Introduction
β-thalassemia is a widespread recessive hereditary
disease characterized by inadequate or ineffective composition of
β-globin, anemia and ineffective erythropoiesis. It has been
reported that 80–90 million individuals have β-thalassemia
worldwide (1). Due to severe
hypoxia caused by anemia, patients with β-thalassemia major (β-TM)
often have hepatosplenomegaly, growth retardation, jaundice, pale
complexion and marrow expansion (2). Current treatment strategies for
β-thalassemia, including thalidomide, deferasirox, deferiprone,
iron chelation, splenectomy, blood transfusion and hematopoietic
stem cell transplantation still have numerous drawbacks including
difficulty in donor matching and graft rejection in addition to a
high cost (3,4). Novel treatment methods, including
gene therapy and gene editing, have been previously investigated,
and relevant clinical trials have shown improvements in anemia in
patients with β-thalassemia (5–8).
Furthermore, it has been demonstrated that the novel activin
receptor ligand trap Luspatercept improves late-stage
erythropoiesis (9,10). Despite the promising nature of
these methods, there remain numerous safety concerns including
off-target activity and chromosomal rearrangement events due to the
small number of clinical trials conducted (11,12).
Therefore, it is crucial to verify novel approaches for managing
β-thalassemia.
Human hemoglobin (Hb) undergoes two switches from
the embryonic to the postnatal period, in which the main Hb changes
from fetal hemoglobin (HbF; α2γ2) to adult
hemoglobin (HbA; α2β2) (13). This process is affected by several
transcription factors, including B-cell lymphoma/leukemia 11A
(BCL11A), activating transcription factor 4, Kruppel-like factor 1,
v-myb avian myeloblastosis viral oncogene homolog, specificity
protein 1 and Ly1 antibody reactive (14–16).
However, the upstream regulators of BCL11A remain incompletely
characterized (4). Currently, a
promising way to treat β-thalassemia is γ-globin reactivation,
which elicits the point mutation of hereditary persistence of HbF
in patients (17).
miRNAs are groups of non-coding RNA molecules and
are essential for various biological functions such as cell
differentiation, maturation and proliferation (18,19).
Certain microRNAs, such as miR-32, influence early erythroid
commitment; miR-22 and miR-28 have been shown to impact the
maturation of erythroid cultures in vitro maturation
(20–22). Normal erythropoiesis is
characterized by a significant rise in miR-155, while the
expression pattern of miR-339 is biphasic (23). By changing the lifespan of globin
chains, these miRNAs control the production of Hb, iron metabolism
and resistance to oxidative stress in red blood cells (24).
miRNA sequence tests were conducted as in a previous
study on the peripheral blood of patients with β-TM and the healthy
controls to detect miRNAs with varying expression levels (23). Out of the differently expressed 196
miRNAs, miR-6747-3p was identified as being notably increased (fold
change, 4.76; P=0.001) and showing a positive association with HbF
(25). Current studies about
miR-6747-3p have focused on the direction of endometriosis
(26), Alzheimer's disease
(27) and small cell lung cancer
(SCLC) (28). However, whether
miR-6747-3p plays a role in hematologic diseases remains unclear.
By conducting in vitro functional experiments, the present
study aims to identify the expression of miR-6747-3p in patients
with β-TM, in addition to seeking its regulatory impacts on red
blood cell lineage development of erythroid precursor cells and
γ-globin expression.
Materials and methods
Patient enrollment
The Ethics Committee of the Fujian Maternity and
Child Health Hospital authorized the present study (Fuzhou, China;
approval no. 2019073), which followed the Helsinki Declaration.
Peripheral blood samples were collected from 20 patients with β-TM
(age, 8.30±1.59 years; female/male, 13/7) and 20 healthy controls
(age, 9.00±2.23 years; female/male, 12/8) before blood transfusion
or hydroxyurea treatment at Fujian Maternity and Child Health
Hospital (Fuzhou, China) between January 2020 to December 2021. For
the group comprised of patients with β-thalassemia, the inclusion
criteria included patients exhibiting anemia symptoms (Hb <90
g/l; normal reference value in children aged 6 months to 6 years,
105–140 g/l; normal reference value in children aged 7–12 years,
110–160 g/l) and carrying β°/β° (n=16), β°/β+ (n=2),
β+/β+ (n=2) genotypes. Control groups were
those age-matched individuals with normal thalassemia gene
diagnosis and peripheral blood indexes. The exclusion criteria
included: i) Patients with asthma, epilepsy and diabetes; ii)
patients with acute and chronic lung infection; iii) patients with
abnormal blood coagulation; and iv) patients with α-thalassemia,
iron deficiency anemia and megaloblastic anemia. All patients or
their guardians provided written informed consent.
Sample collection
Peripheral blood samples were gathered as previously
described (29). Briefly, 5 ml
peripheral blood from the participants was preserved and isolated
using a PAX gene blood RNA kit (Qiagen GmbH). Analysis was
conducted using the Sysmex XN-3000 automated hematology analyzer
(Sysmex Corporation) to evaluate blood cell parameters. An
automated capillary electrophoresis device (version 6.2; Sebia) was
used to analyze the Hb composition and levels.
Cell culture and transfection
The human umbilical cord blood-derived erythroid
progenitor (HUDEP-2) cells were provided by RIKEN BioResource
Centre through the National BioResource Project of the Ministry of
Education, Culture, Sports, Science and Technology (Tsukuba,
Ibaraki, Japan). The cells were cultured in a serum-free StemSpan
SFEM® medium (Stemcell Technologies, Inc.), supplemented
with 3 IU/ml erythropoietin (EPO; Amgen, Inc.), 1 µg/ml doxycycline
(Sigma-Aldrich; Merck KGaA) and 1×10−6 M dexamethasone
(Sigma-Aldrich; Merck KGaA). K562 cells derived from human
erythroleukemia were acquired from Shanghai Anwei Biotechnology
Co., Ltd. All cells were incubated at 37°C in a humidified
atmosphere containing 5% CO2.
The miR-6747-3p mimic (5′-UCCUGCCUUCCUCUGCACCAG-3′)
and its negative control (5′-UUCUCCGAACGUGUCACGUTT-3′), along with
the miR-6747-3p inhibitor (5′-CUGGUGCAGAGGAAGGCAGGA-3′) and its
control inhibitor (5′-CAGUACUUUUGUGUAGUACAA-3′) were obtained from
Shanghai Genepharma Co., Ltd. HUDEP-2/K562 cell transfections were
carried out by Amaxa Nucleofector II Device (Lonza Group, Inc.)
according to the manufacturer's instructions at room temperature.
The reagents used for electroporation of HUDEP-2 and K562 cells
were Cell Line Nucleofector™ Kit (Lonza Group, Inc.),
and the electroporation programs were U-008 and ATCC. After
electroporation with oligonucleotides at 100 nM concentration at
room temperature for 2 sec, the cells were cultured in a 37°C
incubator for 48 h for flow cytometry, cell cycle detection and
cell RNA extraction. The cell protein was extracted after 72 h.
K562 cells were induced by Hemin for 96 h for cell differentiation
detection and benzidine staining. HUDEP-2 cells were cultured in
three-stage medium for 14 days before erythroid differentiation
testing and Wright-Giemsa staining.
RNA extraction and qPCR analysis
Total RNA of peripheral blood samples and
HUDEP-2/K562 cells were gathered by the PAX Gene Blood RNA Kit
(Qiagen GmbH) and the Eastep® Super Total RNA Extraction
Kit (Promega Corporation) following the manufacturer's guidelines.
The RNA was quantified by NanoDrop ND-1000 spectrophotometer
(NanoDrop Technologies). The cDNA of miRNA and mRNA were generated
with Mir-X™ miRNA First-Strand Synthesis Kit and
PrimeScript™ RT reagent Kit with gDNA Eraser (Takara
Bio, Inc.). γ-globin, BCL11A and miR-6747-3p relative expression
levels were computed by applying the comparative cycle threshold
approach. The StepOnePlus™ Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was adopted to perform
miRNA and mRNA qRT-PCR with TB Green®
Advantage® qPCR Premix and TB Green® Premix
Ex Taq™ II Kit (Takara Bio, Inc.,), according to the
manufacturer's instruction. The miRNA RT-qPCR protocols were
demonstrated as follows: After a 10 sec denaturation stage, there
were 40 cycles of incubation at 95°C for 5 sec, 62°C for 20 sec and
55°C for 30 sec. The RT-qPCR detection protocol for mRNA was
conducted as follows: After a 30 sec pre-denaturation phase, a
total of 40 amplification cycles were executed, comprising
denaturation at 95°C for 5 sec, annealing at 60°C for 34 sec, and
extension at 95°C for 15 sec. The 2−ΔΔCq technique was
utilized to determine the relative fold-change of each target gene
in relation to GAPDH and U6 (30).
The primer sequences are available in Table SI.
Cell proliferation assessment
The Cell Counting Kit-8 (CCK8; APeXBIO Technology
LLC) was utilized to quantify cell proliferation ratio. HUDEP-2 and
K562 were seeded in 96-well plates after adding 10 µl CCK-8 reagent
to each well. After 2 h of incubation at 37°C, the cell viability
was assessed using a microtiter reader (Thermo Fisher Scientific,
Inc.) at 450 nm. Proliferation of the cells was assessed at 0, 24,
48, 72 and 96 h. Every experiment was run three times.
Flow cytometry assay
The impacts of miR-6747-3p on cell cycle and
apoptosis were examined using flow cytometry. A total of 5 µl
propidium iodide (PI; BD Biosciences) were added to stain HUDEP-2
and K562 cells after fixation with 75% ethanol at −20°C overnight.
The G0/G1, S and G2/M ratios were analyzed using ModFit software
(V3.2.; Verity Software House, Inc.). Annexin V-FITC/PI (BD
Biosciences) was used to stain cells for 30 min at room temperature
following the manufacturer's guidelines to detect cell apoptosis.
After which, BD LSRFortessa™ X-20 (BD Biosciences) and
FlowJo software V10 (FlowJo LLC) were used to assess the apoptosis
experiments.
Erythroid differentiation test
A three-phase differentiation protocol was used to
differentiate HUDEP-2 cells (31).
The procedure involved three phases including phase 1 (days 1–4),
using Iscove's Modified Dulbecco's Medium (IMDM) supplemented with
100 ng/ml Stem Cell Factor (SCF), 10 µg/ml recombinant human
insulin, 5% human AB serum, 1% L-glutamine, 330 µg/ml
holo-transferrin, 3 U/ml EPO, 1 µg/ml doxycycline, 2 U/ml heparin
and 1% penicillin/streptomycin. Phase 2 (days 5–7) included the
same cytokines as phase 1, except without SCF. During phase 3 (days
9–14), DOX was removed. For K562 cells, 50 µM of hemin was added
and the cells were cultured for another 96 h. The CD71/CD235a kit
(BD Biosciences) was used to detect erythroid lineage
differentiation. All cells were incubated at 37°C in a humidified
atmosphere containing 5% CO2.
Wright-Giemsa staining
To assess the differentiation of different groups,
HUDEP-2 cells were centrifuged for 300 × g at room temperature for
5 min, resuspended using 20 µl FBS (Stemcell Technologies, Inc.)
and coated on the slide. After drying naturally at room
temperature, Giemsa A and B solutions (Zhuhai Beso Biotechnology
Co., Ltd.) were combined at a 1:1 volume ratio. The slide was
washed with running water after staining for 5 min at room
temperature, the cell morphology [including basophilic erythroblast
(Baso-E), polychromatic erythroblast (Poly-E) and orthochromatic
erythroblast (Ortho-E)] was observed and captured using Leica
Aperiod CT6 microscope (Leica Microsystems GmbH).
Benzidine blue staining
The Hb expression level in K562 cells induced to
differentiate by hemin was evaluated by mixing the cell suspension
with a freshly made benzidine-H2O2 solution,
consisting of 50 µl of 3% H2O2
(Sigma-Aldrich; Merck KGaA) and 0.4%/ml benzidine (Merck KGaA).
After a 5-min treatment at room temperature, the cells were
photographed under a light microscope (Olympus Corporation). The
benzidine-positive cells were expressed as a percentage of at least
100 cells.
Fluorescence in situ hybridization
(FISH)
In HUDEP-2 cells, FISH was performed using specific
probes for miR-6747-3p and BCL11A according to the manufacturer's
instructions (Shanghai GenePharma Co., Ltd.). A total of
~5×104 HUDEP-2 cells were seeded on coverslip (NEST,
Inc.; cat. no. 801007) in 24-well plates overnight. After which,
the cells were washed with PBS and fixed in a 4% formaldehyde
solution for 15 min at room temperature. The cells were incubated
at room temperature with 0.1% buffer A for 10 min. After 15 min of
incubation at 37°C in Protein Free Rapid Blocking Buffer (EpiZyme,
Inc.; cat. no. PS108), 1 µl of 1 µM FAM-labeled miR-6747-3p probe
(5′-CTGGTGCAGAGGAAGGCAGGA-3′) or 1 µM cy3-labeled BCL11A probe
(5′-CCTGGTATTCTTAGCAGGTTAAAGG-3′) with 73°C rehydrated buffer E was
added into the cells and incubated at 37°C overnight in darkness.
The next day, the cells were successively washed three times for 10
min: 0.1% buffer F at 37°C, 2X buffer C at 60°C, and 2X buffer C at
37°C. 4′,6′-DAPI was used to dye the cell nuclei for 10 min at room
temperature. A Leica TCS SP8 CARS Confocal Microscope (Leica
Microsystems GmbH) was used to identify the subcellular
localization of miR-6747/BCL11A.
Luciferase reporter assay
The binding spot between miR-6747-3p and BCL11A was
identified using the online tool Targetscan (version 8.0;
https://www.targe tscan.org/vert_80/), miRWalk
(version 3.0; http://mirwalk.umm.uni-heidelberg.de/) and miRDB
(version V6; http://mirdb.org/miRDB/). Luciferase
reporter vector pmiR-RB-REPORT™ (Promega Corporation)
was inserted with wt-BCL11A and mut-BCL11A and co-transfected with
HUDEP-2 cells with a density of 1×105 cells/well using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.).
This includes the miR-6747-3p mimic, negative control, miR-6747-3p
inhibitor and inhibitor negative control. The relative luciferase
activity was calculated by the ratio of Renilla to firefly
luciferase after incubation for 48 h at 37°C. A total of three
replicates were established for the experiment.
Western blotting
Proteins were extracted following the previously
published method (32). Briefly,
protein samples were obtained from HUDEP-2 and K562 cells using
RIPA lysis buffer (Beyotime Institute of Biotechnology) and
quantified by BCA Kit (Beyotime Institute of Biotechnology). After
being separated on a 12.5% SDS-PAGE gel, the protein samples (20
µg) were imprinted on a polyvinylidene difluoride membrane.
Following 2 h of incubation at room temperature in Protein Free
Rapid Blocking Buffer (EpiZyme, Inc.; cat. no. PS108), the
membranes were then exposed to the following primary antibodies
overnight at 4°C: Anti-GAPDH (1:20,000; cat. no. ab8245; Abcam),
anti-γ-globin (1:1,000; cat. no. ab156584; Abcam) and anti-BCL11A
(1:1,000; cat. no. ab19487; Abcam). Horseradish peroxidase
(HRP)-conjugated goat anti-mouse IgG (1:5,000; Santa Cruz
Biotechnology, Inc.; cat. no. SC-2005) and HRP-conjugated goat
anti-rabbit IgG (1:5,000; Santa Cruz Biotechnology, Inc.; cat. no.
SC-2004) were utilized as the secondary antibodies at room
temperature for 2 h. To identify proteins, the Chemiluminescence
Western Blotting Detection system (Thermo Fisher Scientific, Inc.)
was used. Densitometric analysis with Image J software (version
1.5; National Institutes of Health) was used to ascertain the
relative expression of each protein.
Statistical analysis
GraphPad Prism 9.0 (Dotmatics) and SPSS 26.0 (IBM
Corp.) were utilized for data analysis. The Kolmogorov-Smirnov test
was employed to examine the normality of the data distributions.
The differences between groups with normally distributed data
(displayed as mean ± standard deviation) were assessed utilizing
the two-tailed unpaired Student's t-test (2 groups) or the one-way
ANOVA followed by Tukey's test (≥3 groups), as appropriate. The
differences between groups without normally distributed data
[displayed as the median and interquartile range M (P25, P75)] were
tested by the Mann-Whitney U test (2 groups) or the Kruskal-Wallis
test (≥3 groups), as appropriate. The association between
hematological indicators and miR-6747-3p was examined using
Spearman correlation analysis. Mean ± standard deviation was
reported from three separate trials. P<0.05 was considered to
indicate a statistically significant difference.
Results
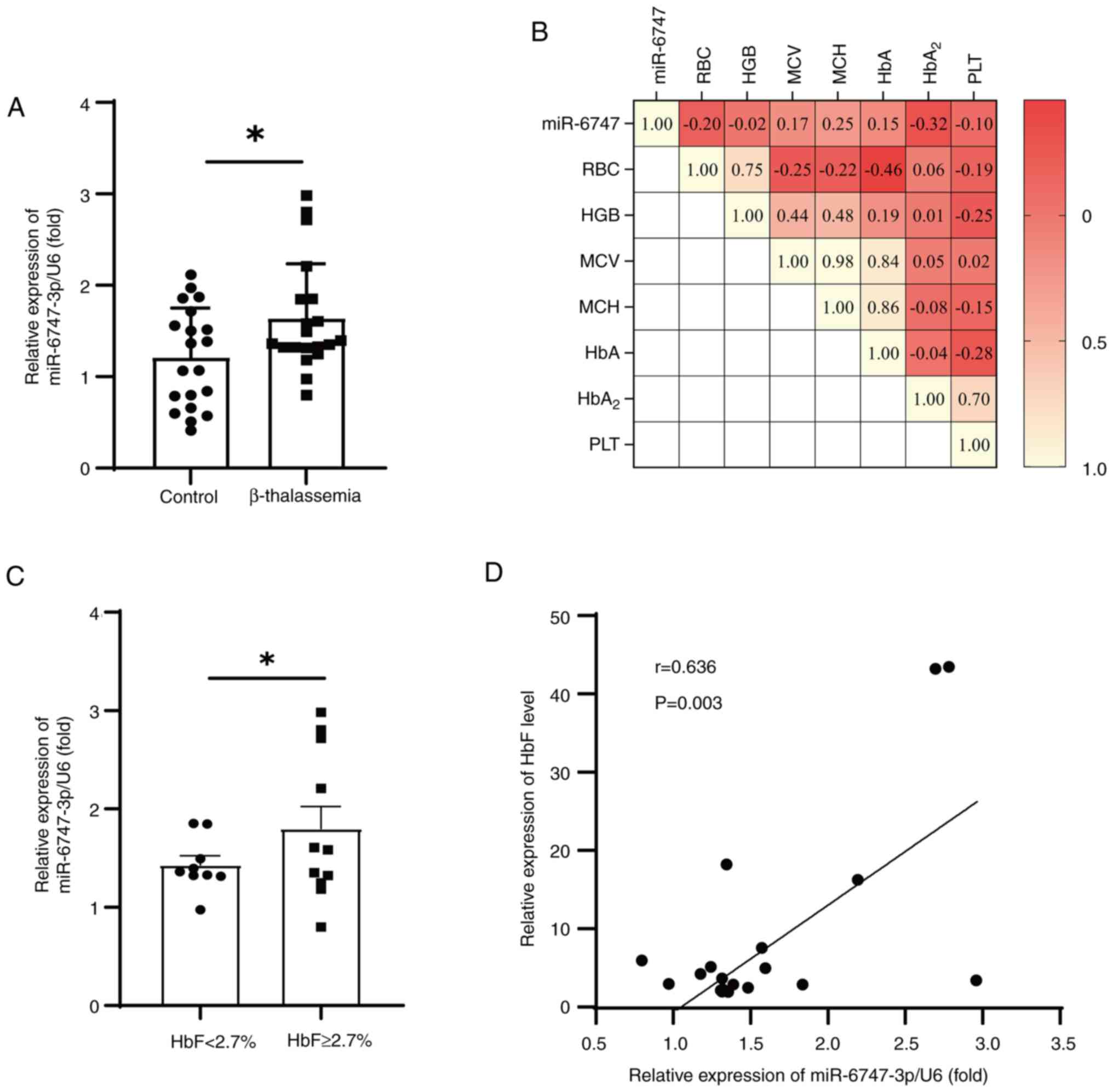
miR-6747-3p is upregulated in β-TM
patients and associated with HbF
In the present study, 20 patients with β-TM and 20
healthy controls were recruited to confirm the expression of
miR-6747-3p and evaluate its clinical significance. Compared with
the healthy participants, patients with β-TM had significantly
decreased Hb levels and higher HbF levels (Table I), which was consistent with the
anemia phenotype of β-TM (33).
There was a notable rise in the expression of MiR-6747-3p in
patients with β-TM (Fig. 1A).
Notably, correlations were evaluated between miR-6747-3p and
hematological parameters (Fig.
1B), and HbA2 was found to be associated with
miR-6747-3p. However, miR-6747-3p was statistically not correlated
with RBC, HGB, MCV, HbA and PLT. By dividing patients into HbF high
(HbF, ≥2.7%) and low (HbF, <2.7%) expressing groups, it was
discovered that patients with HbF ≥2.7% had significantly higher
levels of miR-6747-3p than the patients with <2.7% HbF (Fig. 1C). Additional examinations
uncovered that miR-6747-3p was significantly correlated with HbF
levels in patients with β-TM (r=0.636; P<0.05; Fig. 1D). These results indicate that
miR-6747-3p may play a role in the elevated HbF in patients with
β-TM. Consequently, the effects of miR-6747-3p overexpression and
knockdown in erythroid precursor cells were further
investigated.
 | Table I.Comparison of hematological
parameters and biochemical indicators in patients with β-TM and
healthy controls. |
Table I.
Comparison of hematological
parameters and biochemical indicators in patients with β-TM and
healthy controls.
|
Characteristics | Control group | β-TM group | P-value |
|---|
| Sex
(female/male) | 65 (13/7) | 60 (12/8) | 0.744 |
| Age, yrs | 9.00±2.23 | 8.30±1.59 | 0.053 |
| RBC,
×1012/l | 4.65±0.24 | 3.71±0.42 | <0.01 |
| Hb, g/l | 132.95±6.45 | 85.60±12.21 | <0.01 |
| MCV, fl | 83.13±2.50 | 80.34±5.01 | 0.043 |
| MCH, pg | 28.62±0.91 | 26.33±2.07 | <0.01 |
| HbA, % | 96.78±0.87 | 88.29±12.68 | 0.005 |
| HbA2, % | 2.79±0.04 | 3.49±0.50 | 0.169 |
| HbF, % | 0.15±0.08 | 8.22±2.86 | 0.007 |
| PLT,
×109/l | 291.35±14.43 | 422.13±45.00 | 0.005 |
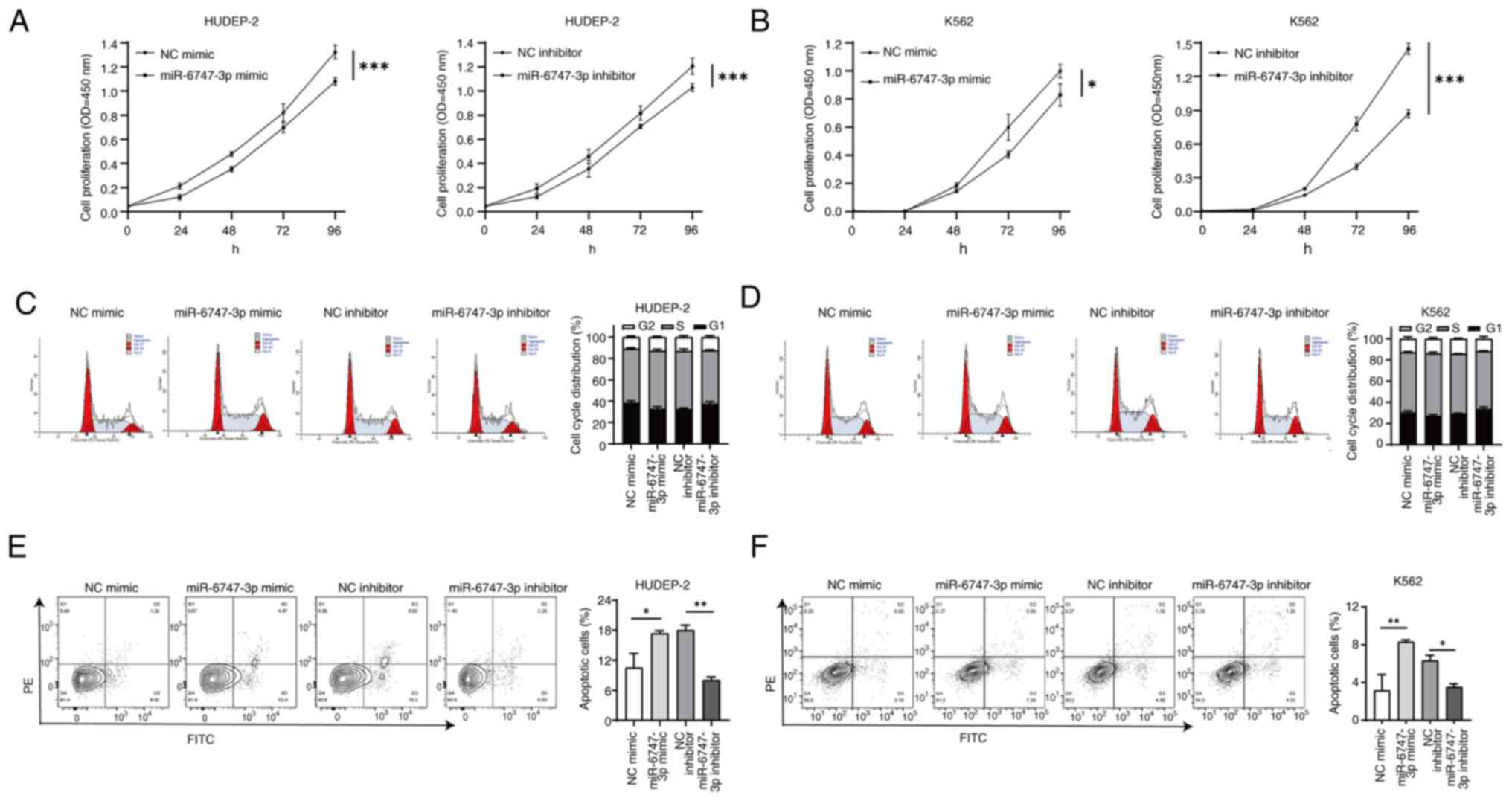
miR-6747-3p regulates cell
proliferation and apoptosis of erythroid precursor cells
Electroporation transfection with overexpression and
knockdown vectors for miR-6747-3p was performed on HUDEP-2 cells
and K562 cells. A validation of cell transfection efficiency was
provided in Fig. S1. The CCK-8
results showed that the absorbance of the miR-6747-3p mimic group
was reduced in HUDEP-2 cells. Conversely, absorbance of the
miR-6747-3p inhibitor group exceeded the NC inhibitor group
(Fig. 2A). Similar results were
detected in K562 cells (Fig. 2B),
inferring that miR-6747-3p expression decreases cell proliferation
in HUDEP-2 cells and K562 cells.
Cell cycle analysis showed that miR-6747-3p mimic
cells led to higher percentages of the S phase, while the inhibitor
group had the opposite effect (Fig. 2C
and D). Consistent with the cell growth findings where
miR-6747-3p demonstrated the capability to reduce cell growth in a
laboratory setting, these findings suggested that overexpression of
miR-6747-3p results in a halt from S to G2/M phase in the cell
cycle.
Apoptosis was examined in HUDEP-2 and K562 cells
with either overexpression or knockdown of miR-6747-3p to explore
its role in cell apoptosis. The results indicated that the
apoptosis rate was increased in the miR-6747-3p mimic group in both
HUDEP-2 and K562 cells (P<0.5). By contrast, the group treated
with the miR-6747-3p inhibitor had a lower rate of apoptosis than
the group treated with the NC inhibitor (P<0.5; Fig. 2E and F). The aforementioned results
indicate that miR-6747-3p induces cell cycle arrest and apoptosis,
thereby reducing cell proliferation.
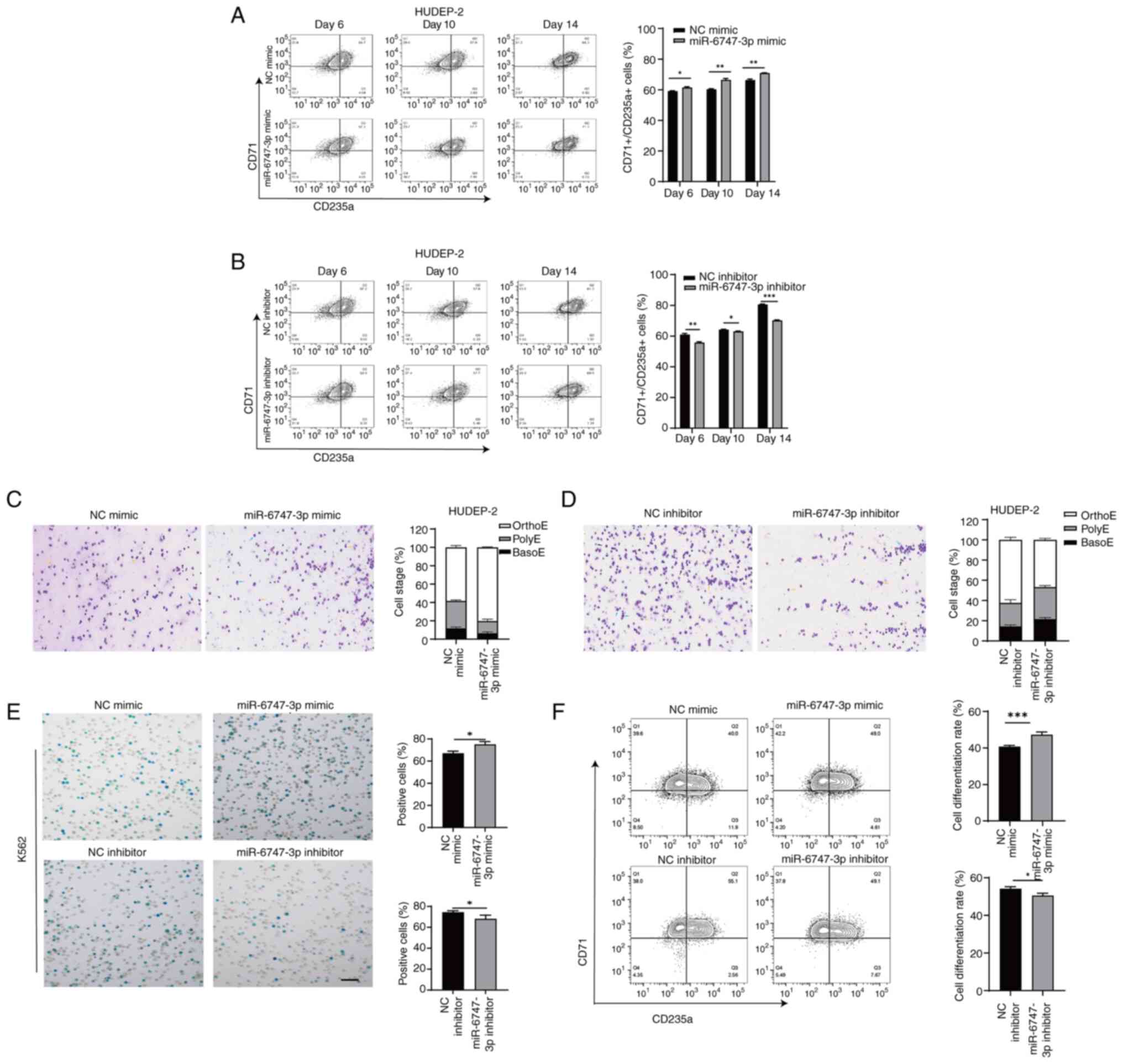
miR-6747-3p controls the maturation of
precursor cells
The transferrin receptor CD71 is abundantly present
in early erythroid cells, whereas the surface marker CD235a becomes
more prominent as erythroblasts mature (34). After 14 days of terminal
differentiation, flow cytometry analysis revealed that 71.3±2.77%
of the differentiated erythroid precursors in the miR-6747-3p mimic
group expressed CD71/CD235a, a notably higher percentage compared
with the NC mimic group (65.3±2.20%; Fig. 3A), while the cell differentiation
rate was reduced in HUDEP-2 cells treated with the miR-6747-3p
inhibitor compared with the NC inhibitor group (69.5±3.35% vs.
81.2±2.66%; Fig. 3B).
Wright-Giemsa was applied to stain cultivated
erythroblasts. Morphological analysis of HUDEP-2 cells using
miR-6747-3p mimics revealed a concomitant increase in Orth-E
(Fig. 3C), whereas the miR-6747-3p
inhibitor group failed to progress beyond the Poly-E stage of
differentiation at day 14 (Fig.
3D).
After 96 h of co-culture with hemin, the miR-6747-3p
mimic group showed a greater percentage of positive K562 cells in
the Benzidine blue staining compared with the NC group. In Fig. 3E, the miR-6747-3p inhibitor group
showed a lower positive rate than the NC inhibitor group. Flow
cytometry results showed a rise in CD71/CD235a+ cells in
the miR-6747-3p mimic group compared with the NC group while the
differentiation rate of the miR-6747-3p inhibitor group was lower
than that of NC inhibitor group (Fig.
3F), indicating that miR-6747-3p speeds up erythroid
differentiation.
miR-6747-3p induces HbF expression of
erythroid precursor cells
It was previously confirmed that miR-6747-3p can
enhance the development of HUDEP-2 and K562 cells, and is
associated with the levels of HbF in patients with β-TM. Next,
F-cell detection in HUDEP-2 cells was performed to further confirm
whether miR-6747-3p could regulate the expression of HbF. The
expression of HbF was subsequently quantified after 14 days of
differentiation in each group. The results indicated that
miR-6747-3p could strongly induce γ-globin in erythroid precursor
cells. The miR-6747-3p mimic group (55.1±0.76%) had significantly
higher HbF expression than the NC group (47.1±0.62%; P<0.05),
while the miR-6747-3p inhibitor group had significantly lower HbF
expression (44.0±0.47% vs. 37.2±1.80%; P<0.05; Fig. S2).
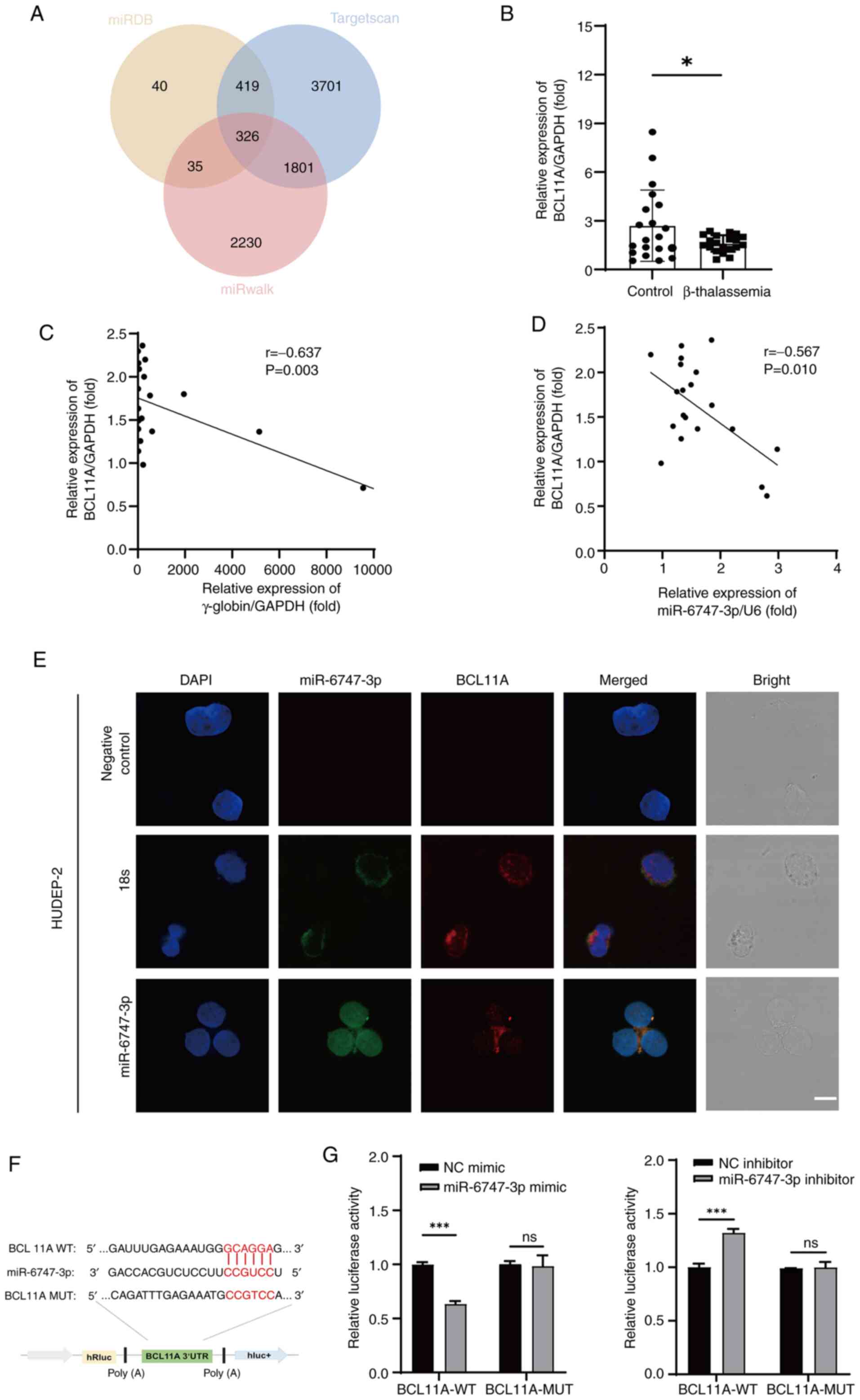
BCL11A is the direct target of
miR-6747-3p
TargetScan, miRwalk and miRDB were employed to
forecast the mRNA targets of miR-6747-3p to illustrate the
molecular mechanism of generating HbF expression. According to the
results, the anticipated target mRNA numbers were 820, 6,247 and
4,392, according to the sequence. Among the three programs, a total
of 326 mRNA targets were shared (Fig.
4A). The erythroid-related transcription factor BCL11A was
selected as a possible target gene for previous publications
showing the negative regulation of HbF (35–39).
RT-qPCR was used to measure BCL11A mRNA levels. The results showed
that, in comparison to normal controls, patients with β-TM had
considerably lower levels of BCL11A mRNA (Fig. 4B). Furthermore, BCL11A mRNA was
found to have a negative correlation with both γ-globin (r=−0.637;
P<0.05; Fig. 4C) and
miR-6747-3p (r=−0.567; P<0.05; Fig.
4D). In addition, the colocalization of miR-6747-3p and BCL11A
in HUDEP-2 cells was confirmed by fluorescence in situ
hybridization assay (Fig. 4E),
suggesting that the target gene of miR-6747-3p was BCL11A.
Whether miR-6747-3p directly interacts with BCL11A
was also examined. The CCGUCC binding site in miR-6747-3p targeting
GCAGGA in BCL11A 3′-UTR was identified using Targetscan (Fig. 4F). Thus, HUDEP-2 cells were
co-transfected with miR-6747-3p mimic and a
pmiR-RB-REPORT™ plasmid with the wild-type BCL11A
3′-UTR. The miR-6747-3p mimic significantly decreased the
luciferase activity, according to the results. This interaction was
further validated by demonstrating that it was eliminated when the
BCL11A 3′-UTR binding region was mutated from GCAGGA to CCGTCC
(Fig. 4G). By contrast, the
luciferase activity of the BCL11A seed region was notably higher in
the miR-6747-3p inhibitor group, with no significant change
observed in the mutant group. Briefly, miR-6747-3p was able to
directly attach to the 546–552 loci of the BCL11A 3′-UTR.
miR-6747-3p targets BCL11A to increase
the expression of γ-globin
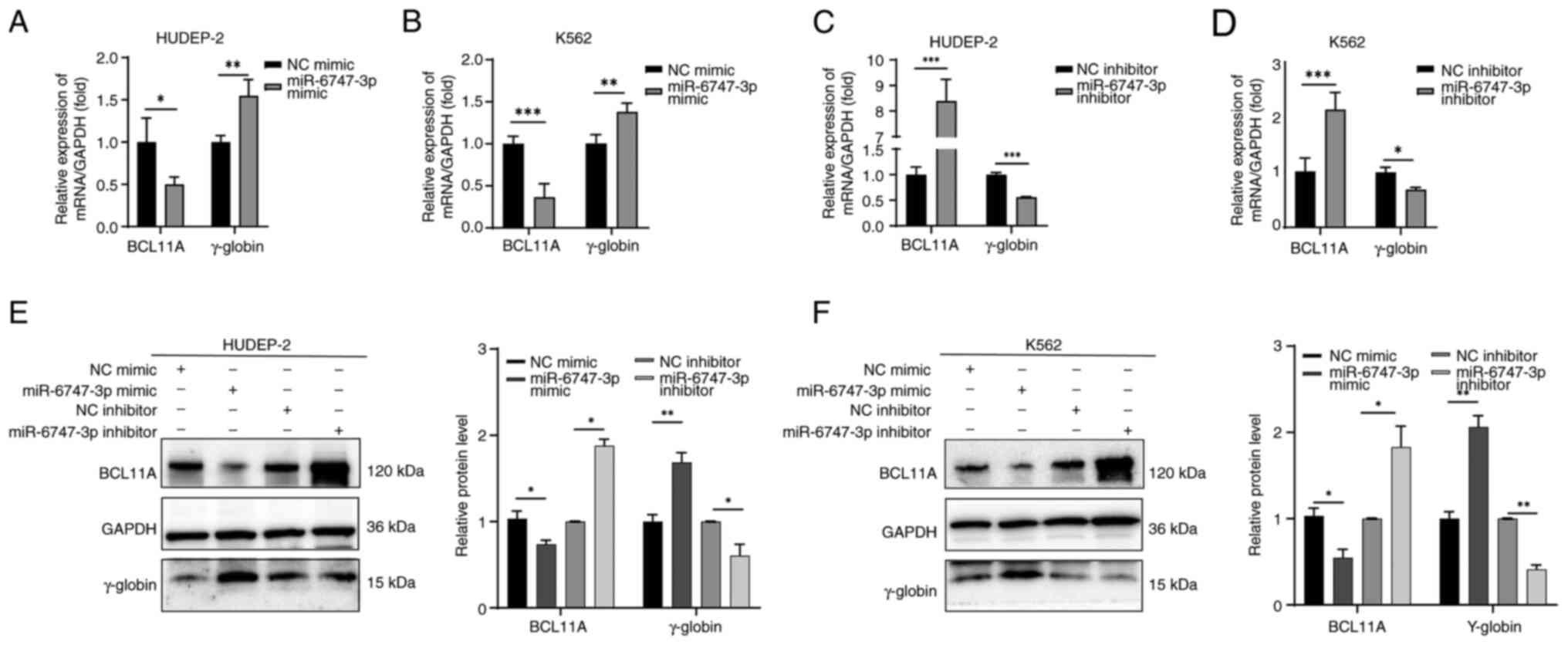
Fig. 5 illustrates
how the transfection of miR-6747-3p mimics into HUDEP-2 (Fig. 5A) and K562 (Fig. 5B) cells reduces BCL11A transcripts
and increases γ-globin mRNA levels. Notably, a >2-fold elevation
of BCL11A mRNA in HUDEP-2 (Fig.
5C) and K562 (Fig. 5D) cells
in the miR-6747-3p inhibitor group was observed, while γ-globin
mRNA expression was significantly decreased. These results were
also corroborated at the protein level. Western blot analysis
showed a notable increase in BCL11A protein levels in the
miR-6747-3p inhibitor group compared with the NC inhibitor group,
while γ-globin levels were significantly decreased (Fig. 5E and F). These findings indicate
that miR-6747-3p can inhibit the expression of BCL11A in both
HUDEP-2 and K562 cells.
Discussion
miRNAs are essential for controlling the expression
of Hb as well as several biological processes, such as
erythropoiesis and cell proliferation (40). In the present study it was first
demonstrated that β-TM had a markedly elevated expression level of
miR-6747-3p. Based on correlation analysis, miR-6747-3p has a
significant positive correlation with HbA2 and HbF.
Overexpression of hsa-miR-6747-3p impedes cell growth by causing
cell cycle arrest, inducing cell apoptosis, accelerating erythroid
differentiation and increasing HbF expression. In addition, it was
shown that miR-6747-3p negatively controls BCL11A by binding to the
546–552 loci of BCL11A mRNA 3′-UTR. The aforementioned results
indicated that miR-6747-3p may be an essential regulator of the HbF
level via modulating BCL11A expression.
Numerous researchers have explored the interaction
between miRNA and HbF reactivation in β-thalassemia. For example,
let-7/LIN28, miR-138 and miR-210 elevate γ-globin
expression, whereas miR-223-3p, miR-150 and miR-146a suppress
γ-globin production (41–43). Single nucleotide polymorphisms in
miRNA target genes may also lead to abnormal Hb expression
(44). These findings imply that
miRNAs may be valuable biomarkers for β-TM diagnosis and prognosis.
miR-6747-3p expression was studied in several diseases. In patients
with endometriosis, miR-6747-3p showed a good diagnostic capability
for infertility combined with ultrasonography (26). Another cohort study demonstrated a
noteworthy correlation between miR-6747-3p and SCLC by targeting
the colony-stimulating factor 3 receptor, a crucial component of
cellular autophagy factor, which was highly associated with myeloid
and lymphoid leukemias (45). In a
previous study, the research team identified upregulated
miR-6747-3p expression in patients with β-TM by microRNA sequencing
(25). However, whether
miR-6747-3p plays a role in β-thalassemia remains unknown. The
present analysis of miR-6747-3p expression levels revealed higher
levels in patients with β-TM (average age, 8.30±1.59 years)
compared with healthy controls (average age, 9.00±2.23 years),
aligning with previous findings (25). Moreover, miR-6747-3p has a
significant positive correlation with HbF.
The effect of miR-6747-3p overexpression/knockdown
on cell cycle, apoptosis, differentiation and proliferation were
examined by CCK8, flow cytometry, Wright-Giemsa and benzidine
staining tests. The results revealed that miR-6747-3p
overexpression inhibited cell growth, accelerated apoptosis and
stimulated cellular erythroid differentiation. Ineffective
erythropoiesis, a prevalent condition in β-thalassemia, is
characterized by high cell proliferation (46). Prior research on miRNA variance
examination in thalassemia indicated that the level of miRNA-101-3p
was notably elevated in CD34+ cells separated from
peripheral blood of patients with thalassemia, with a more
pronounced impact observed in individuals with thalassemia minor
compared with major patients and healthy controls (47). However, in the present results, the
increased expression of miRNA-6747-3p inhibited cell proliferation,
which was speculated to be related to the cell variance, and the
present experimental group were all made up of patients with
thalassemia major. Moreover, it is noteworthy that the change in
450 nm absorbance was more pronounced in K562 cells than in HUDEP-2
cells. The miR-6747-3p mimic group in K562 cells exhibited a
notably increased apoptosis rate compared with the other three
groups. The variable expression patterns of miRNAs and their
capacity to modify physiological processes within cells may be a
contributing factor to this phenomenon. Subsequent experiments with
superior red lineage cells (CD34+) and relevant
subgroups of patients with minor and intermediate thalassemia are
required.
By contrast, inhibition of miR-6747-3p was also
shown to have an impact on erythroid precursor cells, perhaps
reducing cell cycle arrest. This finding needs to be validated by
other experiments involving cell cycle-related proteins.
Furthermore, examining the morphological changes during erythroid
differentiation showed that miR-6747-3p inhibitor cells had a
greater number of basophilic erythroblasts and a lower number of
orthochromatic erythroblasts compared with the control groups at
the end of 14 days in HUDEP-2 cells. The multistep process of
erythropoiesis involves committing multipotent HSCs to develop into
the red blood cell lineage (34).
It is reasonable to infer that miR-6747-3p regulates the
differentiation of erythroid precursor cells and ameliorates
symptoms of anemia. K562 cells, first discovered in a patient with
chronic myeloid leukemia, are frequently utilized as a laboratory
model for studying the molecular processes in human globin gene
expression and assessing the effectiveness of novel medications
that promote differentiation (48,49).
As first demonstrated by Rutherford et al (50), K562 cells have a low potential for
Hb-synthesizing but can undergo erythrocyte differentiation in
response to various compounds, such as hemin. In the present study,
benzidine blue staining results demonstrate that miR-6747-3p
overexpression in K562 cells leads to an apparent increase
proportion of benzidine-positive cells, whereas inhibiting
miR-6747-3p leads to the opposite effect, suggesting that
miR-6747-3p promotes Hb synthesis in K562 cells.
In previous decades, efforts to increase HbF
synthesis have been motivated by the concept that higher HbF
diminishes the severity of β-thalassemia (38,40).
Liu et al (38) found that
overexpression of miR-486-3p could notably decrease BCL11A protein
and enhance the synthesis of γ-globin. Likewise, miR-210 boosts the
synthesis of γ-globin by reducing the levels of BCL11A in erythroid
progenitors derived from patients with β-thalassemia (40). Furthermore, in K562 cells, miR-210
can enhance the suppression of BCL11A induced by mithramycin
(51). The BCL11A gene is mainly
found in the brain and hematological organs, and it is situated on
chromosome 2p16.1 (52,53). Research has indicated that BCL11A
is essential for regulating the transition of Hb and preserving the
inactivity of the γ-globin gene (54,55).
An attempt has been made to reactivate HbF through BCL11A knockdown
by synthesizing BCL11A short hairpin RNA that inserts into the
flanking region of the miRNA precursor. In vitro research
combined with in vivo mouse models have validated the
shRNA-based treatment (56). Based
on the aforementioned research, β-thalassemia may be treated using
miRNA-based targeted therapies (57). In the present study, patients
diagnosed with β-TM exhibited decreased levels of BCL11A mRNA
expression. Moreover, the Pearson correlation test found that
BCL11A levels were negatively correlated with miR-6747-3p and HbF.
Therefore, it was hypothesized that hsa-miR-6747-3p may be involved
in regulating HbF expression by targeting BCL11A.
Through the application of bioinformatics methods,
it was discovered that miR-6747-3p could bind to the 546–552
positions on the 3′-UTR of BCL11A mRNA. This finding was validated
through fluorescence in situ hybridization. In HUDEP-2 cells,
miR-6747-3p and BCL11A were discovered extensively distributed in
the cytoplasm. Furthermore, the luciferase results indicated that
miR-6747-3p could directly interact with the 546–552 region of the
BCL11A mRNA 3′-UTR. Of note, miR-6747-3p was found to decrease
BCL11A levels and increase γ-globin expression in HUDEP-2 and K562
cells, as shown by RT-qPCR and Western blot analyses. These results
confirm that miR-6747-3p in β-TM could target BCL11A directly.
Overall, the results of the present study indicated
that miR-6747-3p has a specific clinical utility for β-TM. The
statistical analysis revealed a notable molecular pathway that
includes miR-6747-3p, BCL11A and γ-globin. Despite the small sample
size recruited in the present study, this newly identified
translational regulatory mechanism may offer a significant target
for synthesizing HbF by mimicking miR-6747-3p functions. Notably,
HbF levels can be influenced by drugs, autoimmune disease,
pregnancy, malignancy, diabetes, genetic modifiers and
hematological disorders and splenic dysfunction (44). Several SNPs in BCL11A, such as
rs4671393, rs4127407 and rs7606173, are associated with decreased
HbF levels but can also lead to elevated HbF levels via
microdeletion (37). Previous
research found no significant variations in hematological
parameters between rs1426407, rs1018987 and rs11886868 (58). However, 3.3% of HbF level variation
in β°-thalassemia/HbE among Thai patients was found to be strongly
correlated with rs6729815 (59).
Additionally, SNPs rs6545816 (A/C), rs6545817 (A/G), rs766432 (A/C)
and rs6729815 (A/G) were linked with high HbF levels (59). Given that mutations at the BCL11A
locus can influence HbF expression, future research should
implement more rigorous inclusion criteria. In the future, the
miR-6747-3p-BCL11A-γ-globin axis in β-thalassemia will be
investigated in more detail by increasing the sample size and
conducting in vivo studies.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Xinhua Zhang from
the Department of Hematology, 923rd Hospital of the People's
Liberation Army (Guangxi, China) for directing the blood sample
collection. The HUDEP-2 cells were provided by RIKEN BioResource
Centre through the National BioResource Project of the Ministry of
Education, Culture, Sports, Science and Technology (Tsukuba,
Japan).
Funding
This work was supported by The Major Scientific Research Program
for Young and Middle-aged Health Professionals of Fujian Province
(grant no. 2023ZQNZD009), The National Natural Science Foundation
of China (grant no. 81970170), Joint Funds for the Innovation of
Science and Technology of Fujian Province (grant nos. 2020Y9150,
2021Y9173, 2021Y9174 and 2023Y9364), Startup Fund for scientific
research, Fujian Medical University (grant no. 2023QH2044), Fujian
Provincial Natural Science Foundation of China (grant no.
2023J011217), Key Project on the Integration of Industry, Education
and Research Collaborative Innovation of Fujian Province (grant no.
2021YZ034011) and Key Project on Science and Technology Program of
Fujian Health Commission (grant no. 2021ZD01002).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
AL, MC, SZ and WZ performed all the experiments and
collected the data. JL was responsible for analyzing the data. SL
and YZ assisted in the cell culture. NL, LX and HH conceived and
designed the study. AL and SZ wrote the main manuscript. LX and HH
supervised the study and confirm the authenticity of all the raw
data. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
This study was conducted following the Declaration
of Helsinki, and approved by The Ethics Committee of The Fujian
Maternity and Child Health Hospital (Fuzhou, China; approval no.
2019073). All patients or their guardians provided written informed
consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Higgs DR, Engel JD and Stamatoyannopoulos
G: Thalassaemia. Lancet. 379:373–383. 2012. View Article : Google Scholar
|
|
2
|
Origa R: β-Thalassemia. Genet Med.
19:609–619. 2017. View Article : Google Scholar
|
|
3
|
Finotti A and Gambari R: Recent trends for
novel options in experimental biological therapy of β-thalassemia.
Expert Opin Biol Ther. 14:1443–1454. 2014. View Article : Google Scholar
|
|
4
|
Wang F, Ling L and Yu D: MicroRNAs in
β-thalassemia. Am J Med Sci. 362:5–12. 2021. View Article : Google Scholar
|
|
5
|
Finotti A, Breda L, Lederer CW, Bianchi N,
Zuccato C, Kleanthous M, Rivella S and Gambari R: Recent trends in
the gene therapy of β-thalassemia. J Blood Med. 6:69–85. 2015.
|
|
6
|
Xu XS, Hong X and Wang G: Induction of
endogenous gamma-globin gene expression with decoy oligonucleotide
targeting Oct-1 transcription factor consensus sequence. J Hematol
Oncol. 2:152009. View Article : Google Scholar
|
|
7
|
Basak A and Sankaran VG: Regulation of the
fetal hemoglobin silencing factor BCL11A. Ann N Y Acad Sci.
1368:25–30. 2016. View Article : Google Scholar
|
|
8
|
Finotti A, Borgatti M, Bianchi N, Zuccato
C, Lampronti I and Gambari R: Orphan drugs and potential novel
approaches for therapies of β-thalassemia: Current status and
future expectations. Expert Opin Orphan Drugs. 4:299–315. 2016.
View Article : Google Scholar
|
|
9
|
Cappellini MD, Viprakasit V, Taher AT,
Georgiev P, Kuo KHM, Coates T, Voskaridou E, Liew HK,
Pazgal-Kobrowski I, Forni GL, et al: A phase 3 trial of
luspatercept in patients with transfusion-dependent β-thalassemia.
N Engl J Med. 382:1219–1231. 2020. View Article : Google Scholar
|
|
10
|
Cazzola M: Ineffective erythropoiesis and
its treatment. Blood. 139:2460–2470. 2022. View Article : Google Scholar
|
|
11
|
Frangoul H, Altshuler D, Cappellini MD,
Chen YS, Domm J, Eustace BK, Foell J, de la Fuente J, Grupp S,
Handgretinger R, et al: CRISPR-Cas9 gene editing for sickle cell
disease and β-thalassemia. N Engl J Med. 384:252–260. 2021.
View Article : Google Scholar
|
|
12
|
Fu B, Liao J, Chen S, Li W, Wang Q, Hu J,
Yang F, Hsiao S, Jiang Y, Wang L, et al: CRISPR-Cas9-mediated gene
editing of the BCL11A enhancer for pediatric β°/β°
transfusion-dependent β-thalassemia. Nat Med. 28:1573–1580. 2022.
View Article : Google Scholar
|
|
13
|
Hariharan P and Nadkarni A: Insight of
fetal to adult hemoglobin switch: Genetic modulators and
therapeutic targets. Blood Rev. 49:1008232021. View Article : Google Scholar
|
|
14
|
Bianchi N, Cosenza LC, Lampronti I,
Finotti A, Breveglieri G, Zuccato C, Fabbri E, Marzaro G, Chilin A,
De Angelis G, et al: Structural and functional insights on an
uncharacterized Aγ-globin-gene polymorphism present in four
β0-thalassemia families with high fetal hemoglobin levels. Mol
Diagn Ther. 20:161–173. 2016. View Article : Google Scholar
|
|
15
|
Sankaran VG and Weiss MJ: Anemia: Progress
in molecular mechanisms and therapies. Nat Med. 21:221–230. 2015.
View Article : Google Scholar
|
|
16
|
Sankaran VG and Orkin SH: The switch from
fetal to adult hemoglobin. Cold Spring Harb Perspect Med.
3:a0116432013. View Article : Google Scholar
|
|
17
|
Venkatesan V, Christopher AC, Rhiel M,
Azhagiri MKK, Babu P, Walavalkar K, Saravanan B, Andrieux G,
Rangaraj S, Srinivasan S, et al: Editing the core region in HPFH
deletions alters fetal and adult globin expression for treatment of
β-hemoglobinopathies. Mol Ther Nucleic Acids. 32:671–688. 2023.
View Article : Google Scholar
|
|
18
|
Bissels U, Bosio A and Wagner W: MicroRNAs
are shaping the hematopoietic landscape. Haematologica. 97:160–167.
2012. View Article : Google Scholar
|
|
19
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View Article : Google Scholar
|
|
20
|
Choong ML, Yang HH and McNiece I: MicroRNA
expression profiling during human cord blood-derived CD34 cell
erythropoiesis. Exp Hematol. 35:551–564. 2007. View Article : Google Scholar
|
|
21
|
Bruchova H, Yoon D, Agarwal AM, Mendell J
and Prchal JT: Regulated expression of microRNAs in normal and
polycythemia vera erythropoiesis. Exp Hematol. 35:1657–1667. 2007.
View Article : Google Scholar
|
|
22
|
Hattangadi SM, Wong P, Zhang L, Flygare J
and Lodish HF: From stem cell to red cell: regulation of
erythropoiesis at multiple levels by multiple proteins, RNAs, and
chromatin modifications. Blood. 118:6258–6268. 2011. View Article : Google Scholar
|
|
23
|
Vasilatou D, Papageorgiou S, Pappa V,
Papageorgiou E and Dervenoulas J: The role of microRNAs in normal
and malignant hematopoiesis. Eur J Haematol. 84:1–16. 2010.
View Article : Google Scholar
|
|
24
|
Chen SY, Wang Y, Telen MJ and Chi JT: The
genomic analysis of erythrocyte microRNA expression in sickle cell
diseases. PLoS One. 3:e23602008. View Article : Google Scholar
|
|
25
|
Wang H, Chen M, Xu S, Pan Y, Zhang Y,
Huang H and Xu L: Abnormal regulation of microRNAs and related
genes in pediatric β-thalassemia. J Clin Lab Anal. 35:e239452021.
View Article : Google Scholar
|
|
26
|
Xu X, Li Z, Liu J, Yu S and Wei Z:
MicroRNA expression profiling in endometriosis-associated
infertility and its relationship with endometrial receptivity
evaluated by ultrasound. J Xray Sci Technol. 25:523–532. 2017.
|
|
27
|
Lu L, Dai WZ, Zhu XC and Ma T: Analysis of
serum miRNAs in Alzheimer's disease. Am J Alzheimers Dis Other
Demen. 36:153331752110217122021. View Article : Google Scholar
|
|
28
|
Wang XJ, Gao J, Yu Q, Zhang M and Hu WD:
Multi-omics integration-based prioritisation of competing
endogenous RNA regulation networks in small cell lung cancer:
Molecular characteristics and drug candidates. Front Oncol.
12:9048652022. View Article : Google Scholar
|
|
29
|
Chen M, Lv A, Zhang S, Zheng J, Lin N, Xu
L and Huang H: Peripheral blood circular RNA circ-0008102 may serve
as a novel clinical biomarker in beta-thalassemia patients. Eur J
Pediatr. 183:1367–1379. 2024. View Article : Google Scholar
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Himadewi P, Wang XQD, Feng F, Gore H, Liu
Y, Yu L, Kurita R, Nakamura Y, Pfeifer GP, Liu J and Zhang X: 3′HS1
CTCF binding site in human β-globin locus regulates fetal
hemoglobin expression. Elife. 10:e705572021. View Article : Google Scholar
|
|
32
|
Chen M, Wang X, Wang H, Zhang M, Chen L,
Chen H, Pan Y, Zhang Y, Xu L and Huang H: The clinical value of
hsa-miR-190b-5p in peripheral blood of pediatric β-thalassemia and
its regulation on BCL11A expression. PLoS One. 18:e02920312023.
View Article : Google Scholar
|
|
33
|
Gao J and Liu W: Advances in screening of
thalassaemia. Clin Chim Acta. 534:176–184. 2022. View Article : Google Scholar
|
|
34
|
Schippel N and Sharma S: Dynamics of human
hematopoietic stem and progenitor cell differentiation to the
erythroid lineage. Exp Hematol. 123:1–17. 2023. View Article : Google Scholar
|
|
35
|
Sankaran VG, Menne TF, Xu J, Akie TE,
Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB and
Orkin SH: Human fetal hemoglobin expression is regulated by the
developmental stage-specific repressor BCL11A. Science.
322:1839–1842. 2008. View Article : Google Scholar
|
|
36
|
Sankaran VG, Xu J, Ragoczy T, Ippolito GC,
Walkley CR, Maika SD, Fujiwara Y, Ito M, Groudine M, Bender MA, et
al: Developmental and species-divergent globin switching are driven
by BCL11A. Nature. 460:1093–1097. 2009. View Article : Google Scholar
|
|
37
|
Basak A, Hancarova M, Ulirsch JC, Balci
TB, Trkova M, Pelisek M, Vlckova M, Muzikova K, Cermak J, Trka J,
et al: BCL11A deletions result in fetal hemoglobin persistence and
neurodevelopmental alterations. J Clin Invest. 125:2363–2368. 2015.
View Article : Google Scholar
|
|
38
|
Liu N, Hargreaves VV, Zhu Q, Kurland JV,
Hong J, Kim W, Sher F, Macias-Trevino C, Rogers JM, Kurita R, et
al: Direct promoter repression by BCL11A controls the fetal to
adult hemoglobin switch. Cell. 173:430–442.e17. 2018. View Article : Google Scholar
|
|
39
|
Martyn GE, Wienert B, Yang L, Shah M,
Norton LJ, Burdach J, Kurita R, Nakamura Y, Pearson RCM, Funnell
APW, et al: Natural regulatory mutations elevate the fetal globin
gene via disruption of BCL11A or ZBTB7A binding. Nat Genet.
50:498–503. 2018. View Article : Google Scholar
|
|
40
|
Gasparello J, Fabbri E, Bianchi N,
Breveglieri G, Zuccato C, Borgatti M, Gambari R and Finotti A:
BCL11A mRNA targeting by miR-210: A possible network regulating
γ-globin gene expression. Int J Mol Sci. 18:25302017. View Article : Google Scholar
|
|
41
|
Basak A, Munschauer M, Lareau CA,
Montbleau KE, Ulirsch JC, Hartigan CR, Schenone M, Lian J, Wang Y,
Huang Y, et al: Control of human hemoglobin switching by
LIN28B-mediated regulation of BCL11A translation. Nat Genet.
52:138–145. 2020. View Article : Google Scholar
|
|
42
|
Lee YT, de Vasconcellos JF, Yuan J, Byrnes
C, Noh SJ, Meier ER, Kim KS, Rabel A, Kaushal M, Muljo SA and
Miller JL: LIN28B-mediated expression of fetal hemoglobin and
production of fetal-like erythrocytes from adult human
erythroblasts ex vivo. Blood. 122:1034–1041. 2013. View Article : Google Scholar
|
|
43
|
Li Y, Bai H, Zhang Z, Li W, Dong L, Wei X,
Ma Y, Zhang J, Yu J, Sun G and Wang F: The up-regulation of
miR-199b-5p in erythroid differentiation is associated with GATA-1
and NF-E2. Mol Cells. 37:213–219. 2014. View Article : Google Scholar
|
|
44
|
Mohammad SNNA, Iberahim S, Wan Ab Rahman
WS, Hassan MN, Edinur HA, Azlan M and Zulkafli Z: Single nucleotide
polymorphisms in XMN1-HBG2, HBS1L-MYB, and BCL11A and their
relation to high fetal hemoglobin levels that alleviate anemia.
Diagnostics (Basel). 12:13742022. View Article : Google Scholar
|
|
45
|
Trottier AM, Druhan LJ, Kraft IL, Lance A,
Feurstein S, Helgeson M, Segal JP, Das S, Avalos BR and Godley LA:
Heterozygous germ line CSF3R variants as risk alleles for
development of hematologic malignancies. Blood Adv. 4:5269–5284.
2020. View Article : Google Scholar
|
|
46
|
Oikonomidou PR and Rivella S: What can we
learn from ineffective erythropoiesis in thalassemia? Blood Rev.
32:130–143. 2018. View Article : Google Scholar
|
|
47
|
Phannasil P, Sukhuma C, Nauphar D, Nuamsee
K and Svasti S: Up-regulation of microRNA 101-3p during
erythropoiesis in β-thalassemia/HbE. Blood Cells Mol Dis.
103:1027812023. View Article : Google Scholar
|
|
48
|
Lozzio CB and Lozzio BB: Human chronic
myelogenous leukemia cell-line with positive Philadelphia
chromosome. Blood. 45:321–334. 1975. View Article : Google Scholar
|
|
49
|
Gambari R and Fibach E: Medicinal
chemistry of fetal hemoglobin inducers for treatment of
beta-thalassemia. Curr Med Chem. 14:199–212. 2007. View Article : Google Scholar
|
|
50
|
Rutherford TR, Clegg JB and Weatherall DJ:
K562 human leukaemic cells synthesise embryonic haemoglobin in
response to haemin. Nature. 280:164–165. 1979. View Article : Google Scholar
|
|
51
|
Bianchi N, Finotti A, Ferracin M,
Lampronti I, Zuccato C, Breveglieri G, Brognara E, Fabbri E,
Borgatti M, Negrini M and Gambari R: Increase of microRNA-210,
decrease of raptor gene expression and alteration of mammalian
target of rapamycin regulated proteins following mithramycin
treatment of human erythroid cells. PLoS One. 10:e01215672015.
View Article : Google Scholar
|
|
52
|
Tsang JCH, Yu Y, Burke S, Buettner F, Wang
C, Kolodziejczyk AA, Teichmann SA, Lu L and Liu P: Single-cell
transcriptomic reconstruction reveals cell cycle and multi-lineage
differentiation defects in Bcl11a-deficient hematopoietic stem
cells. Genome Biol. 16:1782015. View Article : Google Scholar
|
|
53
|
Jawaid K, Wahlberg K, Thein SL and Best S:
Binding patterns of BCL11A in the globin and GATA1 loci and
characterization of the BCL11A fetal hemoglobin locus. Blood Cells
Mol Dis. 45:140–146. 2010. View Article : Google Scholar
|
|
54
|
Sun KT, Huang YN, Palanisamy K, Chang SS,
Wang IK, Wu KH, Chen P, Peng CT and Li CY: Reciprocal regulation of
γ-globin expression by exo-miRNAs: Relevance to γ-globin silencing
in β-thalassemia major. Sci Rep. 7:2022017. View Article : Google Scholar
|
|
55
|
Li H, Lin R, Li H, Ou R, Wang K, Lin J and
Li C: MicroRNA-92a-3p-mediated inhibition of BCL11A upregulates
γ-globin expression and inhibits oxidative stress and apoptosis in
erythroid precursor cells. Hematology. 27:1152–1162. 2022.
View Article : Google Scholar
|
|
56
|
Simbula M, Manchinu MF, Mingoia M, Pala M,
Asunis I, Caria CA, Perseu L, Shah M, Crossley M, Moi P and
Ristaldi MS: miR-365-3p mediates BCL11A and SOX6 erythroid-specific
coregulation: A new player in HbF activation. Mol Ther Nucleic
Acids. 34:1020252023. View Article : Google Scholar
|
|
57
|
Brendel C, Guda S, Renella R, Bauer DE,
Canver MC, Kim YJ, Heeney MM, Klatt D, Fogel J, Milsom MD, et al:
Lineage-specific BCL11A knockdown circumvents toxicities and
reverses sickle phenotype. J Clin Invest. 126:3868–3878. 2016.
View Article : Google Scholar
|
|
58
|
Prasing W, Mekki C, Traisathit P, Pissard
S and Pornprasert S: Genotyping of BCL11A and HBS1L-MYB single
nucleotide polymorphisms in β-thalassemia/HbE and homozygous HbE
subjects with low and high levels of HbF. Walailak J Sci Technol.
15:627–636. 2017. View Article : Google Scholar
|
|
59
|
Nuinoon M, Makarasara W, Mushiroda T,
Setianingsih I, Wahidiyat PA, Sripichai O, Kumasaka N, Takahashi A,
Svasti S, Munkongdee T, et al: A genome-wide association identified
the common genetic variants influence disease severity in
beta0-thalassemia/hemoglobin E. Hum Genet. 127:303–314. 2010.
View Article : Google Scholar
|