Introduction
Lung cancer is one of the most common causes of
cancer-related mortality both in China and globally; according to
statistics, lung cancer accounts for 18.4% of tumor-associated
deaths worldwide (1,2). Non-small cell lung cancer (NSCLC) is
the predominant histopathological subtype of lung cancer, which is
responsible for 80–85% of lung cancer cases (3). Lung adenocarcinoma (LUAD) represents
the most prevalent and the most studied pathological type of NSCLC,
and it accounts for approximately two-fifths of all lung cancer
cases (4). Despite notable efforts
to improve early detection and to develop new treatment methods,
the prognosis of LUAD remains poor, mainly due to late diagnosis,
metastasis and high postoperative recurrence rates (5–7).
Therefore, improving understanding of the potential molecular
mechanisms and the identification of novel candidate biomarkers of
LUAD are critical to the development of novel diagnostic strategies
and targeted therapies.
The S100 protein family, composed of small acidic
proteins with an EF-hand Ca2+ binding motif, has been
reported to perform functions in diverse tumor behaviors, such as
cell proliferation, metastasis, angiogenesis and immune evasion
(8). In particular, S100
calcium-binding protein A16 (S100A16), which is ubiquitously
expressed in human tissues, has been well documented to be
differentially expressed in the majority of human cancer types,
where it functions in tumorigenic processes (9,10).
Notably, S100A16 expression has been reported to be significantly
upregulated in LUAD, and may be associated with poor overall
survival and the efficacy of platinum-based adjuvant chemotherapy
in LUAD (11,12). Moreover, a recent study suggested
that S100A16, targeted by microRNA-508-5p, may participate in the
proliferation and metastasis of LUAD cells (13). However, the mechanism of action of
S100A16 in LUAD has not yet been determined and requires further
evaluation.
Mov10 RNA helicase (MOV10) is a newly discovered
RNA-binding protein (RBP) that belongs to the RNA helicase
superfamily. It has previously been reported that MOV10 expression
is 2–3 times higher in human Burkitt's lymphoma cells and cervical
cancer cells than that in normal cells (14). Furthermore, high MOV10 expression
has been predicted to be related to poor prognosis in LUAD,
according to the Kaplan-Meier plotter database (15). Notably, the ENCORI database
(https://rnasysu.com/encori/) has
predicted that the RBP, MOV10, may target the extracellular matrix
(ECM)-related integrin α3 (ITGA3) mRNA, and ITGA3 may could be
considered an independent prognostic marker of NSCLC (16).
The present study aimed to determine the role of
S100A16 in LUAD, and to identify the S100A16-mediated mechanism in
LUAD. The findings may provide a novel molecular mechanism involved
in LUAD and thus improve understanding of the progression of
LUAD.
Materials and methods
Bioinformatics tools
The Cancer Genome Atlas data in the UALCAN database
(http://ualcan.path.uab.edu/index.html) were used to
analyze S100A16 expression in LUAD tissues and performed survival
analysis using log-rank test (17). BioGRID (https://thebiogrid.org/) and Pathway Commons databases
(http://www.pathwaycommons.org/) were
used to predict the downstream interacting proteins of S100A16.
LinkedOmics database (https://www.linkedomics.org/login.php) was utilized to
study the enrichment of S100A16 in the ECM-receptor interaction
pathway. The ENCORI database (https://rnasysu.com/encori/) was used to predict the
RNA-binding protein that could target MOV10.
Cell culture and treatment
The human bronchial epithelial cell line BEAS2B, the
LUAD cell lines H1975, PC-9, A549 and HCC827, and human umbilical
vein endothelial cells (HUVECs; fourth passage; cat. no.
iCell-h110) were procured from Cellverse Bioscience Technology Co.,
Ltd. The cells were cultured in Dulbecco's Modified Eagle Medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc.), with the exception
of HCC827 cells, which were cultured in Roswell Park Memorial
Institute-1640 medium (Gibco; Thermo Fisher Scientific, Inc.). All
cells were maintained in medium containing 10% fetal bovine serum
(FBS; Gemini Bio Products), 100 U/ml penicillin and 100 µg/ml
streptomycin in a 5% CO2 humidified environment at
37°C.
Plasmid transfection
S100A16 or MOV10 small interfering (si)RNAs
[siRNA-S100A16 (5′-GCCAAATTCCTGCCTGATTCTGG-3′) or siRNA-MOV10
(5′-ATGCTTCTTCAGGGAACAAGTAT-3′)] and the scrambled siRNA
[siRNA-negative control (NC): 5′-GCAACAAGATGAAGAGCACCAA-3′] were
designed and produced by Shanghai Quanyang Biotechnology Co., Ltd.
The MOV10 overexpression vector (Ov-MOV10) and the empty NC vector
(Ov-NC) were synthesized by General Biosystems (Anhui) Corporation
Ltd. The siRNAs or vectors (20 µM) were transfected into H1975
cells (1×105 cells/well) simultaneously using
Lipofectamine® 3000 (Thermo Fisher Scientific, Inc.) for
48 h at 37°C, according to the manufacturer's protocol. Cells were
used in subsequent experiments a total of 48 h
post-transfection.
Cell Counting Kit-8 (CCK-8) assay
H1975 cells were seeded into 96-well plates at a
density of 3×103 cells/well and were incubated at 37°C
for 24 h. The cells were then transfected with siRNA-NC,
siRNA-S100A16, Ov-NC or Ov-MOV10. After incubation for 48 h, 10 µl
CCK-8 solution (Dojindo Laboratories, Inc.) was added to each well.
After a 2-h incubation, the absorbance was measured at 450 nm using
a microplate reader (Shanghai Aolu Biological Technology Co.,
Ltd.).
5-Ethynyl-2′-deoxyuridine (EDU)
staining
Cell proliferation was measured using the iClick™
EDU Andy Fluor 555 Imaging Kit (GeneCopoeia, Inc.). H1975 cells
(5×104 cells/well) were seeded into 96-well plates and
incubated at 37°C for 24 h. The cells were then transfected with
siRNA-NC, siRNA-S100A16, Ov-NC or Ov-MOV10. A total of 48 h after
transfection, the cells in each well were treated with 20 µM EDU
for 2 h, according to the manufacturer's instructions.
Subsequently, the cells were incubated with 4% paraformaldehyde
fixing solution for 30 min followed by 0.5% Triton X-100
permeabilizing solution for 15 min at room temperature. DAPI was
used to stain the nuclei at 37°C for 30 min. The EdU+
cells were finally observed under a fluorescence microscope
(Olympus Corporation).
Wound healing assay
A total of 48 h after transfection, the transfected
H1975 cells were seeded into 6-well plates (5×105
cells/well) and were cultured routinely. After reaching 90%
confluence, a wound was made to the cell monolayer using a 200-µl
micropipette tip. After washing three times with PBS, the cells
were incubated with the serum-free DMEM at 37°C with 5%
CO2 for 24 h. The cell migration distance was documented
under a light microscope (Olympus Corporation). The width of the
scratch at 24 h was calculated as a percentage of the width at 0 h
using ImageJ software (version 1.4; National Institutes of
Health).
Transwell assay
After transfection, H1975 cells (5×104
cells/ well) resuspended in serum-free medium were seeded into the
upper chambers of a 24-well Transwell plate (pore size, 8 µm;
Corning, Inc.), which had been coated with Matrigel (Becton
Dickinson and Company) overnight at 37°C. The lower chambers were
filled with 500 µl medium containing 10% FBS. After 24 h, the
remaining cells in the top surface of the insert were removed with
a cotton swab, whereas the cells that had invaded to the bottom of
the membrane were fixed with 100% methanol for 10 min at 37°C and
stained with crystal violet solution (0.1%) for 15 min at 37°C,
before being subjected to a light microscopic inspection (Olympus
Corporation).
Tube formation assay
Matrigel was added to each well of 96-well plates
and the entire bottom surface of the well was gently covered.
Subsequently, the plates were incubated at 37°C for 30–60 min to
allow the Matrigel to solidify. Finally, eighth passage of HUVECs
(3×104 cells/well) were seeded into the 96-well
Matrigel-coated plates and cultured in the presence of DMEM from
H1975 cells transfected with different siRNAs/Ov plasmids at 37°C
in a 5% CO2 incubator for 24 h. Tubules were observed
under an inverted light microscope (Olympus Corporation).
Reverse transcription-quantitative PCR
(RT-qPCR)
After the total RNA was isolated from H1975 cells
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), cDNA was generated by RT using a ProSTAR
First-Strand RT-PCR kit (Stratagene; Agilent Technologies, Inc.)
according to the manufacturer's protocol. qPCR analysis was
conducted on the ABI 7500 Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.) using SYBR Green PCR Master Mix
Reagents (Takara Bio, Inc.) in accordance with the manufacturer's
protocol. The following thermocycling conditions were used: Initial
denaturation at 95°C for 10 min; followed by 35 cycles of
denaturation at 95°C for 15 sec, annealing at 60°C for 1 min and
extension of 10 min at 65°C. The relative expression levels were
calculated based on the 2−ΔΔCq method (18). GAPDH was used as the housekeeping
control. The following primers were used for qPCR: ITGA3, forward
5′-CCCAGAGGACCAAGGAAACC-3′, reverse 5′-CTCCTGGCTCAGCAAGAACA-3′;
GAPDH, forward 5′-AATGGGCAGCCGTTAGGAAA-3′, reverse
5′-GCGCCCAATACGACCAAATC-3′.
Western blotting
Following the homogenization of BEAS2B cells, LUAD
cells or HUVECs [HUVECs were treated with the conditioned medium
(CM) from H1975 cells transfected with different siRNAs/Ov plasmids
at 37°C for 24 h] in RIPA buffer (Epizyme Biomedical Technology
Co., Ltd.), protein samples were quantified using the BCA method
(Epizyme Biomedical Technology Co., Ltd.) and were separated by
SDS-PAGE on 12% gels, before being transferred to PVDF membranes.
The membranes were then incubated with 5% BSA (Beyotime Institute
of Biotechnology) for 1.5 h at room temperature for non-specific
blocking, followed by immunoblotting with primary antibodies
specific to S100A16 (cat. no. ab240572; 1:1,000; Abcam), matrix
metallopeptidase (MMP)2 (cat. no. ab92536; 1:1,000; Abcam), MMP9
(cat. no. ab76003; 1:1,000; Abcam), vascular endothelial-derived
growth factor (VEGF; cat. no. ab46154; 1:1,000; Abcam), VEGF
receptor-2 (VEGFR2; cat. no. 26415-1-AP; 1:2,000; Proteintech
Group, Inc.), MOV10 (cat. no. ab189919; 1:1,000; Abcam), ITGA3
(cat. no. ab131055; 1:1,000; Abcam), SRC (cat. no. ab133283;
1:1,000; Abcam) and phosphorylated (p)-SRC (cat. no. ab185617;
1:5,000; Abcam) at 4°C overnight. The membranes were then probed
with an HRP-conjugated secondary antibody (cat. no. ab6721;
1:10,000; Abcam) at room temperature for 1.5 h. Anti-GAPDH antibody
(cat. no. ab128915; 1:10,000; Abcam) was used to confirm equal
loading. Blots were at least run in tandem, under the same
conditions. Visualization of the blots was conducted using an ECL
reagent (Epizyme Biomedical Technology Co., Ltd.) and gray value
analysis was performed with ImageJ software (version 1.4).
Co-immunoprecipitation (Co-IP)
assay
The Co-IP assay was conducted utilizing a Co-IP kit
(cat. no. 26149; Pierce; Thermo Fisher Scientific, Inc.). Following
lysis in RIPA lysis buffer (Beyotime Institute of Biotechnology),
the H1975 cells were prepared by centrifugation at 14,000 × g and
4°C for 10 min. The IP was conducted using 2 µg S100A16 (cat. no.
ab240572; Abcam), MOV10 (cat. no. ab80613; Abcam) or anti-rabbit
IgG (cat. no. ab172730; Abcam) antibodies, after which 20 µl
protein A/G agarose beads (Pierce; Thermo Fisher Scientific, Inc.)
were added to isolate the protein complexes. The beads were washed
with PBS and then boiled to release the bound proteins, which were
subjected to SDS-PAGE and western blotting as aforementioned.
RNA immunoprecipitation (RIP)
assay
The RIP assay was conducted utilizing the EZ-Magna
RIP kit (cat. no. 17-701; MilliporeSigma) according to the
manufacturer's protocol. Following lysis in RIPA lysis buffer
(Beyotime Institute of Biotechnology), the H1975 cells were treated
with RIP buffer containing magnetic beads (MilliporeSigma)
conjugated with MOV10 (cat. no. ab80613; Abcam) or IgG (cat. no.
ab172730; Abcam) antibodies. Finally, the isolated RNA complexes
were subjected to qPCR analysis as aforementioned.
Actinomycin D assay
A total of 48 h after transfection, H1975 cells
seeded into 6-well plates (2×105 cells/well) were
treated with 5 µg/ml actinomycin D (GlpBio Technology, Inc.) for 0,
6, 12 and 18 h at 37°C to examine the stability of ITGA3 mRNA. The
remaining ITGA3 mRNA extracted from treated H1975 cells was
determined by qPCR analysis as aforementioned.
Statistical analysis
All data are presented as the mean ± standard error
of mean and were analyzed with GraphPad Prism 8 software
(Dotmatics). An unpaired Student's t-test was used to compare two
groups, whereas one-way ANOVA followed by Tukey's post hoc test was
used to compare three or more groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
S100A16 expression is upregulated in
LUAD tissues and cell lines
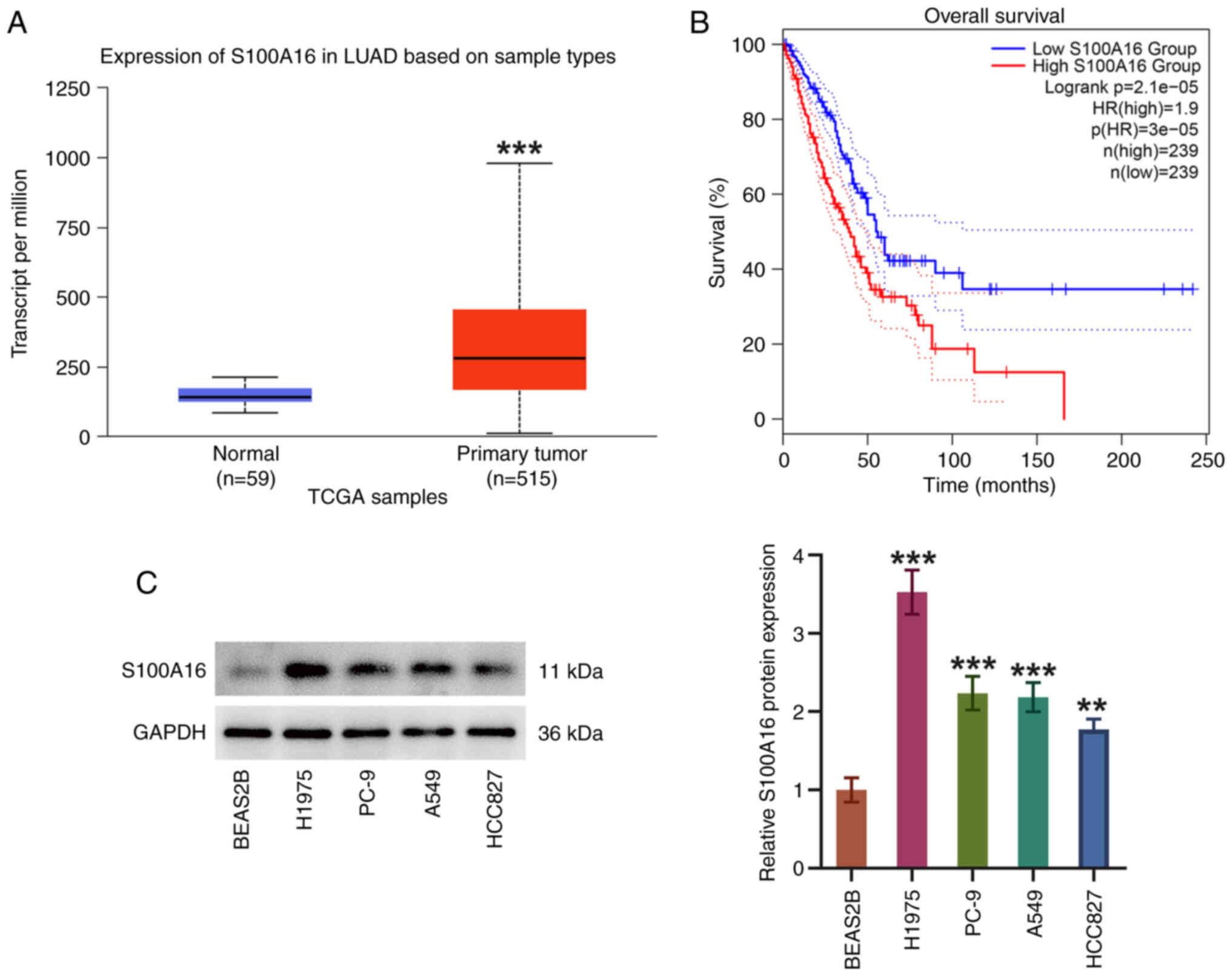
Through UALCAN database analysis, significantly
elevated S100A16 expression was observed in LUAD tissues compared
with in normal tissues from healthy control individuals (Fig. 1A). Moreover, patients with LUAD
were split into high and low expression groups according to median
S100A16 expression. High expression of S100A16 predicted a worse
overall survival in patients with LUAD (Fig. 1B). S100A16 expression was also
examined in cell lines by western blotting, and it was discovered
that S100A16 expression levels were higher in LUAD cell lines
(H1975, PC-9, A549 and HCC827) compared with those in the BEAS2B
cell line (Fig. 1C). H1975 cells
exhibited the highest S100A16 expression and were therefore used in
subsequent experiments. These findings indicated that S100A16
expression was increased in LUAD and it was associated with an
unfavorable outcome in patients with LUAD.
S100A16 knockdown obstructs the
proliferation, migration, invasion and angiogenesis of H1975
cells
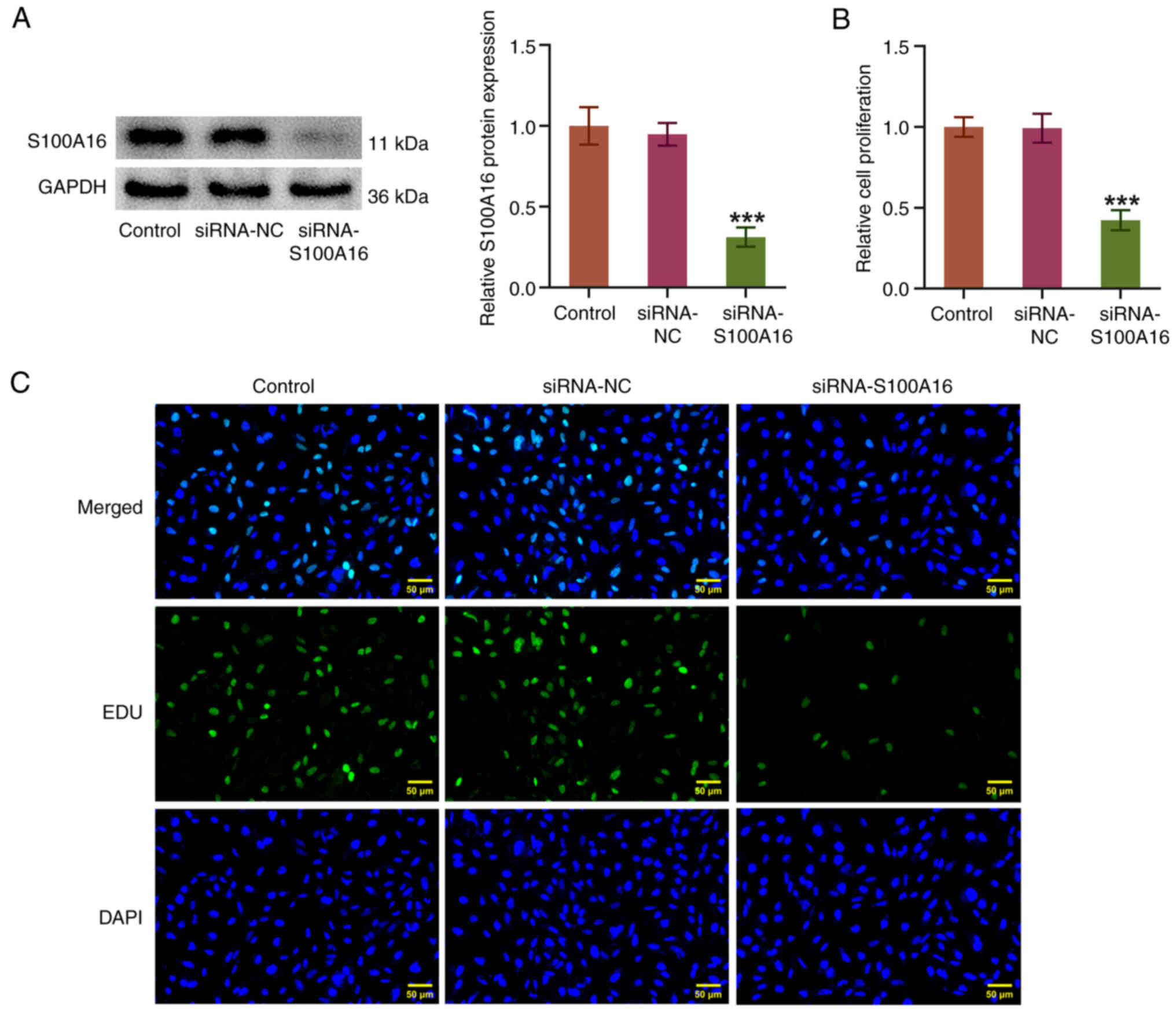
Post-transfection with siRNA-S100A16, S100A16
expression was markedly depleted (Fig.
2A). The experimental results from the CCK-8 and EDU assays
demonstrated that S100A16 knockdown decreased the proliferative
ability and the number of EDU+ H1975 cells compared with
that in the siRNA-NC group (Fig. 2B
and C). In addition, it was demonstrated that the migratory and
invasive capabilities of H1975 cells were reduced when S100A16 was
knocked down (Fig. 3A and B).
Compared with in the siRNA-NC group, knockdown of S100A16 also
resulted in the downregulation of MMP2 and MMP9 expression
(Fig. 3C). Furthermore, as shown
in Fig. 3D, the CM from H1975
cells with S100A16 knockdown markedly reduced the endothelial
capillary-like structures of HUVECs compared with in the siRNA-NC
(CM) + HUVEC group (Fig. 3D).
Furthermore, the protein expression levels of VEGF and VEGFR2 in
HUVECs were downregulated by the CM from H1975 cells with S100A16
knockdown (Fig. 3E). Collectively,
these results indicated that knockdown of S100A16 may slow the
development of LUAD.
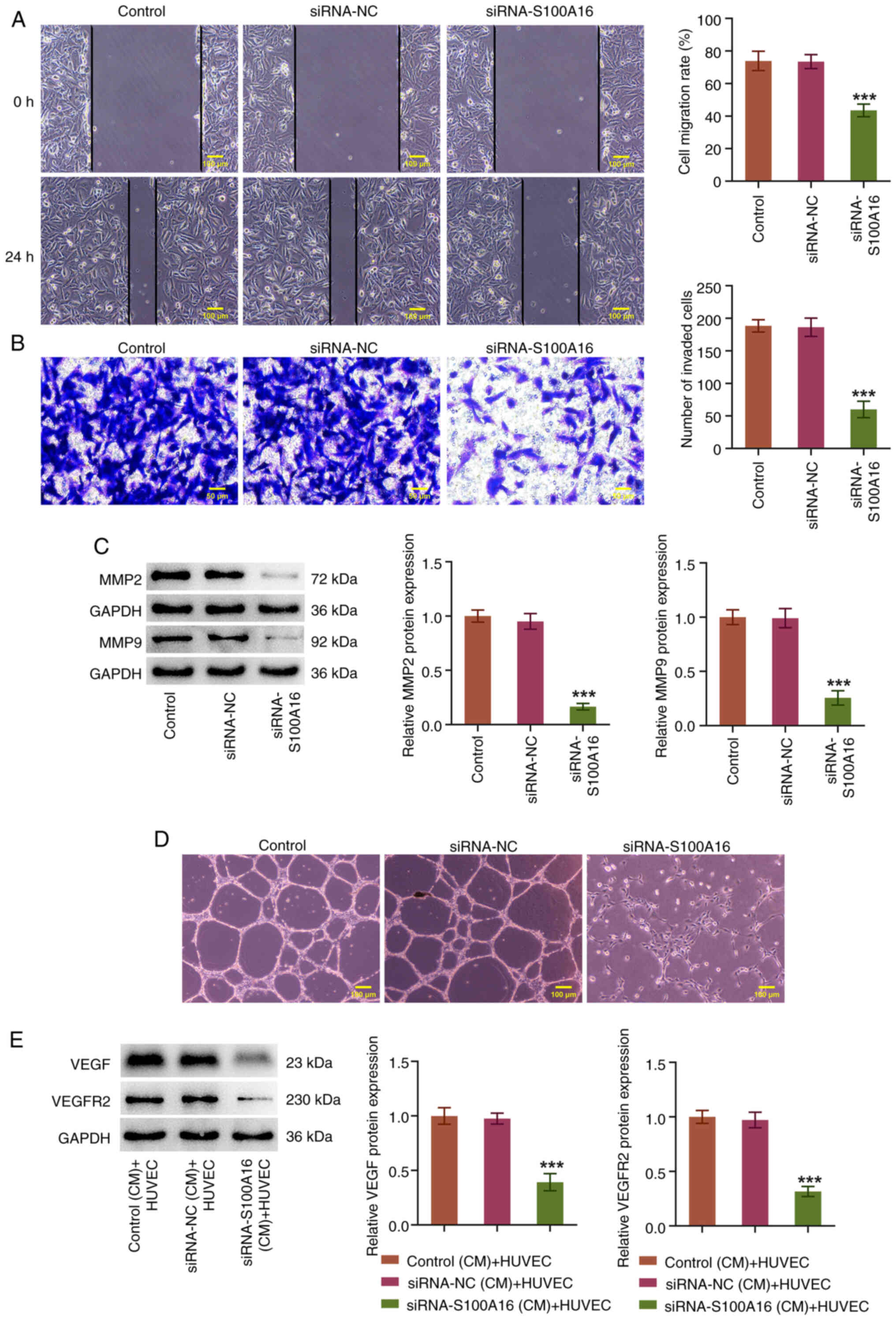 | Figure 3.S100A16 knockdown obstructs the
migration, invasion and angiogenesis of H1975 cells. (A) Wound
healing and (B) Transwell assays were used to evaluate cell
migration and invasion, respectively. (C) Western blotting was used
to examine MMP2 and MMP9 expression. (D) Tube formation assays were
used to estimated cell angiogenesis. ***P<0.001 vs. siRNA-NC.
(E) Western blotting was used to examine VEGF and VEGFR2 expression
in HUVECs. ***P<0.001 vs. siRNA-NC (CM) + HUVEC. CM, conditioned
medium; HUVEC, human umbilical vein endothelial cell; MMP, matrix
metallopeptidase; NC, negative control; S100A16, S100
calcium-binding protein A16; siRNA, small interfering RNA; VEGF,
vascular endothelial growth factor; VEGFR2, VEGF receptor 2. |
S100A16 interacts with MOV10 in H1975
cells
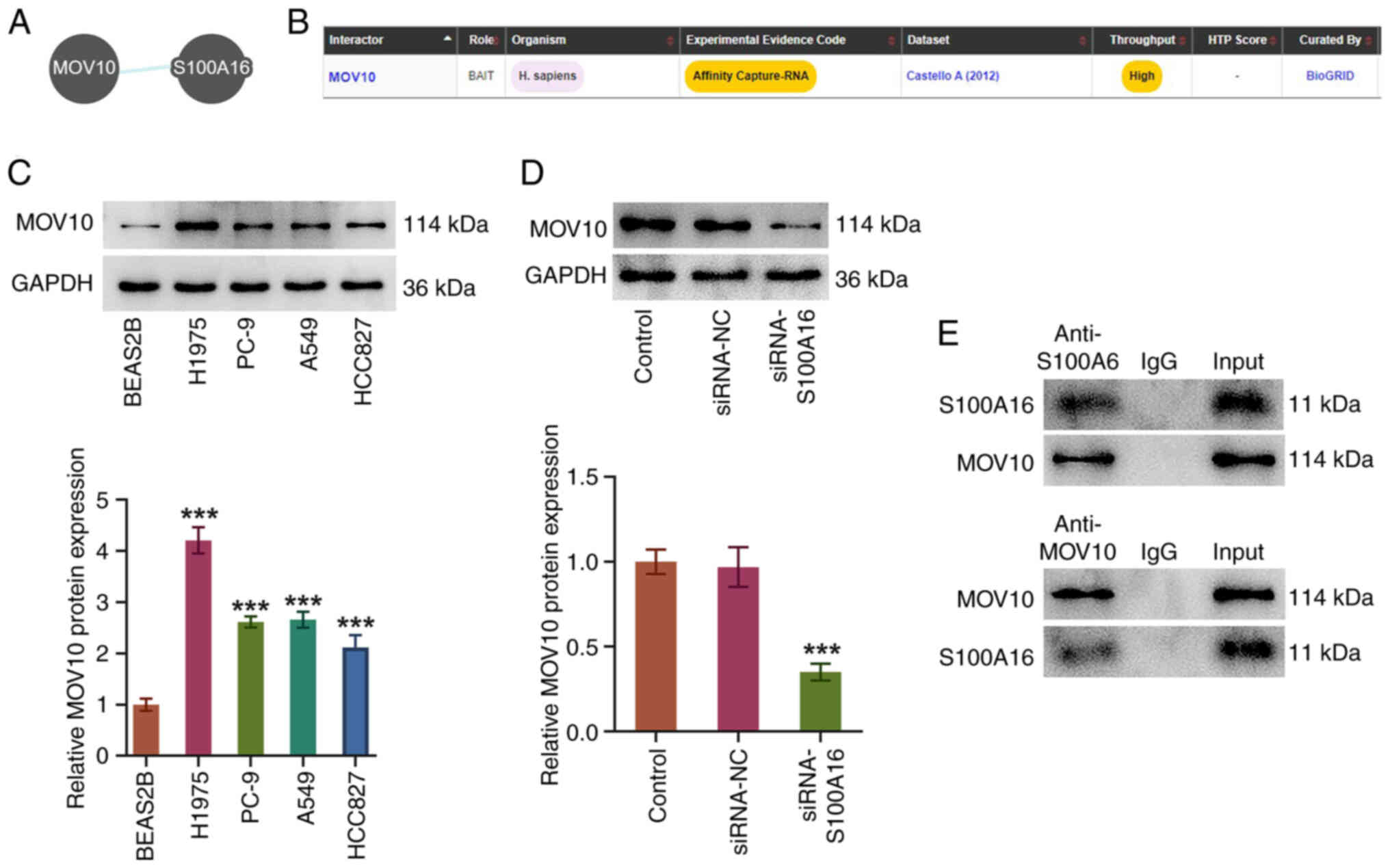
Through the Pathway Commons and BioGRID databases,
it was predicted that MOV10 was a potential target that may bind to
S100A16 (Fig. 4A and B).
Similarly, MOV10 expression in several LUAD cell lines was
assessed, and it was revealed to be elevated in H1975, PC-9, A549
and HCC827 cells compared with that in BEAS2B cells (Fig. 4C). Furthermore, compared with in
the siRNA-NC group, MOV10 protein expression was decreased
following knockdown of S100A16 expression (Fig. 4D). Additionally, Co-IP assays
demonstrated that S100A16 and MOV10 were both co-immunoprecipitated
by MOV10 and S100A16 antibodies, suggesting an interaction between
S100A16 and MOV10 (Fig. 4E).
Knockdown of S100A16 suppresses MOV10
expression to hinder the progression of LUAD
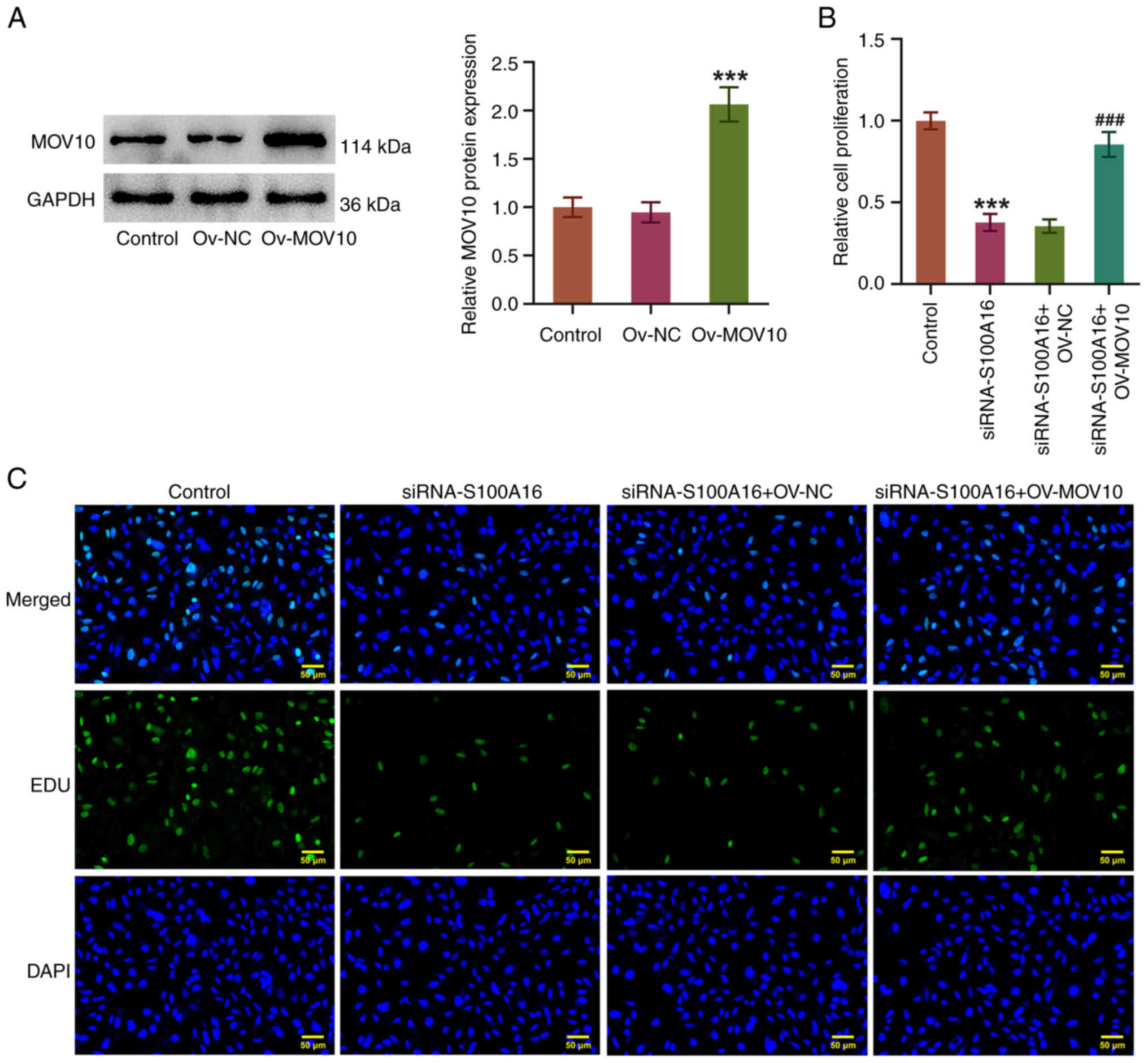
To further confirm the relationship between S100A16
and MOV10 in LUAD cells, the Ov-MOV10 plasmid was transfected into
H1975 cells. Post-transfection with Ov-MOV10, MOV10 expression was
significantly increased compared with that in the Ov-NC group
(Fig. 5A). In addition, it was
demonstrated that the diminished proliferation of H1975 cells
induced by S100A16 knockdown was accelerated again by MOV10
overexpression (Fig. 5B and C).
Moreover, S100A16 knockdown markedly reduced the migration and
invasion of H1975 cells, accompanied by a decrease in MMP2 and MMP9
expression, whereas these effects were reversed by MOV10
overexpression (Fig. 6A-C).
Concurrently, the angiogenic ability of HUVECs was attenuated by
knockdown of S100A16; this was also evidenced by the decreased
expression levels of VEGF and VEGFR2. By contrast, the angiogenic
ability, and the expression levels of VEGF and VEGFR2, were
increased in HUVECs in the siRNA-S100A16 + Ov-MOV10 (CM) + HUVEC
group compared with those in the siRNA-S100A16 + Ov-NC (CM) + HUVEC
group (Fig. 6D and E). In summary,
MOV10 overexpression reversed the suppressive effects of S100A16
knockdown on the aggressiveness of LUAD.
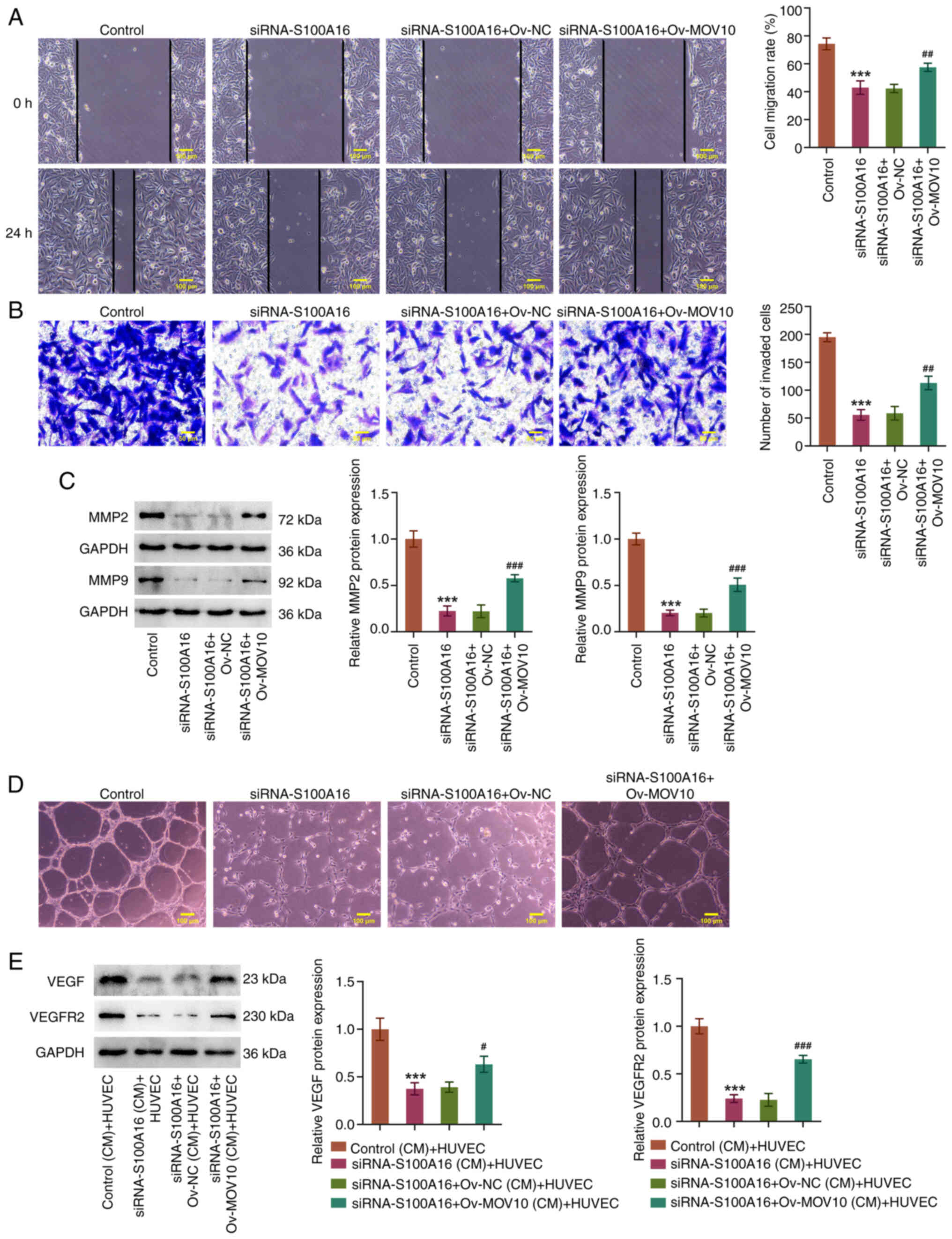 | Figure 6.Knockdown of S100A16 suppresses MOV10
expression to hinder the migration, invasion and angiogenesis of
lung adenocarcinoma cells. (A) Wound healing and (B) Transwell
assays were used to evaluate cell migration and invasion,
respectively. (C) Western blotting was used to examine MMP2 and
MMP9 expression. (D) Tube formation assays were used to estimate
cell angiogenesis. ***P<0.001 vs. Control;
##P<0.01, ###P<0.001 vs. siRNA-S100A16
+ Ov-NC. (E) Western blotting was used to examine VEGF and VEGFR2
expression in HUVECs. ***P<0.001 vs. Control (CM) + HUVEC;
#P<0.05, ###P<0.001 vs. siRNA-S100A16 +
Ov-NC (CM) + HUVEC. CM, conditioned medium; HUVEC, human umbilical
vein endothelial cell; MMP, matrix metallopeptidase; MOV10, Mov10
RNA helicase; NC, negative control; Ov-MOV10, MOV10 overexpression
vector; Ov-NC, empty NC vector; S100A16, S100 calcium-binding
protein A16; siRNA, small interfering RNA; VEGF, vascular
endothelial growth factor; VEGFR2, VEGF receptor 2. |
S100A16 modulates the ECM-receptor
interaction pathway by regulating MOV10-mediated ITGA3 mRNA
stability
Notably, as depicted by the LinkedOmics database,
S100A16 was enriched in the ‘ECM-receptor interaction’ Kyoto
Encyclopedia of Genes and Genomes pathway in LUAD cells (Fig. 7A). S100A16 knockdown significantly
decreased the expression levels of ITGA3 and p-SRC/SRC, which were
both increased by MOV10 overexpression (Fig. 7B). Furthermore, a high abundance of
ITGA3 mRNA was pulled down by the MOV10 antibody (Fig. 7C), implying that MOV10 had a high
affinity for ITGA3 mRNA. Notably, following MOV10 knockdown in
H1975 cells (Fig. 7D), ITGA3 mRNA
expression was decreased, whereas it was increased in response to
MOV10 overexpression (Fig. 7E).
Additionally, the results of the actinomycin D assay demonstrated
that MOV10 or S100A16 knockdown decreased the stability of ITGA3
mRNA (Fig. 7F). However, compared
with in the siRNA-S100A16 + Ov-NC group, MOV10 overexpression
increased the stability of ITGA3 mRNA. Overall, these results
indicated that S100A16 interacted with MOV10 to stabilize ITGA3
mRNA and subsequently regulate the ECM-receptor interaction
pathway.
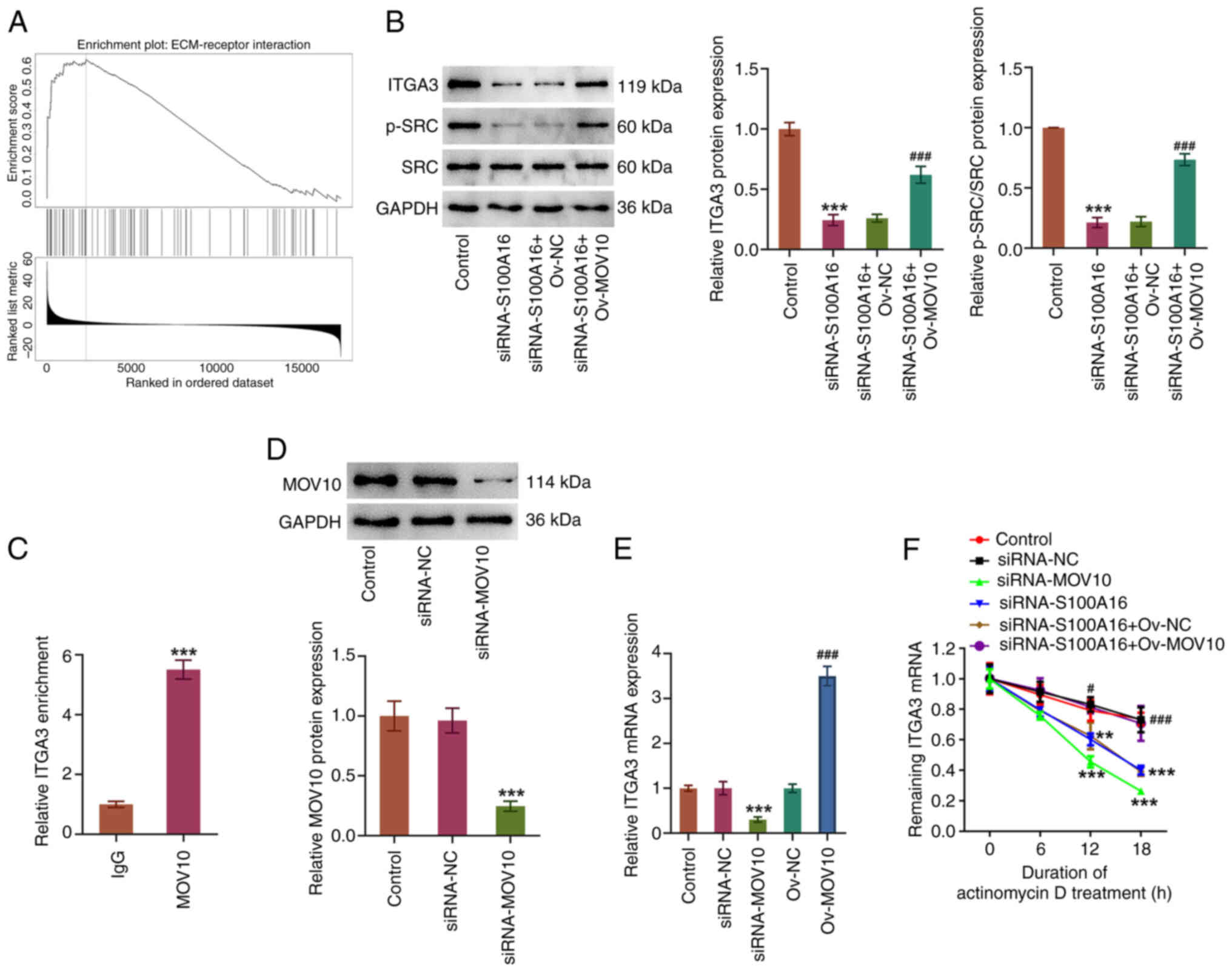 | Figure 7.S100A16 modulates the ECM-receptor
interaction pathway via regulating the MOV10-mediated ITGA3 mRNA
stability. (A) LinkedOmics database was used to predict the
enrichment of S100A16 in the ECM-receptor interaction pathway. (B)
Western blotting was used to examine ITGA3, p-SRC and SRC
expression. ***P<0.001 vs. Control; ###P<0.001 vs.
siRNA-S100A16 + Ov-NC. (C) RNA immunoprecipitation assay was used
to detect the abundance of ITGA3 mRNA pulled down with the MOV10
antibody. ***P<0.001 vs. IgG. (D) Transfection efficacy of
siRNA-MOV10 was detected by western blotting. (E) Reverse
transcription-quantitative PCR analysis of ITGA3 mRNA expression
after MOV10 was knocked down or overexpressed. ***P<0.001 vs.
siRNA-NC; ###P<0.001 vs. Ov-NC. (F) Actinomycin D
assay was used to detect ITGA3 mRNA stability. **P<0.01,
***P<0.001 vs. siRNA-NC; #P<0.05,
###P<0.001 vs. siRNA-S100A16 + Ov-NC. ECM,
extracellular matrix; ITGA3, integrin α3; MOV10, Mov10 RNA
helicase; NC, negative control; Ov-MOV10, MOV10 overexpression
vector; Ov-NC, empty NC vector; p-, phosphorylated; S100A16, S100
calcium-binding protein A16; siRNA, small interfering RNA. |
Discussion
Previous studies have indicated that intricate
biological processes involving genetic and epigenetic alterations
have a key role in tumorigenesis (19,20).
The S100 protein family has a significant role in the regulation of
multiple cancer-related cellular processes, including cell
proliferation, differentiation, migration, invasion and
epithelial-mesenchymal transition (9). Previous studies have reported that
S100A16 may serve as an effective prognostic indicator for various
types of cancer, such as bladder (21), pancreatic (22) and colorectal (10) cancer, and LUAD (11,23).
In the present study, using bioinformatics analysis, S100A16
expression was predicted to be increased in LUAD tissues, which
might be associated with the low overall survival rate of patients
with LUAD. Emerging studies have identified the oncogenic potential
of S100A16 in the majority of cancer types, such as by driving cell
proliferation, migration and invasion (24–26).
In the present study, further investigation showed that S100A16
knockdown hampered the proliferation, migration and invasion of
H1975 cells, which was consistent with the study by Wu et al
(13), in which it was
demonstrated that S100A16 accelerated cell proliferation and
metastasis in LUAD. MMPs are considered critical regulators in
tumor invasion and metastasis, since they are capable of disrupting
the ECM and vascular basement membrane of tumor cells, conferring
the invasive ability of these cells to the basement membrane, and
resulting in tumor cell migration and invasion (27,28).
In the present study, it was discovered that, after S100A16
expression was knocked down, the expression levels of MMP2 and MMP9
were also decreased. Accordingly, the anti-proliferative,
anti-invasive and anti-migratory roles of S100A16 inhibition in
LUAD cells were affirmed.
Angiogenesis is the process by which new blood
vessels form from pre-existing blood vessels (29). Moreover, angiogenesis has been well
established as a prerequisite for tumor progression and metastasis,
through providing oxygen and nutrients to cancer cells and
supplying a route for cancer cell metastasis (30,31).
Additionally, the association of high angiogenic activity with
advanced tumor growth and metastasis in LUAD has been previously
demonstrated (32,33). VEGF is a key regulator of
angiogenesis that typically exerts its biological effects by
binding to its receptors (34).
VEGF and VEGFR2 have been shown to be primarily expressed in lung
cancer cells, and to be closely linked to the occurrence,
development and metastasis of lung cancer (35). Furthermore, previous evidence has
demonstrated that S100A16 knockdown can inhibit angiogenesis, and
decrease VEGF and VEGFR2 expression in renal cancer cells (25). In the present study, the
experimental data highlighted the inhibitory role of S100A16
knockdown in the angiogenesis of HUVECs, as manifested by the
decrease in endothelial capillary-like structures, and the
downregulation of VEGF and VEGFR2 expression.
Interactions among proteins are central to
diversified biological processes, the dysfunction of which is also
implicated in the pathogenesis of cancer (36). In the present study, the Pathway
Commons and BioGRID databases predicted that MOV10 was a potential
protein that may bind to S100A16, which was further verified by
Co-IP assay. In addition, it was observed that MOV10 protein
expression was decreased after S100A16 expression was knocked down.
MOV10 is a novel RBP, the expression of which has been reported to
be 2–3 times higher in cancer cells than that in normal cells
(14). Additionally, it has been
indicated that MOV10 may act as an oncogene in several types of
human cancer (37,38). Particularly, high MOV10 expression
has been implicated in the poor prognosis of LUAD (15), and has been shown to mediate cell
viability, migration and angiogenesis in glioma (39). On this basis, it was further
delineated that MOV10 expression was upregulated in LUAD cells, and
that overexpression of MOV10 partially reversed the effects of
S100A16 knockdown on the proliferation, migration, invasion and
angiogenesis of H1975 cells.
RBPs serve an important role in the regulation of
mRNA stability and translation at the post-transcriptional level
through binding with mRNAs in cancer (40). As predicted by the ENCORI database
in the present study, ITGA3 mRNA may be a downstream target of
MOV10. The findings of the present study demonstrated that a high
abundance of ITGA3 mRNA was pulled down by the MOV10 antibody, and
that MOV10 knockdown reduced both the mRNA stability and expression
of ITGA3 in LUAD cells, suggesting a notable interaction between
MOV10 and ITGA3 mRNA. ITGA3 is a member of the integrin family that
can interact with ECM proteins as a cell surface adhesion molecule
(41). Focal adhesions are
assemblies of cell-ECM linkages mediated by integrin, causing a
cascade activation of focal adhesion kinase and SRC (42). Compelling evidence has suggested
that inactivation of SRC signaling by ING3 can inhibit the
malignant progression of LUAD (43). A recent study has also demonstrated
that early-stage lung cancer may be driven by a transitional cell
state dependent on a KRAS/ITGA3/SRC axis (44). Accumulating reports have indicated
that ITGA3 may be a potential predictor of prognosis in NSCLC
(16), and could contribute to
invasion and angiogenesis in nasopharyngeal carcinoma (45). In the present study, the
LinkedOmics database was used to predict that S100A16 was highly
enriched in the ECM-receptor interaction pathway in LUAD cells, and
knockdown of S100A16 expression was shown to decrease the
expression levels of ITGA3 and p-SRC/SRC, which were increased
following MOV10 overexpression.
Notably, there are some limitations in the present
study. First, only S100A16 knockdown without S100A16 overexpression
was used to detect the effects of S100A16 on MOV10 and the
malignant properties of LUAD cells. Second, only one LUAD cell line
was used to explore the malignant properties of these cells, and
other cell lines should be included in future investigation.
Finally, future studies should aim to analyze the effects of ITGA3
on the proliferation and migration of LUAD cells, which were not
assessed in the present study.
In conclusion, the present study demonstrated that
S100A16 can facilitate cell proliferation, migration, invasion and
angiogenesis in LUAD, most likely as a result of increased ITGA3
mRNA stability and regulation of the ECM-receptor interaction
pathway via binding to MOV10. The present study supports the
significance of S100A16 in LUAD, which may provide value for LUAD
therapy.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
LY and ZZ contributed to the conception and design
of the present study. LY and AS analyzed the data and drafted the
manuscript. AS and RW generated the figures. LY, AS, RW and ZZ
conducted the experiments. ZZ reviewed and edited the manuscript.
All authors read and approved the final version of the manuscript.
LY and ZZ confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Parkin DM, Piñeros M, Znaor A and Bray F: Cancer statistics for the
year 2020: An overview. Int J Cancer. Apr 5–2021.(Epub ahead of
print). View Article : Google Scholar
|
|
2
|
Cao M and Chen W: Epidemiology of lung
cancer in China. Thorac Cancer. 10:3–7. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Skřičková J, Kadlec B, Venclíček O and
Merta Z: Lung cancer. Cas Lek Cesk. 157:226–236. 2018.PubMed/NCBI
|
|
4
|
Myers DJ and Wallen JM: Lung
adenocarcinoma. StatPearls. StatPearls Publishing; Treasure Island,
FL: 2023
|
|
5
|
Blandin Knight S, Crosbie PA, Balata H,
Chudziak J, Hussell T and Dive C: Progress and prospects of early
detection in lung cancer. Open Biol. 7:1700702017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang P, Zhu S, Liang X, Zhang Q, Liu C
and Song L: Revisiting lung cancer metastasis: Insight from the
functions of long non-coding RNAs. Technol Cancer Res Treat.
20:153303382110384882021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Slim A, Kamoun H, Hadidene Y, Smadhi H,
Meddeb A and Megdiche ML: Postoperative recurrence of primary lung
cancer: Anatomo-clinical and therapeutic study. Tunis Med.
99:560–568. 2021.PubMed/NCBI
|
|
8
|
Bresnick AR, Weber DJ and Zimmer DB: S100
proteins in cancer. Nat Rev Cancer. 15:96–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Basnet S, Vallenari EM, Maharjan U, Sharma
S, Schreurs O and Sapkota D: An update on S100A16 in human cancer.
Biomolecules. 13:10702023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sun X, Wang T, Zhang C, Ning K, Guan ZR,
Chen SX, Hong TT and Hua D: S100A16 is a prognostic marker for
colorectal cancer. J Surg Oncol. 117:275–283. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Saito K, Kobayashi M, Nagashio R, Ryuge S,
Katono K, Nakashima H, Tsuchiya B, Jiang SX, Saegusa M, Satoh Y, et
al: S100A16 is a prognostic marker for lung adenocarcinomas. Asian
Pac J Cancer Prev. 16:7039–7044. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Katono K, Sato Y, Kobayashi M, Nagashio R,
Ryuge S, Igawa S, Ichinoe M, Murakumo Y, Saegusa M and Masuda N:
S100A16, a promising candidate as a prognostic marker for
platinum-based adjuvant chemotherapy in resected lung
adenocarcinoma. Onco Targets Ther. 10:5273–5279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu C, Yang J, Lin X, Li R and Wu J:
miR-508-5p serves as an anti-oncogene by targeting S100A16 to
regulate AKT signaling and epithelial-mesenchymal transition
process in lung adenocarcinoma cells. Am J Med Sci. 365:520–531.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nakano M, Kakiuchi Y, Shimada Y, Ohyama M,
Ogiwara Y, Sasaki-Higashiyama N, Yano N, Ikeda F, Yamada E,
Iwamatsu A, et al: MOV10 as a novel telomerase-associated protein.
Biochem Biophys Res Commun. 388:328–332. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mao CG, Jiang SS, Shen C, Long T, Jin H,
Tan QY and Deng B: BCAR1 promotes proliferation and cell growth in
lung adenocarcinoma via upregulation of POLR2A. Thorac Cancer.
11:3326–3336. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Q, Ma W, Chen S, Tian EC, Wei S, Fan
RR, Wang T, Zhou C and Li T: High integrin α3 expression is
associated with poor prognosis in patients with non-small cell lung
cancer. Transl Lung Cancer Res. 9:1361–1378. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Recillas-Targa F: Cancer epigenetics: An
overview. Arch Med Res. 53:732–740. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanwal R, Gupta K and Gupta S: Cancer
epigenetics: An introduction. Methods Mol Biol. 1238:3–25. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Katsumata H, Matsumoto K, Yanagita K,
Shimizu Y, Hirano S, Kitajima K, Koguchi D, Ikeda M, Sato Y and
Iwamura M: Expression of S100A16 is associated with biological
aggressiveness and poor prognosis in patients with bladder cancer
who underwent radical cystectomy. Int J Mol Sci. 24:145362023.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen T, Xia DM, Qian C and Liu SR:
Integrated analysis identifies S100A16 as a potential prognostic
marker for pancreatic cancer. Am J Transl Res. 13:5720–5730.
2021.PubMed/NCBI
|
|
23
|
Chen D, Luo L and Liang C: Aberrant
S100A16 expression might be an independent prognostic indicator of
unfavorable survival in non-small cell lung adenocarcinoma. PLoS
One. 13:e01974022018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fang D, Zhang C, Xu P, Liu Y, Mo X, Sun Q,
Abdelatty A, Hu C, Xu H, Zhou G, et al: S100A16 promotes metastasis
and progression of pancreatic cancer through FGF19-mediated AKT and
ERK1/2 pathways. Cell Biol Toxicol. 37:555–571. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang N, Wang R, Tang J, Gao J, Fang Z,
Zhang M, Shen X, Lu L and Chen Y: Calbindin S100A16 promotes renal
cell carcinoma progression and angiogenesis via the VEGF/VEGFR2
signaling pathway. Contrast Media Mol Imaging. 2022:56020112022.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
You X, Li M, Cai H, Zhang W, Hong Y, Gao
W, Liu Y, Liang X, Wu T, Chen F and Su D: Calcium binding protein
S100A16 expedites proliferation, invasion and
epithelial-mesenchymal transition process in gastric cancer. Front
Cell Dev Biol. 9:7369292021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jabłońska-Trypuć A, Matejczyk M and
Rosochacki S: Matrix metalloproteinases (MMPs), the main
extracellular matrix (ECM) enzymes in collagen degradation, as a
target for anticancer drugs. J Enzyme Inhib Med Chem. 31:177–183.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cabral-Pacheco GA, Garza-Veloz I,
Castruita-De la Rosa C, Ramirez-Acuña JM, Perez-Romero BA,
Guerrero-Rodriguez JF, Martinez-Avila N and Martinez-Fierro ML: The
roles of matrix metalloproteinases and their inhibitors in human
diseases. Int J Mol Sci. 21:97392020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dudley AC and Griffioen AW: Pathological
angiogenesis: Mechanisms and therapeutic strategies. Angiogenesis.
26:313–347. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rajabi M and Mousa SA: The role of
angiogenesis in cancer treatment. Biomedicines. 5:342017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Majidpoor J and Mortezaee K: Angiogenesis
as a hallmark of solid tumors-clinical perspectives. Cell Oncol
(Dordr). 44:715–737. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou Q, Chen X, Chen Q and Hao L: Analysis
of angiogenesis-related signatures in the tumor immune
microenvironment and identification of clinical prognostic
regulators in lung adenocarcinoma. Crit Rev Eukaryot Gene Expr.
33:1–16. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Onn A, Bar J and Herbst RS: Angiogenesis
inhibition and lung-cancer therapy. Lancet Oncol. 15:124–125. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Melincovici CS, Boşca AB, Şuşman S,
Mărginean M, Mihu C, Istrate M, Moldovan IM, Roman AL and Mihu CM:
Vascular endothelial growth factor (VEGF)-key factor in normal and
pathological angiogenesis. Rom J Morphol Embryol. 59:455–467.
2018.PubMed/NCBI
|
|
35
|
Chen H, Cong Q, Du Z, Du Z, Liao W, Zhang
L, Yao Y and Ding K: Sulfated fucoidan FP08S2 inhibits lung cancer
cell growth in vivo by disrupting angiogenesis via targeting
VEGFR2/VEGF and blocking VEGFR2/Erk/VEGF signaling. Cancer Lett.
382:44–52. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cheng SS, Yang GJ, Wang W, Leung CH and Ma
DL: The design and development of covalent protein-protein
inter-action inhibitors for cancer treatment. J Hematol Oncol.
13:262020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang D, Hu Z, Xu J, Tang Y, Wang Y, Cai Q
and Zhu Z: MiR-760 enhances sensitivity of pancreatic cancer cells
to gemcitabine through modulating Integrin β1. Biosci Rep.
39:BSR201923582019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
El Messaoudi-Aubert S, Nicholls J,
Maertens GN, Brookes S, Bernstein E and Peters G: Role for the
MOV10 RNA helicase in polycomb-mediated repression of the INK4a
tumor suppressor. Nat Struct Mol Biol. 17:862–868. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
He Q, Zhao L, Liu X, Zheng J, Liu Y, Liu
L, Ma J, Cai H, Li Z and Xue Y: MOV10 binding circ-DICER1 regulates
the angiogenesis of glioma via miR-103a-3p/miR-382-5p mediated ZIC4
expression change. J Exp Clin Cancer Res. 38:92019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li W, Deng X and Chen J: RNA-binding
proteins in regulating mRNA stability and translation: roles and
mechanisms in cancer. Semin Cancer Biol. 86:664–677. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Shiota T, Li TC, Nishimura Y, Yoshizaki S,
Sugiyama R, Shimojima M, Saijo M, Shimizu H, Suzuki R, Wakita T, et
al: Integrin α3 is involved in non-enveloped hepatitis E virus
infection. Virology. 536:119–124. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hamidi H and Ivaska J: Every step of the
way: integrins in cancer progression and metastasis. Nat Rev
Cancer. 18:533–548. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cheng S, Li M, Zheng W, Li C, Hao Z, Dai
Y, Wang J, Zhuo J and Zhang L: ING3 inhibits the malignant
progression of lung adenocarcinoma by negatively regulating ITGB4
expression to inactivate Src/FAK signaling. Cell Signal.
117:1110662024. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Moye AL, Dost AF, Ietswaart R, Sengupta S,
Ya V, Aluya C, Fahey CG, Louie SM, Paschini M and Kim CF:
Early-stage lung cancer is driven by a transitional cell state
dependent on a KRAS-ITGA3-SRC axis. EMBO J. 43:2843–2861. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tang XR, Wen X, He QM, Li YQ, Ren XY, Yang
XJ, Zhang J, Wang YQ, Ma J and Liu N: MicroRNA-101 inhibits
invasion and angiogenesis through targeting ITGA3 and its systemic
delivery inhibits lung metastasis in nasopharyngeal carcinoma. Cell
Death Dis. 8:e25662017. View Article : Google Scholar : PubMed/NCBI
|