cancer
meta-analysis identified the methylation site associated with
metastasis of breast cancer and CRC (82). Therefore, the potential of
methylated DNA as a marker for prognosis, diagnosis and metastasis
is an area to be further explored.
Histologically, normal colonic mucosal tissues can
be distinguished from colonic tissue with precancerous lesions at
the molecular level. Therefore, the change in DNA methylation can
be used as a marker of colon cancer. However, the reversibility of
DNA methylation also provides a theoretical basis for the treatment
of CRC from the perspective of epigenetics, and research revealed
that the analyses of the DNA methylation results of specific genes
are expected to provide clinicians with useful information such as
early diagnosis and disease stages. Although these DNA methylation
inhibitors have a potent demethylation effect, they can be
accompanied by strong cytotoxicity and lack of specificity in the
selection of action sites. Therefore, more studies are needed to
solve these problems to facilitate the widespread use of DNA
methylation inhibitors in the clinic (Table II).
 | Table II.DNA methylation is an important
therapeutic target for CRC. |
Table II.
DNA methylation is an important
therapeutic target for CRC.
| A, DNA
methyltransferase inhibitors |
|---|
|
|---|
| Drugs | Mechanisms | Clinical
applications | (Refs.) |
|---|
| ZCyd
(5-azacytidine, 5-AzaC) | It downregulates
the expression of DNMTs in CRC cells. | AML, MDS, CML, CLL,
pancreatic cancer, breast cancer, CRC. It is often used in
combination with other drugs. | (74,75) |
| DZCyd
(5,6-dihydro-5-azacytidine, DHAC) | DZCyt functions as
a transition-state inhibitor by forming a tight, non-covalent
complex with DNA C5-MTases. | Leukemia, breast
cancer, bronchial carcinoma, pancreatic cancer, CRC. | (88) |
| ZdCyd
(decitabine) | It induces CpG
demethylation. | AML, MDS, CML,
pancreatic cancer, breast cancer, CRC. | (74,75) |
| RG108 | It binds to the SAM
binding site on DNMT1 with its indole moiety. | Hearing loss, lung
adenocarcinoma, epilepsy, CRC. | (89,90) |
| Zebularine | It inhibits DNA
methyltransferases. | Diabetes mellitus,
liver cancer, stroke, breast cancer, lung cancer, CRC. | (91) |
|
| B, Herbal
drugs |
|
| Ginseng | It has anti-tumor
ability by down-regulating DNMTs to regulate cell apoptosis and
reversing the methylation status of transcriptionally silenced
genes in CRC. | Diabetes mellitus,
depression, epilepsy, liver cancer, CRC. | (67) |
| Curcumin | It inhibits colon
cancer cell growth by up-regulating DLEC1 and reducing CpG
methylation. | Lung cancer, bowel
cancer, stomach cancer, breast cancer, leukemia, CRC. | (92) |
| EGCG | It directly
inhibits DNMT1. | RA, Alzheimer's
disease, breast cancer, CRC, lung cancer, prostate cancer, oral
cancer. | (93) |
Conclusions and outlook
The research prospect of DNA methylation and CRC is
promising. It is considered by the authors that DNA methylation in
genome sequences can be an important marker to predict the
incidence and molecular typing of CRC and the prognosis of patients
with cancer. Further experiments are required to make improved use
of the DNA methylation spectrum. DNA methylation is expected to be
an important site for CRC screening. High sensitivity is its
biggest advantage, which helps to reduce the misdiagnosis rate.
Therefore, it is important to clarify the methylation markers
corresponding to different types of CRC through further in-depth
research. Using methylation biomarkers to identify cancer cells
should be a trend in the future. Reducing the damage caused by
invasive examination of the body is also a major research
direction.
Studies have revealed that CRC is a highly
heterogeneous disease. For example, MLH1 gene hypermethylation is
only discovered in a small proportion of sporadic CRC cases
(83). This enables the
individualized treatment of patients. However, each patient has
considerable differences in drug sensitivity, effect of treatment
and mortality. However, since epigenetics is reversible, that is,
DNA methylation is reversible, it offers new strategies for CRC
treatment. DNMT inhibitors are expected to become targeted drugs
for the treatment of CRC, which can be used to target patients with
hypermethylated CRC subtypes. For example, azacytidine, decitabine,
temozolomide and other specific drugs can reverse the methylation
state of DNA to achieve the purpose of targeted treatment of CRC;
however, more clinical studies are required to demonstrate the
sensitivity and effectiveness of these drugs on various CRC
subtypes. Furthermore, the side effects of these drugs also have a
significant impact on the prognosis. Clinically, according to the
different symptoms of patients, a more appropriate treatment plan
can be formulated, and symptomatic drugs can be used to improve the
treatment effect. The best possible strategy would be to adapt the
drugs to local conditions, reduce side effects, and improve tumor
prognosis. The identification of metastatic potential signatures is
complicated by the heterogeneity among tumor cells, and there is
still a long way from this.
Immunotherapy is an emerging field in the treatment
of CRC, and DNA methylation is likely to be related to the effect
of immunotherapy. It has been reported that m6 methylation-mediated
intercellular communication in the tumor microenvironment plays an
antitumor immunomodulatory role (84); and ALKBH5, an RNA
N6-methyladenosine eraser, can be used as a target to promote CRC
immunotherapy (85). A recent
study revealed that upregulated THBS2 expression in CRC cells
inhibits antitumor immunity through HIF1A/lactic acid/GPR132
pathway. In addition, THBS2 expression is also related to PFI,
immune cell infiltration and immune regulation (86). Moreover, some mouse experiments
have revealed that the combination of epigenetics and immunotherapy
is more effective than the use of blockers alone, which further
demonstrates the feasibility of methylation immunotherapy (87). Scientists also continue to actively
explore the role of methylation with immune escape and
immunotherapy resistance to find new immunotherapy strategies. This
helps to improve the cure rate of patients.
DNA methylation has a broad application prospect in
the field of CRC research. Through in-depth study of the mechanism
and regulatory network of methylation in CRC, it is expected to
provide new breakthrough points for early diagnosis, individualized
treatment and immunotherapy of CRC, thereby improving the prognosis
and survival rate of patients.
According to the present study, DNA methylation can
be used as a biomarker for early diagnosis of CRC. By detecting the
methylation status of DNA in feces, minimal residual disease can be
detected, which is helpful for early detection of the disease. In
addition, DNA methylation also plays an important role in the
prognosis of CRC. The survival rate and quality of life of the
patients were tested by detecting the methylation status. With the
development of sequencing technology and bioinformatics, the study
of DNA methylation in CRC will be more in-depth. In conclusion, DNA
methylation still has great potential in the study of CRC and
provides more possibilities for the treatment of CRC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Dalian Medical University
Affiliated Second Hospital Subspecialty Fund (grant no.
84671291).
Availability of data and materials
Not applicable.
Authors' contributions
LW and LS conceptualized, wrote, reviewed and edited
the manuscript. YW, CW and RZ prepared and wrote the original
draft. Data authentication is not applicable. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
CMS
|
common molecular subtypes
|
|
CIMP
|
CpG island methylation phenotype
|
|
SAM
|
s-adenosylmethionine
|
|
DNMT
|
DNA methyltransferase
|
|
CIN
|
chromosomal instability
|
|
MSI
|
microsatellite instability
|
|
COX-2
|
cyclooxygenase 2
|
|
TCM
|
Traditional Chinese Medicine
|
References
|
1
|
Migliore L, Migheli F, Spisni R and
Coppedè F: Genetics, cytogenetics, and epigenetics of colorectal
cancer. J Biomed Biotechnol. 2011:7923622011.PubMed/NCBI
|
|
2
|
Araghi M, Soerjomataram I, Bardot A,
Ferlay J, Cabasag CJ, Morrison DS, De P, Tervonen H, Walsh PM,
Bucher O, et al: Changes in colorectal cancer incidence in seven
high-income countries: A population-based study. Lancet
Gastroenterol Hepatol. 4:511–518. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reilly NM, Novara L, Di Nicolantonio F and
Bardelli A: Exploiting DNA repair defects in colorectal cancer. Mol
Oncol. 13:681–700. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liebl MC and Hofmann TG: The role of p53
signaling in colorectal cancer. Cancers (Basel). 13:21252021.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bose S, Saha S, Goswami H, Shanmugam G and
Sarkar K: Involvement of CCCTC-binding factor in epigenetic
regulation of cancer. Mol Biol Rep. 50:10383–10398. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jamai D, Gargouri R, Selmi B and Khabir A:
ERCC1 and MGMT methylation as a predictive marker of relapse and
FOLFOX response in colorectal cancer patients from South Tunisia.
Genes (Basel). 14:14672023. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Okada Y, Peng F, Perea J, Corchete L,
Bujanda L, Li W and Goel A: Genome-wide methylation profiling
identifies a novel gene signature for patients with synchronous
colorectal cancer. Br J Cancer. 128:112–120. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Okano M, Bell DW, Haber DA and Li E: DNA
methyltransferases Dnmt3a and Dnmt3b are essential for de novo
methylation and mammalian development. Cell. 99:247–257. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhai P, Zhang H, Li Q, Yang M, Guo Y and
Xing C: DNMT1-mediated NR3C1 DNA methylation enables transcription
activation of connexin40 and augments angiogenesis during
colorectal cancer progression. Gene. 892:1478872024. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Christman JK, Sheikhnejad G, Dizik M,
Abileah S and Wainfan E: Reversibility of changes in nucleic acid
methylation and gene expression induced in rat liver by severe
dietary methyl deficiency. Carcinogenesis. 14:551–557. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goll MG, Kirpekar F, Maggert KA, Yoder JA,
Hsieh CL, Zhang X, Golic KG, Jacobsen SE and Bestor TH: Methylation
of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science.
311:395–398. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smith ZD and Meissner A: DNA methylation:
Roles in mammalian development. Nat Rev Genet. 14:204–220. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cho HY, Wang X, Campbell MR, Panduri V,
Coviello S, Caballero MT, Bennett BD, Kleeberger SR, Polack FP,
Ofman G and Bell DA: Prospective epigenome and transcriptome
analyses of cord and peripheral blood from preterm infants at risk
of bronchopulmonary dysplasia. Sci Rep. 13:122622023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guinney J, Dienstmann R, Wang X, de
Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda
G, Angelino P, et al: The consensus molecular subtypes of
colorectal cancer. Nat Med. 21:1350–1356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Powell SM, Zilz N, Beazer-Barclay Y, Bryan
TM, Hamilton SR, Thibodeau SN, Vogelstein B and Kinzler KW: APC
mutations occur early during colorectal tumorigenesis. Nature.
359:235–237. 1992. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cancer Genome Atlas Network, .
Comprehensive molecular characterization of human colon and rectal
cancer. Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lin CY, Shen MY, Chen WTL and Yang CA:
Evaluation of the prognostic value of low-frequency KRAS mutation
detection in circulating tumor DNA of patients with metastatic
colorectal cancer. J Pers Med. 13:10512023. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rajagopalan H, Bardelli A, Lengauer C,
Kinzler KW, Vogelstein B and Velculescu VE: Tumorigenesis: RAF/RAS
oncogenes and mismatch-repair status. Nature. 418:9342002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tsujii M, Kawano S and DuBois RN:
Cyclooxygenase-2 expression in human colon cancer cells increases
metastatic potential. Proc Natl Acad Sci USA. 94:3336–3340. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hidalgo-Estévez AM, Stamatakis K,
Jiménez-Martínez M, López-Pérez R and Fresno M: Cyclooxygenase
2-regulated genes an alternative avenue to the development of new
therapeutic drugs for colorectal cancer. Front Pharmacol.
11:5332020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Samowitz WS, Slattery ML, Sweeney C,
Herrick J, Wolff RK and Albertsen H: APC mutations and other
genetic and epigenetic changes in colon cancer. Mol Cancer Res.
5:165–170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pruitt K and Der CJ: Ras and Rho
regulation of the cell cycle and oncogenesis. Cancer Lett.
171:1–10. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tozaki Y, Aoki H, Kato R, Toriuchi K,
Arame S, Inoue Y, Hayashi H, Kubota E, Kataoka H and Aoyama M: The
combination of ATM and Chk1 inhibitors induces synthetic lethality
in colorectal cancer cells. Cancers (Basel). 15:7352023. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lai WL, Lee SC, Chang KF, Huang XF, Li CY,
Lee CJ, Wu CY, Hsu HJ and Tsai NM: Juniperus communis extract
induces cell cycle arrest and apoptosis of colorectal
adenocarcinoma in vitro and in vivo. Braz J Med Biol Res.
54:e108912021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhou Y, Dai W, Wang H, Pan H and Wang Q:
Long non-coding RNA CASP5 promotes the malignant phenotypes of
human glioblastoma multiforme. Biochem Biophys Res Commun.
500:966–972. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gönenc II, Wolff A, Schmidt J, Zibat A,
Müller C, Cyganek L, Argyriou L, Räschle M, Yigit G and Wollnik B:
Single-cell transcription profiles in Bloom syndrome patients link
BLM deficiency with altered condensin complex expression
signatures. Hum Mol Genet. 31:2185–2193. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bader S, Walker M, Hendrich B, Bird A,
Bird C, Hooper M and Wyllie A: Somatic frameshift mutations in the
MBD4 gene of sporadic colon cancers with mismatch repair
deficiency. Oncogene. 18:8044–8047. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
De Palma FDE, D'Argenio V, Pol J, Kroemer
G, Maiuri MC and Salvatore F: The molecular hallmarks of the
serrated pathway in colorectal cancer. Cancers (Basel).
11:10172019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Rajamäki K, Taira A, Katainen R, Välimäki
N, Kuosmanen A, Plaketti RM, Seppälä TT, Ahtiainen M, Wirta EV,
Vartiainen E, et al: Genetic and epigenetic characteristics of
inflammatory bowel disease-associated colorectal cancer.
Gastroenterology. 161:592–607. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pajares MJ, Palanca-Ballester C, Urtasun
R, Alemany-Cosme E, Lahoz A and Sandoval J: Methods for analysis of
specific DNA methylation status. Methods. 187:3–12. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Gaudet F, Hodgson JG, Eden A,
Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H and Jaenisch R:
Induction of tumors in mice by genomic hypomethylation. Science.
300:489–492. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Takeuchi C, Yamashita S, Liu YY, Takeshima
H, Sasaki A, Fukuda M, Hashimoto T, Naka T, Ishizu K, Sekine S, et
al: Precancerous nature of intestinal metaplasia with increased
chance of conversion and accelerated DNA methylation. Gut.
73:255–267. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Oshima M, Murai N, Kargman S, Arguello M,
Luk P, Kwong E, Taketo MM and Evans JF: Chemoprevention of
intestinal polyposis in the Apcdelta716 mouse by rofecoxib, a
specific cyclooxygenase-2 inhibitor. Cancer Res. 61:1733–1740.
2001.PubMed/NCBI
|
|
34
|
Xu X, Nie J, Lu L, Du C, Meng F and Song
D: LINC00337 promotes tumor angiogenesis in colorectal cancer by
recruiting DNMT1, which suppresses the expression of CNN1. Cancer
Gene Ther. 28:1285–1297. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hu YH, Ma S, Zhang XN, Zhang ZY, Zhu HF,
Ji YH, Li J, Qian XL and Wang YX: Hypermethylation of ADHFE1
promotes the proliferation of colorectal cancer cell via modulating
cell cycle progression. Onco Targets Ther. 12:8105–8115. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Suzuki H, Watkins DN, Jair KW, Schuebel
KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van
Engeland M, et al: Epigenetic inactivation of SFRP genes allows
constitutive WNT signaling in colorectal cancer. Nat Genet.
36:417–422. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jafarpour S, Yazdi M, Nedaeinia R,
Vatandoost N, Ferns GA and Salehi R: Status of integrin subunit
alpha 4 promoter DNA methylation in colorectal cancer and other
malignant tumors: A systematic review and meta-analysis. Res Pharm
Sci. 18:231–243. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Y, Zhou J, Zhang J, Cao H, Han F,
Zhang H and Xu E: The expression of ADAMTS14 is regulated by
promoter DNA methylation and is associated with poor prognosis in
colorectal cancer. Exp Cell Res. 410:1129532022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kim YI, Pogribny IP, Basnakian AG, Miller
JW, Selhub J, James SJ and Mason JB: Folate deficiency in rats
induces DNA strand breaks and hypomethylation within the p53 tumor
suppressor gene. Am J Clin Nutr. 65:46–52. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kim YI, Christman JK, Fleet JC, Cravo ML,
Salomon RN, Smith D, Ordovas J, Selhub J and Mason JB: Moderate
folate deficiency does not cause global hypomethylation of hepatic
and colonic DNA or c-myc-specific hypomethylation of colonic DNA in
rats. Am J Clin Nutr. 61:1083–1090. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Marugame T, Tsuji E, Kiyohara C, Eguchi H,
Oda T, Shinchi K and Kono S: Relation of plasma folate and
methylenetetrahydrofolate reductase C677T polymorphism to
colorectal adenomas. Int J Epidemiol. 32:64–66. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Othman R, Mohtarrudin N, Ahmad Zubir NM,
Seow HF, Ngan KW and Osman M: HER3 overexpression and
hypomethylation in colorectal adenocarcinoma. Malays J Pathol.
44:67–74. 2022.PubMed/NCBI
|
|
43
|
Timar J and Kashofer K: Molecular
epidemiology and diagnostics of KRAS mutations in human cancer.
Cancer Metastasis Rev. 39:1029–1038. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Santini D, Loupakis F, Vincenzi B,
Floriani I, Stasi I, Canestrari E, Rulli E, Maltese PE, Andreoni F,
Masi G, et al: High concordance of KRAS status between primary
colorectal tumors and related metastatic sites: Implications for
clinical practice. Oncologist. 13:1270–1275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lièvre A, Bachet JB, Boige V, Cayre A, Le
Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C, et al: KRAS
mutations as an independent prognostic factor in patients with
advanced colorectal cancer treated with cetuximab. J Clin Oncol.
26:374–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wong CC, Xu J, Bian X, Wu JL, Kang W, Qian
Y, Li W, Chen H, Gou H, Liu D, et al: In colorectal cancer cells
with mutant KRAS, SLC25A22-mediated glutaminolysis reduces DNA
demethylation to increase wnt signaling, stemness, and drug
resistance. Gastroenterology. 159:2163–2180.e6. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mangelinck A and Mann C: DNA methylation
and histone variants in aging and cancer. Int Rev Cell Mol Biol.
364:1–110. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Sakai E, Nakajima A and Kaneda A:
Accumulation of aberrant DNA methylation during colorectal cancer
development. World J Gastroenterol. 20:978–987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kasprzak A: Prognostic biomarkers of cell
proliferation in colorectal cancer (CRC): From immunohistochemistry
to molecular biology techniques. Cancers (Basel). 15:45702023.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hinoue T, Weisenberger DJ, Lange CPE, Shen
H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk
CM, et al: Genome-scale analysis of aberrant DNA methylation in
colorectal cancer. Genome Res. 22:271–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jensen SØ, Øgaard N, Ørntoft MW, Rasmussen
MH, Bramsen JB, Kristensen H, Mouritzen P, Madsen MR, Madsen AH,
Sunesen KG, et al: Novel DNA methylation biomarkers show high
sensitivity and specificity for blood-based detection of colorectal
cancer-a clinical biomarker discovery and validation study. Clin
Epigenetics. 11:1582019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li Y, Li B, Jiang R, Liao L, Zheng C, Yuan
J, Zeng L, Hu K, Zhang Y, Mei W, et al: A novel screening method of
DNA methylation biomarkers helps to improve the detection of
colorectal cancer and precancerous lesions. Cancer Med.
12:20626–20638. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Dai Y, Li H, Wu Q, Wang J, Wang K, Fei S,
Pei B, Song L, Chen G, Ma Y, et al: A sensitive and robust
plasma-based DNA methylation panel for early detection of target
gastrointestinal cancers. Neoplasia. 46:1009412023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Benatti P, Gafà R, Barana D, Marino M,
Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L,
Menigatti M, et al: Microsatellite instability and colorectal
cancer prognosis. Clin Cancer Res. 11:8332–8340. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Toh JWT, Phan K, Reza F, Chapuis P and
Spring KJ: Rate of dissemination and prognosis in early and
advanced stage colorectal cancer based on microsatellite
instability status: Systematic review and meta-analysis. Int J
Colorectal Dis. 36:1573–1596. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Popat S, Hubner R and Houlston RS:
Systematic review of microsatellite instability and colorectal
cancer prognosis. J Clin Oncol. 23:609–618. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Klump B, Nehls O, Okech T, Hsieh CJ, Gaco
V, Gittinger FS, Sarbia M, Borchard F, Greschniok A, Gruenagel HH,
et al: Molecular lesions in colorectal cancer: Impact on prognosis?
Original data and review of the literature. Int J Colorectal Dis.
19:23–42. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Koyama M, Ito M, Nagai H, Emi M and
Moriyama Y: Inactivation of both alleles of the DPC4/SMAD4 gene in
advanced colorectal cancers: Identification of seven novel somatic
mutations in tumors from Japanese patients. Mutat Res. 406:71–77.
1999.PubMed/NCBI
|
|
59
|
Song JH, Oh TJ, An S, Lee KH, Kim JY and
Kim JS: Comparative detection of syndecan-2 methylation in
preoperative and postoperative stool DNA in patients with
colorectal cancer. World J Gastrointest Surg. 15:2032–2041. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Shima K, Nosho K, Baba Y, Cantor M,
Meyerhardt JA, Giovannucci EL, Fuchs CS and Ogino S: Prognostic
significance of CDKN2A (p16) promoter methylation and loss of
expression in 902 colorectal cancers: Cohort study and literature
review. Int J Cancer. 128:1080–1094. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Carragher LAS, Snell KR, Giblett SM,
Aldridge VS, Patel B, Cook SJ, Winton DJ, Marais R and Pritchard
CA: V600EBraf induces gastrointestinal crypt senescence and
promotes tumour progression through enhanced CpG methylation of
p16INK4a. EMBO Mol Med. 2:458–471. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
van der Weyden L, Arends MJ, Dovey OM,
Harrison HL, Lefebvre G, Conte N, Gergely FV, Bradley A and Adams
DJ: Loss of rassf1a cooperates with Apc(Min) to accelerate
intestinal tumourigenesis. Oncogene. 27:4503–4508. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Melling N, Muth J, Simon R, Bokemeyer C,
Terracciano L, Sauter G, Izbicki JR and Marx AH: Cdc7
overexpression is an independent prognostic marker and a potential
therapeutic target in colorectal cancer. Diagn Pathol. 10:1252015.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
el-Deiry WS, Nelkin BD, Celano P, Yen RW,
Falco JP, Hamilton SR and Baylin SB: High expression of the DNA
methyltransferase gene characterizes human neoplastic cells and
progression stages of colon cancer. Proc Natl Acad Sci USA.
88:3470–3474. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lu ZH, Ding Y, Wang YJ, Chen C, Yao XR,
Yuan XM, Bu F, Bao H, Dong YW, Zhou Q, et al: Early administration
of Wumei Wan inhibit myeloid-derived suppressor cells via PI3K/Akt
pathway and amino acids metabolism to prevent colitis-associated
colorectal cancer. J Ethnopharmacol. 333:1182602024.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen J, Zheng X, Xu G, Wang B, Hu L, Mao
J, Lu X, Cai Y, Chai K and Chen W: Sini decoction inhibits tumor
progression and enhances the anti-tumor immune response in a murine
model of colon cancer. Comb Chem High Throughput Screen.
26:2517–2526. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Okuno K, Pratama MY, Li J, Tokunaga M,
Wang X, Kinugasa Y and Goel A: Ginseng mediates its anticancer
activity by inhibiting the expression of DNMTs and reactivating
methylation-silenced genes in colorectal cancer. Carcinogenesis.
44:394–403. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Boughanem H, Kompella P, Tinahones FJ and
Macias-Gonzalez M: An overview of vitamins as epidrugs for
colorectal cancer prevention. Nutr Rev. 81:455–479. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Brockmueller A, Sajeev A, Koklesova L,
Samuel SM, Kubatka P, Büsselberg D, Kunnumakkara AB and Shakibaei
M: Resveratrol as sensitizer in colorectal cancer plasticity.
Cancer Metastasis Rev. 43:55–85. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Link A, Balaguer F, Shen Y, Lozano JJ,
Leung HC, Boland CR and Goel A: Curcumin modulates DNA methylation
in colorectal cancer cells. PLoS One. 8:e577092013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Lopez M, Gilbert J, Contreras J, Halby L
and Arimondo PB: Inhibitors of DNA methylation. Adv Exp Med Biol.
1389:471–513. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Fabianowska-Majewska K, Kaufman-Szymczyk
A, Szymanska-Kolba A, Jakubik J, Majewski G and Lubecka K: Curcumin
from turmeric rhizome: A potential modulator of DNA methylation
machinery in breast cancer inhibition. Nutrients. 13:3322021.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Sharma M and Tollefsbol TO: Combinatorial
epigenetic mechanisms of sulforaphane, genistein and sodium
butyrate in breast cancer inhibition. Exp Cell Res. 416:1131602022.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Futterman B, Derr J, Beisler JA, Abbasi MM
and Voytek P: Studies on the cytostatic action, phosphorylation and
deamination of 5-azacytidine and 5,6-dihydro-5-azacytidine in HeLa
cells. Biochem Pharmacol. 27:907–909. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ghanim V, Herrmann H, Heller G, Peter B,
Hadzijusufovic E, Blatt K, Schuch K, Cerny-Reiterer S, Mirkina I,
Karlic H, et al: 5-Azacytidine and decitabine exert proapoptotic
effects on neoplastic mast cells: role of FAS-demethylation and FAS
re-expression, and synergism with FAS-ligand. Blood. 119:4242–4252.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang Y, Chen FR, Wei CC, Sun LL, Liu CY,
Yang LB and Guo XY: Zinc finger protein 671 has a cancer-inhibiting
function in colorectal carcinoma via the deactivation of Notch
signaling. Toxicol Appl Pharmacol. 458:1163262023. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Bhullar DS, Barriuso J, Mullamitha S,
Saunders MP, O'Dwyer ST and Aziz O: Biomarker concordance between
primary colorectal cancer and its metastases. EBioMedicine.
40:363–374. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Guo H, Vuille JA, Wittner BS, Lachtara EM,
Hou Y, Lin M, Zhao T, Raman AT, Russell HC, Reeves BA, et al: DNA
hypomethylation silences anti-tumor immune genes in early prostate
cancer and CTCs. Cell. 186:2765–2782.e28. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Gkountela S, Castro-Giner F, Szczerba BM,
Vetter M, Landin J, Scherrer R, Krol I, Scheidmann MC, Beisel C,
Stirnimann CU, et al: Circulating tumor cell clustering shapes DNA
methylation to enable metastasis seeding. Cell. 176:98–112.e14.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Padmanaban V, Krol I, Suhail Y, Szczerba
BM, Aceto N, Bader JS and Ewald AJ: E-cadherin is required for
metastasis in multiple models of breast cancer. Nature.
573:439–444. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Warton K and Samimi G: Methylation of
cell-free circulating DNA in the diagnosis of cancer. Front Mol
Biosci. 2:132015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhou X, Yu L, Wang L, Xiao J, Sun J, Zhou
Y, Xu X, Xu W, Spiliopoulou A, Timofeeva M, et al: Alcohol
consumption, blood DNA methylation and breast cancer: A Mendelian
randomisation study. Eur J Epidemiol. 37:701–712. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Gaiani F, Marchesi F, Negri F, Greco L,
Malesci A, de'Angelis GL and Laghi L: Heterogeneity of colorectal
cancer progression: Molecular gas and brakes. Int J Mol Sci.
22:52462021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Bao Y, Zhai J, Chen H, Wong CC, Liang C,
Ding Y, Huang D, Gou H, Chen D, Pan Y, et al: Targeting
m6A reader YTHDF1 augments antitumour immunity and
boosts anti-PD-1 efficacy in colorectal cancer. Gut. 72:1497–1509.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zhai J, Chen H, Wong CC, Peng Y, Gou H,
Zhang J, Pan Y, Chen D, Lin Y, Wang S, et al: ALKBH5 drives immune
suppression via targeting AXIN2 to promote colorectal cancer and is
a target for boosting immunotherapy. Gastroenterology. 165:445–462.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Liu Y, Jiang C, Xu C and Gu L: Systematic
analysis of integrated bioinformatics to identify upregulated THBS2
expression in colorectal cancer cells inhibiting tumour immunity
through the HIF1A/Lactic Acid/GPR132 pathway. Cancer Cell Int.
23:2532023. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Yang L, Chen X, Lee C, Shi J, Lawrence EB,
Zhang L, Li Y, Gao N, Jung SY, Creighton CJ, et al: Functional
characterization of age-dependent p16 epimutation reveals
biological drivers and therapeutic targets for colorectal cancer. J
Exp Clin Cancer Res. 42:1132023. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Sheikhnejad G, Brank A, Christman JK,
Goddard A, Alvarez E, Ford H Jr, Marquez VE, Marasco CJ, Sufrin JR,
O'Gara M and Cheng X: Mechanism of inhibition of DNA (cytosine
C5)-methyltransferases by oligodeoxyribonucleotides containing
5,6-dihydro-5-azacytosine. J Mol Biol. 285:2021–2034. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Zheng Z, Zeng S, Liu C, Li W, Zhao L, Cai
C, Nie G and He Y: The DNA methylation inhibitor RG108 protects
against noise-induced hearing loss. Cell Biol Toxicol. 37:751–771.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Ou Y, Zhang Q, Tang Y, Lu Z, Lu X, Zhou X
and Liu C: DNA methylation enzyme inhibitor RG108 suppresses the
radioresistance of esophageal cancer. Oncol Rep. 39:993–1002.
2018.PubMed/NCBI
|
|
91
|
Tanaka S, Hosokawa M, Matsumura J,
Matsubara E, Kobori A, Ueda K and Iwakawa S: Effects of zebularine
on invasion activity and intracellular expression level of let-7b
in colorectal cancer cells. Biol Pharm Bull. 40:1320–1325. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Guo Y, Shu L, Zhang C, Su ZY and Kong ANT:
Curcumin inhibits anchorage-independent growth of HT29 human colon
cancer cells by targeting epigenetic restoration of the tumor
suppressor gene DLEC1. Biochem Pharmacol. 94:69–78. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Singh BN, Shankar S and Srivastava RK:
Green tea catechin, epigallocatechin-3-gallate (EGCG): Mechanisms,
perspectives and clinical applications. Biochem Pharmacol.
82:1807–1821. 2011. View Article : Google Scholar : PubMed/NCBI
|