Introduction
The global prevalence of myopia has risen in recent
decades and is estimated to increase from 30 to 50% (1), with high myopia expected to affect
10–20% of the population by 2050. Myopia often progresses to high
myopia, which involves notable elongation of the eyeball and higher
refractive error, leading to an increased risk of complications
such as retinal detachment. Blindness caused by high myopia affects
3.8% of adults in Singapore and 3.1% of adults in Beijing, China,
highlighting the significant public health impact of this condition
in these regions (2,3). In early life, myopia prevalence
varies by age, typically worsening from adolescence to adulthood
and continuing to progress into high myopia later in life (4,5).
High myopia is a key risk factor for eye diseases such as
open-angle glaucoma, cataracts and myopic macular degeneration, and
also leads to irreversible loss of vision such as retinal
detachment and choroidal neovascularization, both of which are key
causes of visual impairment (6–8).
Previous studies have suggested that myopia is
associated with metabolic disorders that cause changes in the
composition of aqueous humor (9–16).
N-3 polyunsaturated fatty acids, which serve as vasodilators with
an anti-inflammatory effect, may influence the progression of
myopia by suppressing choroidal thinning (9). Hypoxia activates the
hypoxia-inducible factor (HIF)-1α signalling pathway, which
promotes scleral fibroblast-myofibroblast transition and
remodelling of extracellular matrix (10). Circular RNA zinc finger protein 609
acting as microRNA (miRNA or miR)-615 sponge to regulate retinal
neurodegeneration, and thus influencing refractive state, is a
promising target for the treatment of retinal neurodegeneration
(11). Recent studies have found
that abnormal metabolism of thyroid hormones, β/γ-crystallin, MMPs,
tissue inhibitors of metalloproteases and transforming growth
factor-β (TGF-β) in the aqueous humor of patients with diabetes
mellitus or highly myopic cataracts affects the refractive state by
influencing ocular axis length, causing the onset and progression
of myopia (12–14). Additionally, as collagen decreases
with aging and scleral tensile strength diminishes, this affects
ocular accommodation, causing changes in refractive error over
time. Altered hyaluronic acid metabolism in vitreous and aqueous
humor also contributes to vitreous opacity and myodesopsia of
myopia in older adults compared with younger individuals (15,16).
Although studies have explored age-related pathways associated with
high myopia, such as TGF-β1 and scleral HIF-1α signalling pathways,
to the best of our knowledge, comparative analyses of
metabolism-associated genes between young and old adults with high
myopia are rare (13,17). Studies have identified miRNAs, a
class of small, non-coding RNAs, as key modulators of gene
expression with sequence-specific function (18,19).
These molecules serve essential roles in regulating key metabolic
pathways, including those involved in glucose, lipid and
high-density lipoprotein metabolism (20,21)
and are ubiquitously expressed in the ocular tissues of humans and
other mammals (22). Notably,
certain miRNAs, such as miR-3144-3p, miR-320a, miR-9 and miR-22,
have been linked to the onset and progression of ocular disease
such as glaucoma and age-related macular degeneration (23,24).
Previous studies on miRNA sequencing using aqueous humor have used
patients with age-associated cataracts as normal controls (25–27).
However, the miRNAs uniquely associated with high myopia,
particularly those whose expression is independent of
cataract-associated factors, remain underexplored.
Numerous studies have investigated pathogenesis and
treatment of myopia, such as outdoor activity (28), as well as various treatments for
high myopia such as surgery, medication and optical interventions
that can help control its progression (29–32).
However, to minimize the incidence of high myopia in the future, it
is necessary to explore the mechanisms of myopia development at the
molecular level. The present study used bioinformatics to compare
miRNAs in patients with age-related cataracts with high myopia and
cataracts without myopia, as well as compare high myopia in young
adults and age-related cataracts without myopia, and to screen for
common miRNAs to obtain predicted target genes. Based on
metabolism-related genes and target genes, hub genes related to
both high myopia and metabolism were identified and the miRNAs that
regulate them characterized to offer new insights into the
treatment of high myopia.
Materials and methods
Subjects
Aqueous humor samples from 54 patients, aged 17–79
years, with a male-to-female ratio of 1:2, were collected from the
First Affiliated Hospital of Chongqing Medical University
(Chongqing, China) between June 2022 and September 2023. Patients
aged ≤30 years were assigned to the younger group, and those aged
≥60 to the older group. Samples from younger patients were obtained
during implantable collamer lens surgery, while those from older
patients were collected during cataract surgery. To ensure adequate
volume for sequencing accuracy while minimizing individual
variability, aqueous humor samples were carefully collected during
the surgery, with each sample size ≤150 µl to maintain normal
intraocular pressure. Samples were combined from samples of three
patients matched according to age, sex, presence or absence of
cataracts and refractive status, resulting in a total of 18 mixed
samples. These samples were then divided into three groups
(n=6/group): Young with high myopia (YH), age-related cataracts
with high myopia (AH) and age-related cataracts without high myopia
(AN). A total of three mixed samples from each group was taken for
sequencing and the remaining mixed samples used for subsequent
validation. High myopia inclusion criteria were spherical
equivalent (SE) ≤-6.0 diopters or an axial length (AL) >26 mm in
either eye (33). The diagnosis of
age-associated cataracts followed internationally recognized
criteria from the American Academy of Ophthalmology's Preferred
Practice Pattern guidelines (34,35).
Inclusion criteria were patients with a need for vision correction
or age-related cataracts who exhibited clinical signs of nuclear,
cortical or posterior subcapsular lens opacities. Exclusion
criteria were patients who had undergone vitreous cavity
injections, anterior chamber paracentesis or any treatments that
could potentially alter the aqueous humor environment prior to
surgery. Additionally, patients with history of hypertension,
abnormal blood glucose levels, autoimmune disease or
immunodeficiency were excluded.
miRNA sequencing and differential
analysis
Total RNA was extracted using Total RNA Purification
Kit (cat. no. TRK1001; LC Sciences, Houston, USA). Bioanalyzer 2100
and RNA 6000 Nano LabChip kit (Agilent Technologies, Inc.) were
utilized to analyse the quantity and purity of total RNA. A total
of ~1 µg total RNA was used to construct a small RNA library, using
TruSeq Small RNA Library Prep kits (cat. no. RS-200-0012; Illumina,
Inc.) according to the manufacturer's instructions. HiSeq SBS Kit
v4 (250 cycles; cat. no. FC-401-4003; Illumina, San Diego, USA) was
used for sequencing. The final library loading concentration was
3–4 nM, calculated based on the actual quality of each sample.
Single-end sequencing (50 bp) was performed on an Illumina
Hiseq2500 at Lc-Bio Technologies according to the manufacturer's
protocol. For the miRNA-seq data, Reads with reference sequences of
human miRNA (miRBase mature human.fa). Differentially expressed
miRNAs1 (DEmiRNA1), DEmiRNA2, and DEmiRNA3 between AH and AN, YH
and AN, YH and AH groups were sifted out by DESeq2 package (version
1.32.0) setting |Log2FC|>1 and P<0.05,
respectively. In order to explore the miRNAs that played a role in
both AH and AN, YH and AN, we intersected DEmiRNA1 and DEmiRNA2 to
yield intersected miRNAs, and the remained miRNAs were treated as
non-intersected miRNAs. The intersected DEmiRNAs were considered
high myopia characteristic miRNAs and their target genes were
predicted via mirtarbase data of mirNet database (https://www.mirnet.ca).
Screening of metabolism-related hub
genes for high myopia
Metabolism-related genes were downloaded from MsigDB
database (https://www.gsea-msigdb.org/gsea/msigdb) (36), and the genes intersecting with
miRNA target genes were considered metabolism-related high myopia
genes. Functional enrichment analysis of target genes using
Metascape (http://metascape.org/gp/index.html#/main/step1) and
prediction of disease targets using DisGeNET database (https://disgenet.com) was performed. The
metabolism-associated high myopia genes were enriched into the
Reactome passages, and the network was visualized using Cytoscape
software (version 3.10.1, http://cytoscape.org). Protein interaction (PPI)
network construction was performed for using the STRING (https://string-db.org) website with a confidence level
of 0.1 to screen the genes. Next, the cytohubba function of
Cytoscape (version 3.10.1) was used for identifying hub genes and
the top ten metabolism-related high myopia hub genes were obtained
by sorting degree function.
Network construction of hub genes
The GeneMANIA database (https://genemania.org/) was used for the analysis of
hub genes and prediction of their function. The comparative
toxicogenomics database (https://ctdbase.org) was utilized to predict which
drugs hub genes would be affected by. The mirNet website was used
to predict long non-coding (lnc)RNAs of miRNAs related to the hub
genes. Regulatory relationships of characteristic miRNA-hub genes
and predicted transcription factors (TFs) of hub genes via ChEA3
website (https://maayanlab.cloud/chea3). All network diagrams
were visualized with Cytoscape software (version 3.10.1).
Reverse-transcription quantitative
(RT-q)PCR
Total RNA was isolated from intraocular aqueous
humor using a miEASY microRNA Serum/Plasma kit (cat. no. RN4601;
Aidlab Biotechnologies, Ltd.). miRNA was reverse-transcribed into
cDNA using the miRNA 1st Strand cDNA Synthesis kit (Stem-loop)
[cat. no. AG11743; Hunan Aikerui Bioengineering Co., Ltd.)
according to the manufacturer's protocol. qPCR was performed using
the 2X Universal SYBR Green Fast qPCR Mix (cat. no. RK21203;
ABclonal Biotech Co., Ltd.) according to the manufacturer's
instructions. The thermocycling conditions comprised initial
denaturation at 95°C for 10 min, followed by 40 cycles of
denaturation at 92°C for 30 sec and annealing/extension at 60°C for
35 sec. The relative expression levels were ascertained by the
2−ΔΔCq method (37),
with the expression levels normalized to U6. The primer sequences
were as follows: miR-490 forward, 5′-CAACCTGGAGGACTCCATGC-3′ and
reverse, 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCAGCAT-3′;
miR-4423 forward, 5′-CGCGATAGGCACCAAAAAG-3′ and reverse,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGGATACGACTTGTTG-3′ and U6
forward, 5′-GGAACGATACAGAGAAGATTAGC-3′ and reverse
5′-TGGAACGCTTCACGAATTTGCG-3′.
Statistical analysis
Statistical analysis and diagram generation were
conducted using SPSS (version 26.0, IBM Corp.), TBtools (version
2.047, http://github.com/CJ-Chen/TBtools-Manual) and GraphPad
Prism (version 10.1.2, Dotmatics). Data are presented as the mean ±
SD, with three independent experimental repeats. Shapiro-Wilk test
was employed to assess normality. Comparisons of >2 groups were
performed by one-way ANOVA and Bonferroni post hoc tests. When
comparing two groups, data following a normal distribution were
analysed using a t-test, while the Mann-Whitney U test was employed
for non-normally distributed data. P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
There were significant differences in age,
refraction or ocular axis length between each group (Table I). AN and AH groups were
significantly older than the YH group. For SE, both the YH and AH
groups showed significant differences compared with the AN group.
For AL, both the YH and AH groups had significantly longer AL
compared with the AN group.
 | Table I.Basic information of enrolled
patients. |
Table I.
Basic information of enrolled
patients.
| Group | Sample | Age, years | Sex | SE, diopter | AL, mm |
|---|
| YH | YH1 | 23.33±5.51 | F | −10.92±1.81 | 29.57±2.32 |
|
| YH2 | 20.33±2.89 | F | −10.83±1.23 | 28.67±1.55 |
|
| YH3 | 24.33±6.03 | M | −12.17±2.01 | 30.37±1.82 |
| AH | AH1 | 72.67±3.06 | F | −9.00±2.00 | 28.77±1.61 |
|
| AH2 | 65.67±4.93 | F | −13.08±3.47 | 30.03±2.70 |
|
| AH3 | 65.00±4.00 | M | −13.83±5.01 | 30.07±1.89 |
| AN | AN1 | 70.67±2.45 | F | −1.42±2.45 | 24.23±0.72 |
|
| AN2 | 72.33±1.00 | F | +1.00±1.00 | 23.87±0.71 |
| | AN3 | 68.33±0.58 | M | +0.33±0.58 | 23.50±1.04 |
Sequencing data and differential
expression analysis
For sequencing data of transcriptome miRNAs, reads
were compared with the reference sequence of human miRNA
(miRBase_mature_human.fa). Count values for 2,656 miRNAs were
obtained from 9 samples. A total of 18 miRNAs were significantly DE
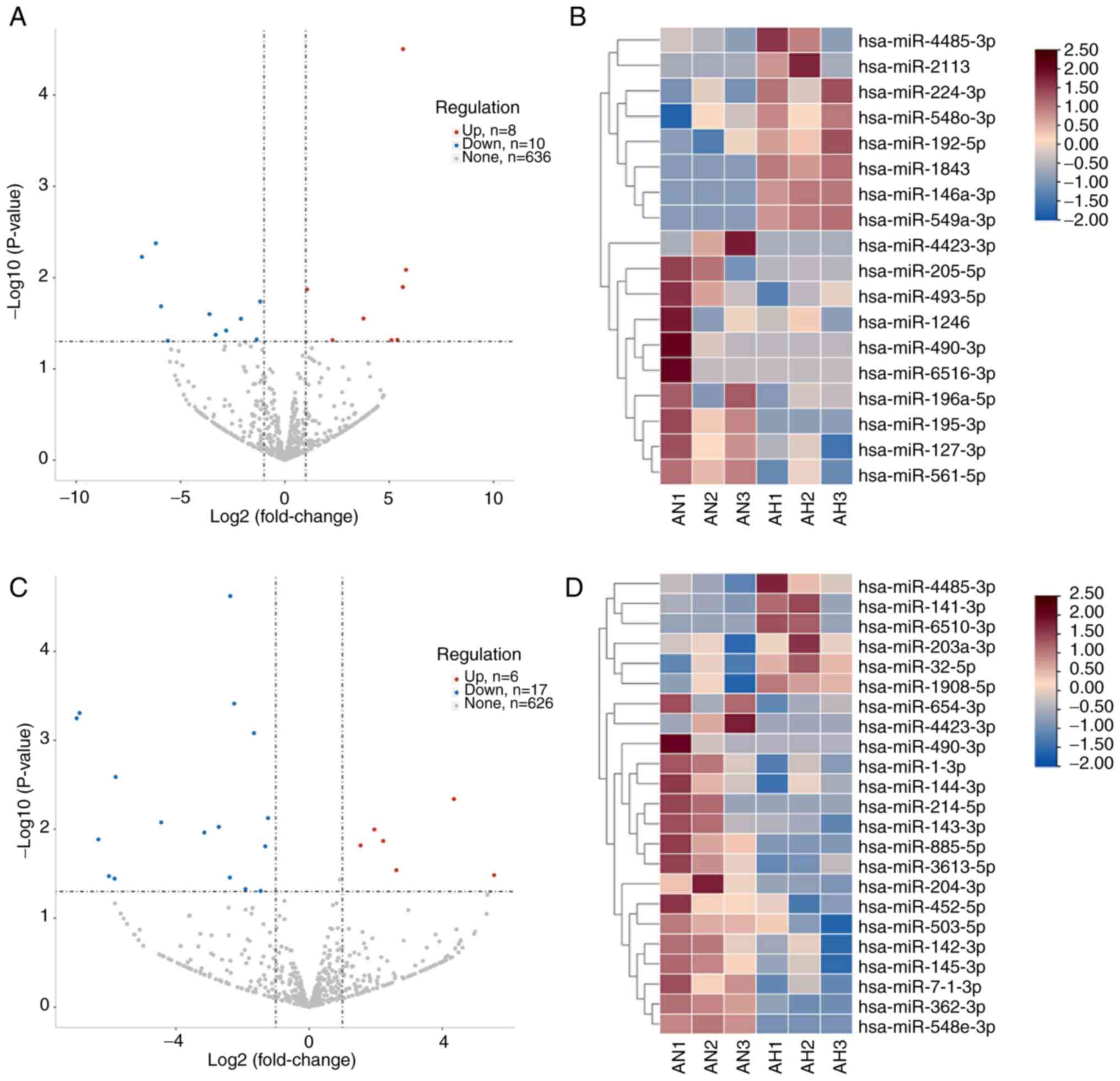
between AH and AN group, of which eight were up- and 10 were
downregulated (Fig. 1A and B). A
total of 23 miRNAs were significantly DE between the YH and AN
group, of which six were up- and 17 were downregulated (Fig. 1C and D). Differences between YH and
AH group are shown in Fig.
S1.
Analysis of high myopic characterized
miRNAs and their target genes
Intersecting DE miRNAs in AH and YH were
hsa-miR-490-3p, hsa-miR-4423-3p and hsa-miR-4485-3p, which were
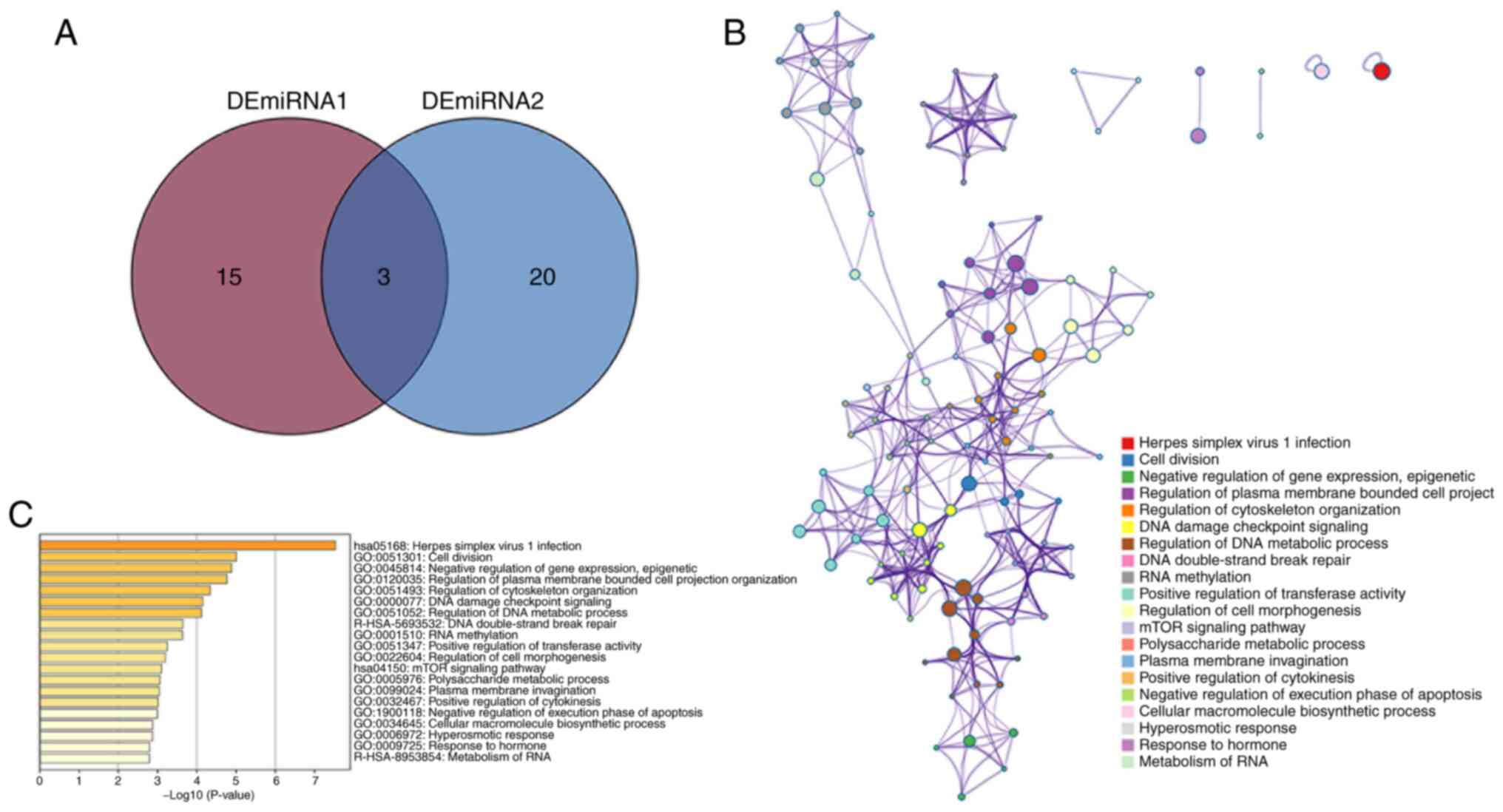
defined as the high myopic miRNAs (Fig. 2A). Enrichment of non-intersecting
miRNAs is shown in Fig. S2. The
target genes of the three characterized miRNAs were predicted from
the mirtarbase data of mirNet database and 289 target genes were
obtained. Functional enrichment analysis of the 289 genes was
performed using Metascape and a total of 169 terms was enriched
(Fig. 2B and C).
Metabolism-related target gene
screening and analysis
A total of 1,717 metabolism-related genes were
obtained, included 19 that intersected with the aforementioned 289
target genes (Fig. 3A). Functional
enrichment analysis showed 22 terms were enriched (Fig. 3B-D). DisGeNET database for disease
target prediction revealed 24 terms were enriched, of which the top
20 diseases related to target genes were highlighted (Fig. 3E).
Screening and analysis of hub target
genes
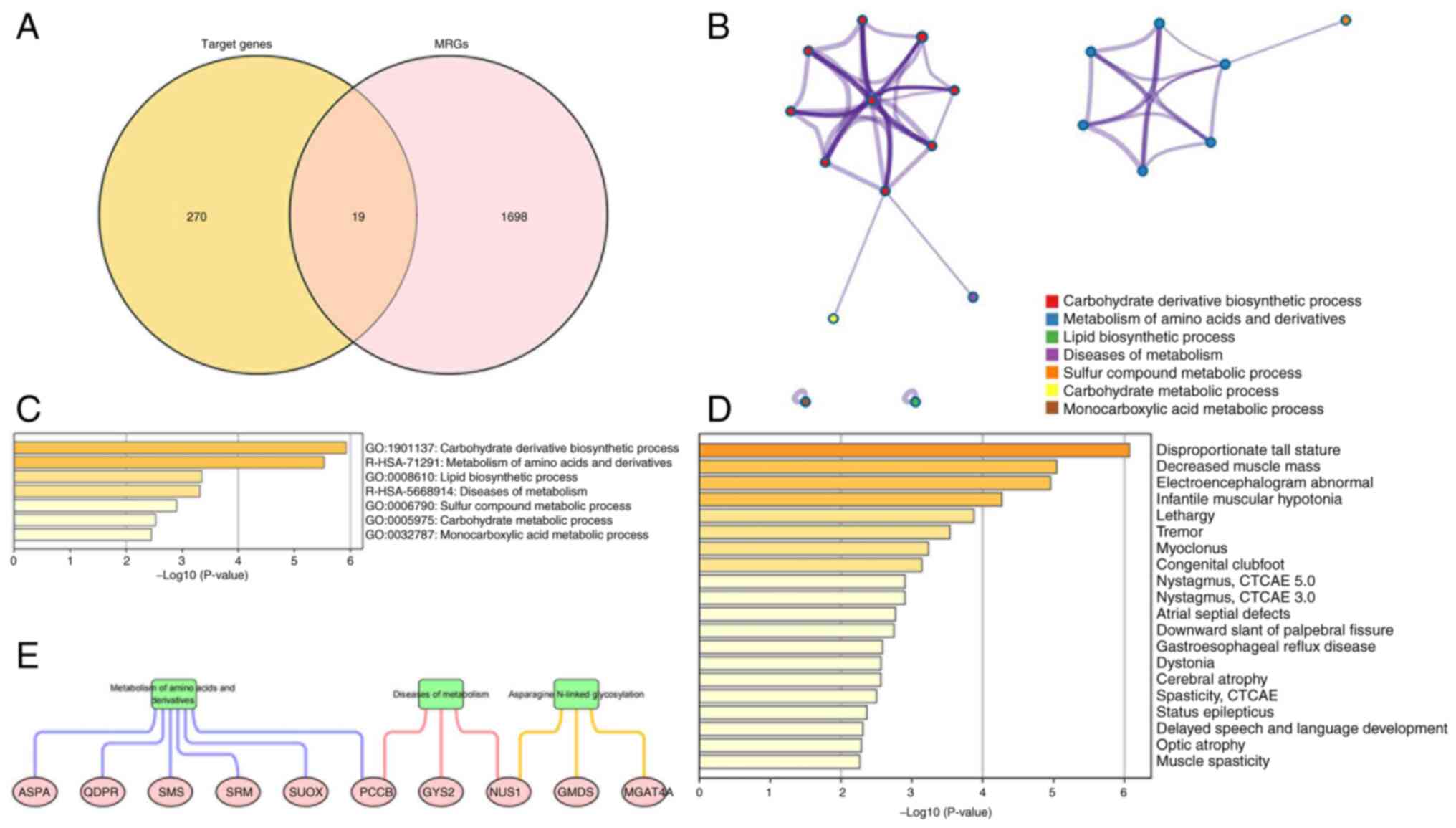
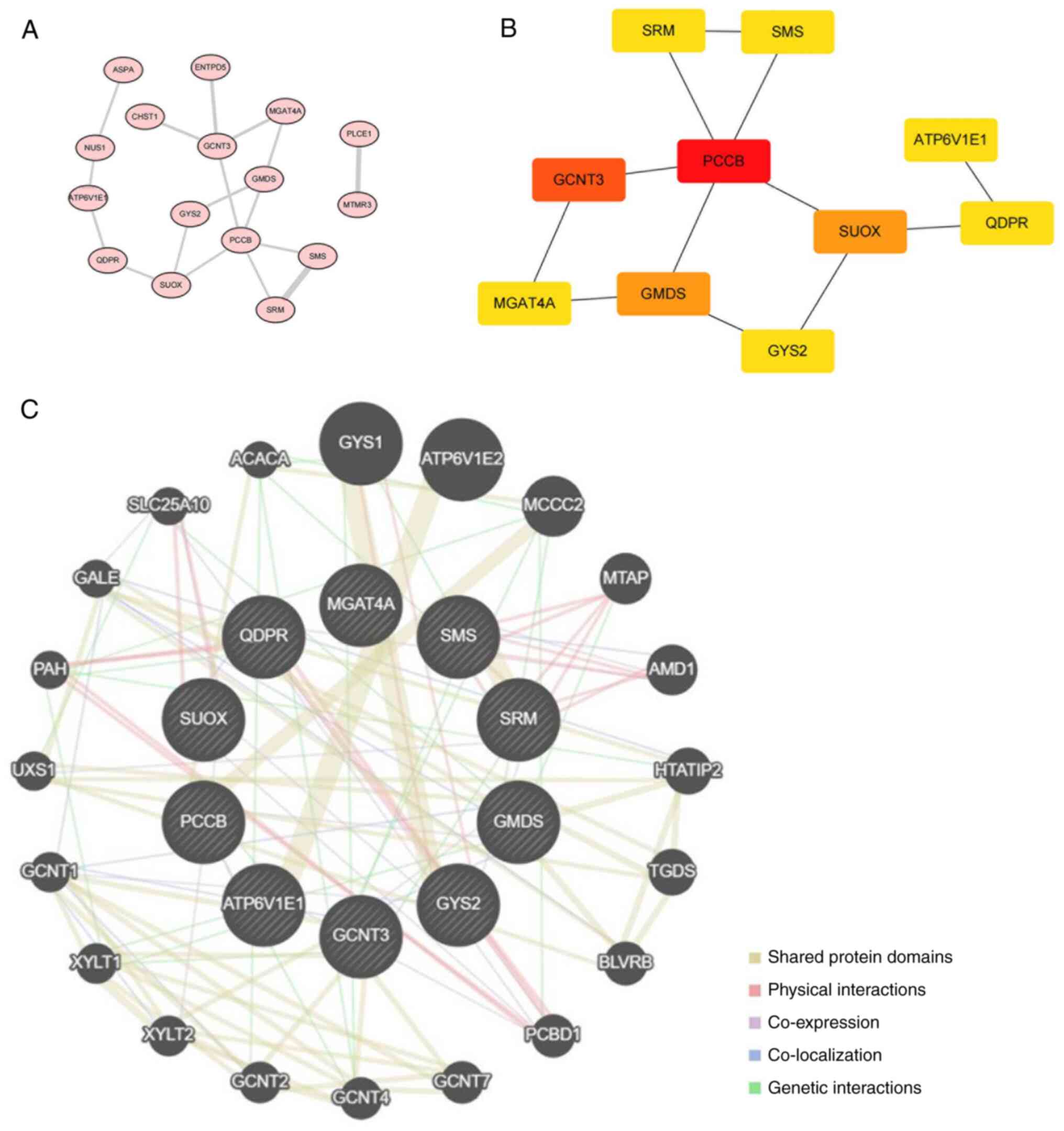
STRING was utilized to explore the interactions
between 19 metabolism-related target genes. During PPI network
construction, three discrete proteins were revealed without any
edges or sub-networks, so only 16 proteins were displayed in the
interaction network (Fig. 4A). The
top 10 hub target genes, propionyl-CoA Carboxylase Subunit β
(PCCB), Glucosaminyl (N-Acetyl) Transferase 3 (GCNT3), GDP-Mannose
4,6-Dehydratase (GMDS), sulfite Oxidase (SUOX), Spermidine Synthase
(SRM), Spermine Synthase (SMS), ATPase H+ Transporting V1 Subunit
E1 (ATP6V1E1), Quinoid dihydropteridine Reductase (QDPR), Glycogen
Synthase 2 (GYS2), Alpha-1,3-Mannosyl-Glycoprotein
4-Beta-N-Acetylglucosaminyltransferase A (MGAT4A), were obtained
(Fig. 4B). The GeneMANIA database
was used to obtain 20 genes that may share protein structural
domains, have physical interactions, co-expression, co-localization
and gene interactions with these hub target genes (Fig. 4C).
Drug prediction of hub target
genes
After obtaining hub target genes, CTD database was
used to predict drugs that would affect their function. Drugs
applicable to the human species that would cause an increase or
decrease in the expression of the corresponding gene were selected
(Table II). A total of 17 drugs
was identified. Based on the number of genes affected,
cyclosporine, tretinoin, acetaminophen, tetrachlorodibenzodioxin,
and benzopyrene had the most extensive impact.
 | Table II.Drug predictions of the hub
genes. |
Table II.
Drug predictions of the hub
genes.
| Drug/ID | Gene | Regulation |
|---|
|
Cyclosporine/D016572 | SUOX | Down |
|
| QDPR | Down |
|
| GCNT3 | Up |
|
| SRM | Up |
|
Tretinoin/D014212 | SRM | Down |
|
| SUOX | Up |
|
| MGAT4A | Up |
|
Acetaminophen/D000082 | PCCB | Down |
|
| GCNT3 | Down |
|
| GMDS | Down |
|
Estradiol/D004958 | GCNT3 | Up |
|
| SRM | Up |
|
Perfluoro-n-nonanoic acid/C101816 | SUOX | Down |
|
Dihydrotestosterone/D013196 | SMS | Up |
| Bisphenol
A/C006780 | SMS | Up |
| Cobaltous
chloride/C018021 | SRM | Down |
| Valproic
acid/D014635 | GMDS | Up |
|
| QDPR | Up |
|
Cisplatin/D002945 | GMDS | Down |
|
Tetrachlorodibenzodioxin/D013749 | GMDS | Down |
|
| GYS2 | Down |
|
| GCNT3 | Up |
|
Benzo(a)pyrene/D001564 | GMDS | Down |
|
| GYS2 | Down |
|
| GCNT3 | Up |
| Nickel/D009532 | SRM | Up |
|
| MGAT4A | Up |
| Aflatoxin
B1/D016604 | GMDS | Down |
| Trichostatin
A/C012589 | MGAT4A | Down |
| Zoledronic
acid/D000077211 | GCNT3 | Up |
|
Azathioprine/D001379 | GCNT3 | Up |
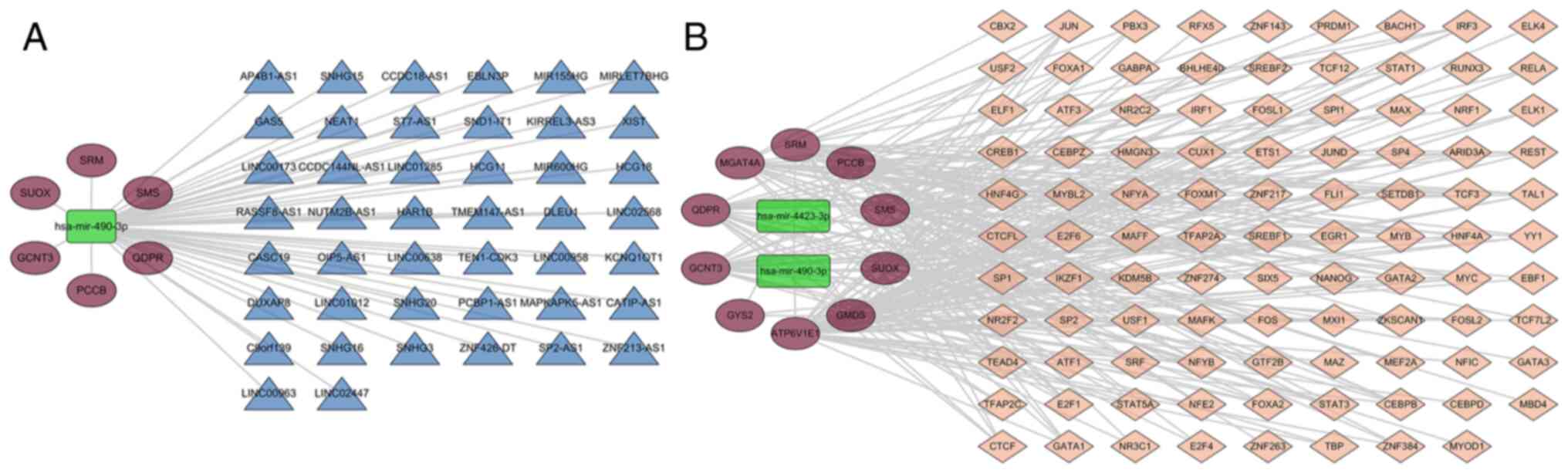
Characterized miRNA-hub target
gene-network construction and validation
A network was constructed around the hub target
gene, based on the three characterized miRNAs screened; two miRNAs
corresponding to the hub target gene were extracted, and then the
mirNet website was used to predict lncRNAs of the miRNAs. A total
of six genes, one miRNA and 44 lncRNAs were screened (Fig. 5A). TFs of 10 hub target genes were
analysed by ChEA3 and a total of 98 TFs were predicted to be
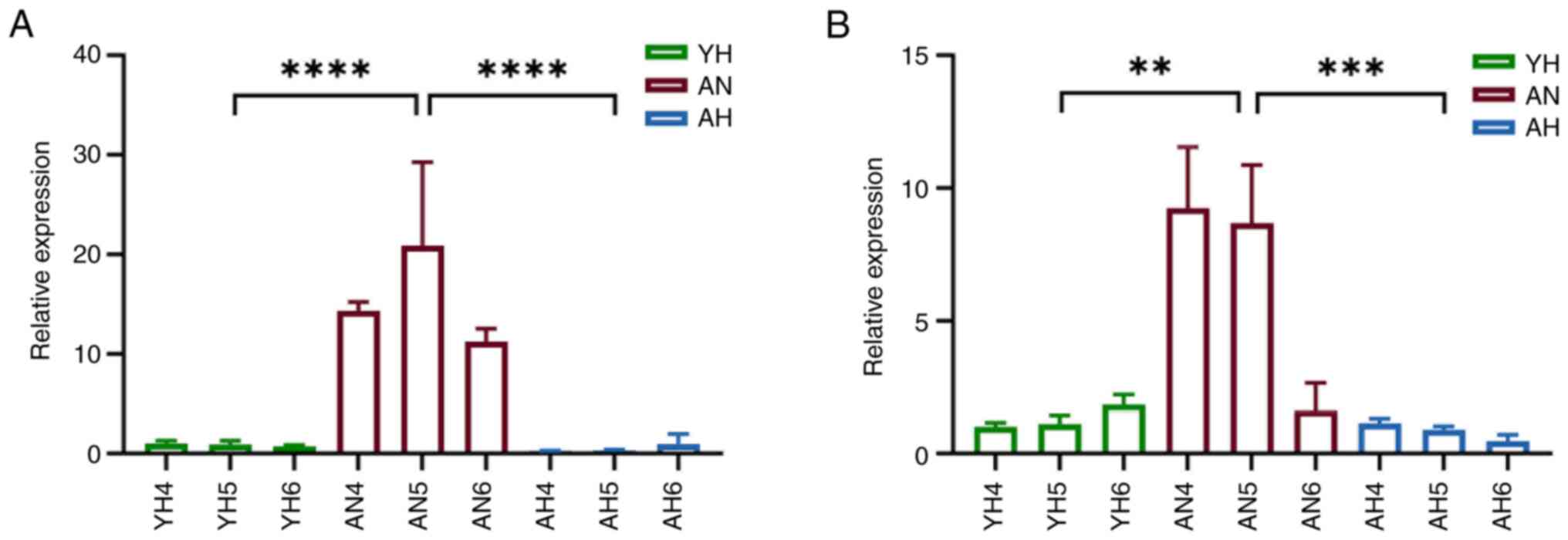
associated (Fig 5B; Table SI). Expression levels of the
primary characterized miRNAs in the network, hsa-miR-490-3p and
hsa-miR-4423-3p, in aqueous humor were assessed. Consistently,
miR-490 and miR-4423 expression levels were decreased in both AH
and YH groups in comparison with AN group (Fig. 6A and B).
Discussion
The present study used miRNA sequencing in patients
with AH, AN and YH and found three shared miRNAs, hsa-miR-490-3p,
hsa-miR-4423-3p and hsa-miR-4485-3p, which were defined as high
myopic miRNAs. Furthermore, 19 target genes were associated with
metabolism and high myopia were explored for interactions via
STRING website and a PPI network was constructed. Potential drugs
affecting 10 hub genes function were predicted and a characterized
miRNA/hub gene/TFs network was visualized. DE genes were associated
with high myopia; intersecting DE miRNAs were identified to exclude
the influence of age, which, to the best of our knowledge, has not
been performed previously. Adults without cataracts and high myopia
are not indicated for surgery and juvenile cataracts are mostly due
to lens opacity caused by other genetic disorder (38), therefore these patients were not
included.
Among the characterized miRNAs, hsa-miR-490-3p and
hsa-miR-4423-3p were downregulated in both AH and YH groups. In
previous studies, hypoxia was an important pathological mechanism
of high myopia, especially the scleral hypoxia and changes in
reactive oxygen species-associated metabolites in aqueous humor
(39,40). In agreement with the present
results, hsa-miR-490-3p is significantly downregulated in patients
with squamous lung carcinoma, a disease associated with systemic
hypoxia (41). miR-4423 is a
regulator of airway epithelial differentiation and its diminished
function contributes to development of lung cancer (42), suggesting this miRNA may affect the
myopic process by regulating hypoxic mechanisms. However,
hsa-miR-4485-3p was upregulated in both AH and YH groups. It has
been shown that the source of miR-4485-3p is one of the transcripts
of antisense nc mitochondrial RNA (ASncmtRNA), ASncmtRNA-2, which
is derived from the mitochondrial 16S gene (43). Increase in miR-4485 induces
downregulation of cell cycle proteins cyclin B1 and D1 (44). These two proteins are key
regulators of the cell cycle, with cyclin D1 serving a key role in
G1 to S phase transition and cyclin B1 affecting G2 to M phase
transition; their downregulation may lead to decreased cell
proliferation, suggesting that high myopia may be associated with
changes in intraocular cell cycle regulation. In addition to the
intersecting miRNAs, there were non-intersecting miRNAs related to
high myopia. The mechanism of high myopia is complex and requires
further study.
A total of 289 target genes were predicted by three
characterized miRNAs and functional enrichment yielded 169 terms.
Among them, ‘human herpes simplex virus 1 infection’ (HSV-1) was
the most enriched KEGG pathway. HSV-1 is a common human virus and
it can infect the eye, especially the cornea, resulting in herpes
simplex keratitis and uveitis. HSV-1 can infect trabecular meshwork
cells in rats, causing viral anterior uveitis, which causes
elevated intraocular pressure, tissue damage in the anterior
chamber angle and inflammatory cell infiltration (45). It is suggested that high myopia may
also contribute to the pathological state by affecting aqueous
humor circulation pathway and inducing inflammatory processes
(41). In 19 metabolism-related
target genes, ‘carbohydrate derivative biosynthetic process’ was
the GO pathway with most pronounced enrichment, which is consistent
with earlier studies (46,47). A total of 12 altered metabolic
pathways are identified in patients with myopia combined with
choroidal neovascularization, five of which are related to
carbohydrate derivative metabolism, suggesting an important role
for their involvement in disease development (48). There are also articles suggesting
the carbohydrate derivative biosynthetic process is associated with
a variety of diseases, including Alzheimer's disease, diabetic
retinopathy and glaucoma, largely due to accumulation of advanced
glycation end products (AGEs) (49,50).
Hyperglycaemia, alterations in oxidative environment and cell
proliferation status all affect the formation of AGEs, suggesting
development of high myopia may be related to ocular tissue being
affected by damage similar to that caused by toxic products such as
AGEs. The disease targets predicted by these 19 genes were
primarily associated with musculoskeletal, neurological,
cardiovascular, digestive and developmental disorder; ‘decreased
muscle mass’ was associated with the metabolism-associated high
myopia target genes. Muscle status serves a key role in maintaining
the normal physiological function of the eye. For example, patients
with myasthenia gravis have ocular symptoms such as extraocular
muscle weakness and eye pain (51,52).
In people with high myopia, abnormalities in accommodation are
prevalent (53), which are mainly
influenced by structures such as the lens, Zinn's zonule and
ciliary muscles; visual function training targeting the muscles and
ligaments inside and outside the eye may restore the eye to a
healthy state. The identified diseases, such as nystagmus, cerebral
atrophy, and optic nerve atrophy, may be eye-related but are not
directly linked to high myopia. Future studies could explore their
potential associations with high myopia.
Among 10 ranked hub genes, the most widely
associated gene was PCCB, which, to the best of our knowledge, has
not been described in high myopia to date. The protein encoded by
PCCB gene is reported to be a subunit of propionyl coenzyme A
carboxylase (PCC), which is associated with metabolism of fatty
acids, amino acids and other metabolites in mitochondria (54). Decreased expression of the PCCB
gene leads to impairments in the γ-aminobutyric acid (GABA)
signalling pathway. Specifically, mutations in PCCB gene may lead
to loss of function of PCC, which affects the production of
succinic acid semialdehyde, the precursor of GABA, and indirectly
affects synthesis of GABA (55). A
previous study has shown that the administration of baclofen
(GABABR agonist) intravenously to the eyes of chicks significantly
decreases myopic excursion and AL growth in eyes with deprivation
and lens-induced myopia (56). In
myopic guinea pigs, it was found that compared with normal
controls, retinal concentrations of dopamine and GABA are
decreased, glutamate, 3-methoxytyramine and glycine are increased
and myopic refractive error and AL increase (57). This suggests that by interfering
with PCCB, GABA concentration is regulated to maintain the balance
between excitatory and inhibitory neurotransmitters in the eye,
which may be effective in slowing development of high myopia.
In drug analysis, cyclosporine involved four hub
genes. Cyclosporine is an immunosuppressant that suppresses
activity of the immune system primarily by decreasing the activity
and proliferation of T lymphocytes. Cyclosporine binds to
cyclophilin, which decreases the transcriptional activation of
cytokine genes such as IL-2, TNF-α, IL-3 and IL-4 to reduce the
proliferation of T lymphocytes (58). This medication is currently used as
a first-line agent for uveitis, especially in patients with
Behcet's disease who need to take it orally on a regular basis in
conjunction with hormonal medication (59), but it is more commonly used in
myopia as an anti-inflammatory topical agent following
photorefractive keratectomy or laser in situ keratomileusis
(60). The screening of
cyclosporine, acetaminophen and other medications may suggest that
inflammation exists in high myopia, providing a novel direction for
treatment; larger studies are needed to validate these
findings.
lncRNAs can act as sponges to bind miRNAs, thus
preventing miRNAs from binding to target mRNAs, and can also
influence the expression of their target genes by controlling miRNA
expression (61). TFs control RNA
transcription, localization and stability through binding (62). The present data demonstrated
lncRNAs and TFs corresponding to the characterized miRNAs that may
serve as potential biomarkers of therapeutic response. DE miRNA
expression pattern was also observed in the aqueous humor of the
patients. The present study results indicated that the selected
characterized miRNAs were associated with high myopia, as well as
with common viral infections, metabolic processes, muscle
alterations and upstream genetic factors related to ophthalmology.
Notably, cyclosporine was a key drug linked to the hub genes of
interest. Cyclosporine is widely used in the treatment of various
ocular diseases (63,64), underscoring the validity of the
present analysis and the potential feasibility of future
therapeutic applications. Numerous metabolism-related characterized
miRNAs and hub genes were involved in the pathological progression
of high myopia. However, the present study has limitations. First,
due to the difficulty of obtaining individual aqueous humor
samples, miRNAs were extracted by mixed sampling. Second,
non-metabolism-associated genetic alterations in samples may also
cause different expression changes in the screened genes. Third,
patients with AH represent a minority of patients with cataracts;
to match age and sex between the groups, the present sample size
was relatively small.
To the best of our knowledge, the present study is
the first to perform miRNA sequencing for the exploration of
age-related cataracts and high myopia in patients. hsa-miR-490-3p,
hsa-miR-4423-3p, and hsa-miR-4485-3p may serve as characterized
miRNAs in high myopia. Prediction of metabolism-associated target
genes and functional enrichment analysis indicate the biosynthetic
process of carbohydrate derivatives may serve a pivotal role during
the development and progression of high myopia. Furthermore,
decreased muscle mass might be implicated in the aetiology of high
myopia. PCCB could serve as a potential biomarker in management of
high myopia. The hub genes affected by cyclosporine provide new
avenues for therapeutic strategies in high myopia.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
The authors would like to thank Dr Yujing Li (The
First Affiliated Hospital of Chongqing Medical University,
Chongqing, China) for assisting with topic selection and providing
feedback on image revisions, and to Dr Xueqin Zhou (The First
Affiliated Hospital of Chongqing Medical University, Chongqing,
China) for clinical assistance in collecting aqueous humor samples
from patients.
Funding
The present study was supported by National Natural Science
Foundation of China (grant nos. 81870650, 81900885, 81970832 and
82371098), Project Foundation of Chongqing Science and Technology
Commission of China (grant nos. CSTC2021jscx-gksb-N0017,
cstc202ljcyj-msxm3178, CSTB2022NSCQ-MSX1561 and
cstc2021jcyj-msxmX0967), Chongqing Talent Plan ‘Contract program’
(grant no. cstc2022ycjh-bgzxm0121), First Clinical College of
Chongqing Medical University (grant no. 472020320220007), Chongqing
Science and Health Joint Key Project (grant no. 2024ZDXM033) and
Chongqing Young and Middle-aged High-end Talent Project (grant no.
2024GDRC005).
Availability of data and materials
The data generated in the present study may be found
in the Genome Sequence Archive for Human (https://ngdc.cncb.ac.cn/gsa-human/) using the
accession number HRA009123.
Authors' contributions
FH, WW and KH conceived and designed the study. FH,
YC, SZ and RH performed experiments. JW acquired data. FH and YC
analyzed data. FH wrote the manuscript. WW and KH revised the
manuscript. FH, WW and KH confirm the authenticity of all the raw
data. All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the First Affiliated Hospital of Chongqing Medical
University (approval no. 2023-1; Chongqing, China) and written
informed consent was obtained from patients or their
parent/guardian before sample collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Tran HDM, Tran YH, Tran TD, Jong M,
Coroneo M and Sankaridurg P: A review of myopia control with
atropine. J Ocul Pharmacol Ther. 34:374–379. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morgan IG, French AN, Ashby RS, Guo X,
Ding X, He M and Rose KA: The epidemics of myopia: Aetiology and
prevention. Prog Retin Eye Res. 62:134–149. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baird PN, Saw SM, Lanca C, Guggenheim JA,
Iii EL, Zhou X, Matsui KO, Wu PC, Sankaridurg P, Chia A, et al:
Myopia. Nat Rev Dis Primers. 6:992020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee SS and Mackey DA: Prevalence and risk
factors of myopia in young adults: Review of findings from the
raine study. Front Public Health. 10:8610442022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park DJ and Congdon NG: Evidence for an
‘epidemic’ of myopia. Ann Acad Med Singap. 33:21–26. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ha A, Kim SJ, Shim SR, Kim YK and Jung JH:
Efficacy and safety of 8 atropine concentrations for myopia control
in children: A network meta-analysis. Ophthalmology. 129:322–333.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Holden BA, Fricke TR, Wilson DA, Jong M,
Naidoo KS, Sankaridurg P, Wong TY, Naduvilath TJ and Resnikoff S:
Global prevalence of myopia and high myopia and temporal trends
from 2000 through 2050. Ophthalmology. 123:1036–1042. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Modjtahedi BS, Abbott RL, Fong DS, Lum F
and Tan D; Task Force on Myopia, : Reducing the global burden of
myopia by delaying the onset of myopia and reducing myopic
progression in children: The academy's task force on myopia.
Ophthalmology. 128:816–826. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mori K, Kuroha S, Hou J, Jeong H, Ogawa M,
Ikeda SI, Kang JX, Negishi K, Torii H, Arita M, et al: Lipidomic
analysis revealed n-3 polyunsaturated fatty acids suppressed
choroidal thinning and myopia progression in mice. FASEB J.
36:e223122022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pan M, Zhao F, Xie B, Wu H, Zhang S, Ye C,
Guan Z, Kang L, Zhang Y, Zhou X, et al: Dietary ω-3 polyunsaturated
fatty acids are protective for myopia. Proc Natl Acad Sci USA.
118:e21046891182021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang Q, Su DY, Wang ZZ, Liu C, Sun YN,
Cheng H, Li XM and Yan B: Retina as a window to cerebral
dysfunction following studies with circRNA signature during
neurodegeneration. Theranostics. 11:1814–1827. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yang C, Miao A, Yang C, Huang C, Chen H,
Jiang Y, Deng C and Sun N: Precise detection of cataracts with
specific high-risk factors by layered binary co-ionizers assisted
aqueous humor metabolic analysis. Adv Sci (Weinh). 9:e21059052022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu X, Du Y, Li D, Xu J, Wu Q, He W, Zhang
K, Zhu J, Guo L, Qi M, et al: Aberrant TGF-β1 signaling activation
by MAF underlies pathological lens growth in high myopia. Nat
Commun. 12:21022021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chou T, Huang X, Liu J, Liu X, Zeng K, Yan
Z, Mei S, Su L, Xi W, Ni J, et al: First evidence indicates the
physiology- and axial-myopia-dependent profiles of steroid hormones
in aqueous humor. Metabolites. 12:12202022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sebag J: Vitreous and vision degrading
myodesopsia. Prog Retin Eye Res. 79:1008472020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Consejo A, Radhakrishnan H and Iskander
DR: Scleral changes with accommodation. Ophthalmic Physiol Opt.
37:263–274. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao F, Zhang D, Zhou Q, Zhao F, He M,
Yang Z, Su Y, Zhai Y, Yan J, Zhang G, et al: Scleral HIF-1α is a
prominent regulatory candidate for genetic and environmental
interactions in human myopia pathogenesis. EBioMedicine.
57:1028782020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Boehm M and Slack FJ: MicroRNA control of
lifespan and metabolism. Cell Cycle. 5:837–840. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu TX and Rothenberg ME: MicroRNA. J
Allergy Clin Immunol. 141:1202–1207. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Agbu P and Carthew RW: MicroRNA-mediated
regulation of glucose and lipid metabolism. Nat Rev Mol Cell Biol.
22:425–438. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rayner KJ and Moore KJ: MicroRNA control
of high-density lipoprotein metabolism and function. Circ Res.
114:183–192. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Xu S: microRNA expression in the eyes and
their significance in relation to functions. Prog Retin Eye Res.
28:87–116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Greene KM, Stamer WD and Liu Y: The role
of microRNAs in glaucoma. Exp Eye Res. 215:1089092022. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hyttinen JMT, Blasiak J, Felszeghy S and
Kaarniranta K: MicroRNAs in the regulation of autophagy and their
possible use in age-related macular degeneration therapy. Ageing
Res Rev. 67:1012602021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Duan X, Lu Q, Xue P, Zhang H, Dong Z, Yang
F and Wang N: Proteomic analysis of aqueous humor from patients
with myopia. Mol Vis. 14:370–377. 2008.PubMed/NCBI
|
|
26
|
Zhu XJ, Chen MJ, Zhang KK, Yang J and Lu
Y: Elevated TGF-β2 level in aqueous humor of cataract patients with
high myopia: Potential risk factor for capsule contraction
syndrome. J Cataract Refract Surg. 42:232–238. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan W, Zhang Y, Cao J and Yan H: TGF-β2
levels in the aqueous humor are elevated in the second eye of high
myopia within two weeks after sequential cataract surgery. Sci Rep.
12:179742022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jiang X, Pardue MT, Mori K, Ikeda SI,
Torii H, D'Souza S, Lang RA, Kurihara T and Tsubota K: Violet light
suppresses lens-induced myopia via neuropsin (OPN5) in mice. Proc
Natl Acad Sci USA. 118:e20188401182021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ikeda SI, Kurihara T, Jiang X, Miwa Y, Lee
D, Serizawa N, Jeong H, Mori K, Katada Y, Kunimi H, et al: Scleral
PERK and ATF6 as targets of myopic axial elongation of mouse eyes.
Nat Commun. 13:58592022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yam JC, Zhang XJ, Zhang Y, Yip BHK, Tang
F, Wong ES, Bui CHT, Kam KW, Ng MPH, Ko ST, et al: Effect of
low-concentration atropine eyedrops vs placebo on myopia incidence
in children: The LAMP2 randomized clinical trial. JAMA.
329:472–481. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong J, Zhu Z, Xu H and He M: Myopia
control effect of repeated low-level red-light therapy in chinese
children: A randomized, double-blind, controlled clinical trial.
Ophthalmology. 130:198–204. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dishler JG, Slade S, Seifert S and
Schallhorn SC: Small-incision lenticule extraction (SMILE) for the
correction of myopia with astigmatism: Outcomes of the United
States food and drug administration premarket approval clinical
trial. Ophthalmology. 127:1020–1034. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
He X, Deng J, Xu X, Wang J, Cheng T, Zhang
B, Zhao H, Luan M, Fan Y, Xiong S, et al: Design and pilot data of
the high myopia registration study: Shanghai child and adolescent
large-scale eye study (SCALE-HM). Acta Ophthalmol. 99:e489–e500.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shiels A and Hejtmancik JF: Mutations and
mechanisms in congenital and age-related cataracts. Exp Eye Res.
156:95–102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Asbell PA, Dualan I, Mindel J, Brocks D,
Ahmad M and Epstein S: Age-related cataract. Lancet. 365:599–609.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang S, Luo Q, Huang J, Wei J, Wang S,
Hong C, Qiu P and Li C: A cluster of metabolic-related genes serve
as potential prognostic biomarkers for renal cell carcinoma. Front
Genet. 13:9020642022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Di Lazzaro Filho R, Yamamoto GL, Silva TJ,
Rocha LA, Linnenkamp BDW, Castro MAA, Bartholdi D, Schaller A, Leeb
T, Kelmann S, et al: Biallelic variants in DNA2 cause poikiloderma
with congenital cataracts and severe growth failure reminiscent of
Rothmund-Thomson syndrome. J Med Genet. 60:1127–1132. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lin X, Lei Y, Pan M, Hu C, Xie B, Wu W, Su
J, Li Y, Tan Y, Wei X, et al: Augmentation of scleral glycolysis
promotes myopia through histone lactylation. Cell Metab.
36:511–525. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu H, Chen W, Zhao F, Zhou Q, Reinach PS,
Deng L, Ma L, Luo S, Srinivasalu N, Pan M, et al: Scleral hypoxia
is a target for myopia control. Proc Natl Acad Sci USA.
115:E7091–E7100. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wu D, Huo C, Jiang S, Huang Y, Fang X, Liu
J, Yang M, Ren J, Xu B and Liu Y: Exostosin1 as a novel prognostic
and predictive biomarker for squamous cell lung carcinoma: A study
based on bioinformatics analysis. Cancer Med. 10:2787–2801. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Perdomo C, Campbell JD, Gerrein J, Tellez
CS, Garrison CB, Walser TC, Drizik E, Si H, Gower AC, Vick J, et
al: MicroRNA 4423 is a primate-specific regulator of airway
epithelial cell differentiation and lung carcinogenesis. Proc Natl
Acad Sci USA. 110:18946–18951. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Farfan N, Sanhueza N, Briones M, Burzio LO
and Burzio VA: Antisense noncoding mitochondrial RNA-2 gives rise
to miR-4485-3p by Dicer processing in vitro. Biol Res. 54:332021.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fitzpatrick C, Besndek MF, Briones M,
Farfán N, Silva VA, Nardocci G, Montecino M, Boland A, Deleuze JF,
Villegas J, et al: Mitochondrial ncRNA targeting induces cell cycle
arrest and tumor growth inhibition of MDA-MB-231 breast cancer
cells through reduction of key cell cycle progression factors. Cell
Death Dis. 10:4232019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y, Ke W, Liu X, Zhang Q, Yu N, Wang K
and Chen M: Comparison between two types of viral-induced anterior
uveitis in vitro and in Vivo: A stronger response in herpes simplex
virus type 1 than in murine cytomegalovirus. Invest Ophthalmol Vis
Sci. 64:202023. View Article : Google Scholar
|
|
46
|
Kim TG, Kim W, Choi S and Jin KH: Effects
of scleral collagen crosslinking with different carbohydrate on
chemical bond and ultrastructure of rabbit sclera: Future treatment
for myopia progression. PLoS One. 14:e02164252019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang S, Wang T, Wang H, Gao B and Sun C:
Identification of potential biomarkers of myopia based on machine
learning algorithms. BMC Ophthalmol. 23:3882023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wei Q, Yu Z, Zhou X, Gong R, Jiang R, Xu G
and Liu W: Metabolomic profiling of aqueous humor from pathological
myopia patients with choroidal neovascularization. Metabolites.
13:9002023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bejarano E and Taylor A: Too sweet:
Problems of protein glycation in the eye. Exp Eye Res. 178:255–262.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ryan P, Patel B, Makwana V, Jadhav HR,
Kiefel M, Davey A, Reekie TA, Rudrawar S and Kassiou M: Peptides,
peptidomimetics, and carbohydrate-peptide conjugates as
amyloidogenic aggregation inhibitors for Alzheimer's Disease. ACS
Chem Neurosci. 9:1530–1551. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Akan O and Baysal-Kirac L: Ophthalmologic
manifestations in myasthenia gravis: Presentation and prognosis.
Acta Neurol Belg. 121:1131–1140. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Guyuron B, Bokhari F, Galloway DV and
Thomas T: Oculonasal synkinesis. Plast Reconstr Surg. 94:251–253.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hughes RPJ, Read SA, Collins MJ and
Vincent SJ: Changes in ocular biometry during short-term
accommodation in children. Ophthalmic Physiol Opt. 40:584–594.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wongkittichote P, Ah Mew N and Chapman KA:
Propionyl-CoA carboxylase-A review. Mol Genet Metab. 122:145–152.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zhang W, Zhang M, Xu Z, Yan H, Wang H,
Jiang J, Wan J, Tang B, Liu C, Chen C, et al: Human forebrain
organoid-based multi-omics analyses of PCCB as a schizophrenia
associated gene linked to GABAergic pathways. Nat Commun.
14:51762023. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu H, Schaeffel F, Yang Z and Feldkaemper
MP: GABA(B) receptor activation affects eye growth in chickens with
visually induced refractive errors. Biomolecules. 13:4342023.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wei P, Han G, He M and Wang Y: Retinal
neurotransmitter alteration in response to dopamine D2 receptor
antagonist from myopic guinea pigs. ACS Chem Neurosci.
14:3357–3367. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Bennett WM and Norman DJ: Action and
toxicity of cyclosporine. Annu Rev Med. 37:215–224. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhong Z, Su G and Yang P: Risk factors,
clinical features and treatment of Behçet's disease uveitis. Prog
Retin Eye Res. 97:1012162023. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Hessert D, Tanzer D, Brunstetter T, Kaupp
S, Murdoch D and Mirzaoff M: Topical cyclosporine A for
postoperative photorefractive keratectomy and laser in situ
keratomileusis. J Cataract Refract Surg. 39:539–547. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Puvvula PK: LncRNAs regulatory networks in
cellular senescence. Int J Mol Sci. 20:26152019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lambert SA, Jolma A, Campitelli LF, Das
PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR and Weirauch MT:
The human transcription factors. Cell. 175:598–599. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Karadag O and Bolek EC: Management of
Behcet's syndrome. Rheumatology (Oxford). 59:iii108–iii117. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bruschi G, Ghiglioni DG, Cozzi L, Osnaghi
S, Viola F and Marchisio P: Vernal Keratoconjunctivitis: A
systematic review. Clin Rev Allergy Immunol. 65:277–329. 2023.
View Article : Google Scholar : PubMed/NCBI
|