Introduction
Oral cancer is one of the most common cancers
worldwide (1,2). The 2021 cancer registry annual report
from the Ministry of Health and Welfare of Taiwan revealed that the
age-adjusted incidence and mortality rates of oral cancer in males
were 40.38/100,000 and 16.38/100,000, respectively, making it the
fourth most common cause of cancer-related mortality among males in
Taiwan (3). Other than cigarette
smoking and alcohol drinking, betel nut chewing is the main risk
factor for oral cancer and ~90% of patients in Taiwan are habitual
betel nut chewers (4). Oral
squamous cell carcinoma (OSCC) is the main subset or oral cancer,
accounting for >90% of cases and has a poor prognosis (5). The standard treatments for OSCC
include surgery, chemotherapy and radiotherapy (6). However, the 5-year survival rate of
OSCC is ~50% due to the poor responses to chemotherapy and
radiotherapy resulting in recurrence and worse outcomes (7,8).
However, the survival rate could be improved to ~80% if patients
identify the symptoms and seek medical advice early, allowing them
to be diagnosed in the initial stage and undergo the standard
treatments (9,10). It is therefore important to
identify reliable biomarkers for detecting OSCC.
Epigenetic alterations, particularly
hypermethylation in the promoter region of tumor suppressor genes,
have been identified as biomarkers for a number of cancers
including OSCC and play a critical role in oral cancer development
(11). A number of genes
responsible for cell-cycle events, apoptosis, cell-to-cell adhesion
and DNA repair are found to be hypermethylated and silenced in OSCC
(12). We have previously used the
Illumina GoldenGate Assay to identify the DNA methylation status of
1,505 CpG sites encompassing 807 genes in 40 OSCC tissue and 15
normal samples. The methylation array data was used to identify the
gene Homeobox A5 (HOXA5), which is hypermethylated in OSCC
samples (13).
HOXA5 is a member of the Hox gene
family that plays an important role in embryonic development due to
its transcriptional activation ability (14,15).
A previous study found that HOXA5 protein is a positive regulator
of p53 transcription and function in breast cancer cells,
indicating that reduced HOXA5 expression is an important
step in tumorigenesis (16).
Moreover, the effects of HOXA5 in various cancers have
gradually been elucidated over the past two decades. HOXA5
is downregulated by epigenetic alterations in breast cancer
(17,18), lung adenocarcinoma (19) and xanthoastrocytoma (20), but upregulated in hepatocellular
carcinoma (21). Aberrant
expression of HOXA5 was also observed in OSCC; however, the
epigenetic regulation and role of HOXA5 in OSCC has not been
fully investigated (22).
The present study first examined the methylation
status of HOXA5 in OSCC tissues by using techniques such as
the bisulfite sequencing assay and Illumina Infinium
MethylationEPIC BeadChip analysis (Illumina, Inc.). The expression
level of HOXA5 was also analyzed to determine whether it
differs between normal oral and OSCC tissues in patients with OSCC.
The gene expression of HOXA5 in OSCC cell lines was then
restored by using epigenetic drugs and employing lentivirus
vector-mediated gene transfer of HOXA5. Restoration of
HOXA5 significantly upregulated HOXA5 and p53 expression and
induced OSCC cell death. The results strongly suggested that
HOXA5 was a proapoptotic gene that is epigenetically
downregulated in OSCC.
Materials and methods
Specimen collection and bisulfite
conversion of genomic DNA
The 25 paired OSCC and adjacent normal tissues used
in the present study were collected from the tissue bank of China
Medical University Hospital in Taiwan. The median age at surgery of
the 25 male patients with OSCC from whom tissue was obtained for
analysis was 52.0 years (range: 36–63 years). Tissue samples were
collected from patients after obtaining written informed consent in
accordance with a protocol approved by the Institutional Review
Board of China Medical University Hospital (IRB no.
CMUH102-REC1-054). Genomic DNA of these tissues and cultured cell
lines was extracted using Gentra Puregene Tissue Kit (Qiagen GmbH)
and bisulfite conversion of genomic DNA was performed using EZ DNA
methylation kit (Zymo Research Corp.). Bisulfite conversed
Universal Methylated Genomic DNA (MilliporeSigma) was used as in
vitro methylated DNA (IVD) control for the methylation
analysis.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was extracted from cell lines (70–80%
confluence) or tissue specimens using REzol C & T reagent
(Protech Technology Enterprise Co., Ltd.) according to the
manufacturer's protocol. The aliquots of RNA were then used to
synthesize complementary DNA (cDNA) using SuperScript III
First-Strand Synthesis System (Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. RT-qPCR was performed
using SYBR Green Realtime PCR Master Mix (Toyobo Life Science) and
ABI StepOne real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) as the following steps: 95°C for 5 min, followed
by 50 cycles of successive incubation at 95°C for 30 sec, 62°C for
30 sec and 72°C for 45 sec. The primers used to amplify HOXA5 and
p53 cDNA were: HOXA5 forward, 5′-GCGCAAGCTGCACATAAG-3′ and reverse,
5′-CGGTTGAAGTGGAACTCCTT-3′; p53 forward, 5′-CCGCAGTCAGATCCTAGCG-3′
and reverse, 5′-AATCATCCATTGCTTGGGACG-3′. GAPDH was also amplified
as an internal control with primers 5′-TTGACGGTGCCATGGAATTT-3′ and
5′-GCCATCAATGACCCCTTCATT-3′. All samples were analyzed in
triplicate. The expression levels of HOXA5 and p53 were calculated
using the 2−∆∆Cq method (23) and normalized to that of GAPDH.
Bisulfite sequencing assay and
quantitative methylation-specific PCR
Bisulfite conversed DNA from tissues was amplified
by two pairs of HOXA5 promoter-specific primers which
targeted one HOXA5 promoter region from −1423 to −841
(forward, 5′-AGGAATAAAGGGGGTTTTAATAGAG-3′; and reverse,
5′-TCCAACCTAAAAAATCTTCATCAC-3′) and another promoter region from
−505 to −31 (forward, 5′-ATTTTTAAAATTTAGAGTTGTTGGTAGGA-3′; and
reverse, 5′-CTAAAACATATACTTAATTCCCTCCTA-3′) upstream of the
HOXA5 transcription start site (TSS). As treatment of DNA
with sodium bisulfite converts unmethylated C to U, which is
subsequently converted to T during PCR amplification, the forward
primers for bisulfite sequencing assay are devoid of C, while the
reverse primers are devoid of G. The PCR products were separated by
gel electrophoresis, purified with a QIAquick gel extraction kit
(Qiagen GmbH) and cloned into the yT&A cloning vector (Yeastern
Biotech Co., Ltd.) followed by DNA sequencing. For quantitative
methylation-specific PCR (MSP), the region from-268 to −59 upstream
of the TSS of HOXA5 was amplified for bisulfite converted
DNA from cultured cells with the primers as followed: Forward,
5′-AGTTTTGTTTTTAGCGGGTGGC-3′; and reverse,
5′-GTAAACACCCAAATATAAAATACGAC-3′. Quantitative MSP was performed
using SYBR Green Realtime PCR Master Mix (Toyobo Life Science) and
ABI StepOne real-time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.) using the following steps: 95°C for 5 min,
followed by 45 cycles of successive incubation at 95°C for 30 sec,
58°C for 30 sec and 72°C for 45 sec. A DNA fragment devoid of any
CpG dinucleotide in COL2A1 was amplified as an input control
for quantitative MSP with the primers as followed: forward,
5′-GGGAAGATGGGATAGAAGGGAATAT-3′; reverse,
5′-TCTAACAATTATAAACTCCAACCACCAA-3′. For each sample, the threshold
cycle number of methylated HOXA5 and COL2A1 were
determined. The percentage of HOXA5 methylation was
calculated using the 2−ΔΔCq method as the ratio of
HOXA5 to COL2A1 of a sample divided by the same ratio
of IVD.
Cell culture
The OSCC cell lines OC2 and OCSL (obtained from Dr
Yong-Kie Wong, Department of Dentistry, Taichung Veterans General
Hospital, Taichung, Taiwan) (24,25),
established from surgical specimens of buccal mucosa squamous
carcinoma from two Taiwanese male patients who had the habits of
betel nut chewing, alcohol drinking and cigarette smoking, were
maintained in RPMI 1640 medium (Gibco; Thermo Fisher Scientific,
Inc.) supplemented with 10% fetal bovine serum (FBS, Invitrogen;
Thermo Fisher Scientific, Inc.). They are immortalized cancer cells
with unlimited growth potential and have been characterized by
tumorigenesis in nude mice. The transformed human embryonic kidney
cell line 293T (ATCC; CRL-3216) was grown in DMEM medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
Mycoplasma negativity was confirmed before all cell
experiments.
5-aza-2′-deoxycytidine treatment
Cells were seeded at a density of 1×106
cells in 10-cm culture dishes and treated with 0.5 µM or 5 µM
5-aza-2′-deoxycytidine (5-Aza-dC; (MilliporeSigma). At 72 h
post-treatment, cells were harvested and their DNA, RNA and
proteins were extracted.
Western blot analysis
Cells were lysed by RIPA Lysis & Extraction
Buffer with protease inhibitor cocktail (Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. A BCA protein assay
kit (Thermo Fisher Scientific, Inc.) was used to measure the
protein concentration in cell lysates. Proteins (20 µg/lane) in
cell lysates were resolved using SDS-10% polyacrylamide gel
electrophoresis and transferred onto polyvinylidene difluoride
membranes (Bio-Rad Laboratories, Inc.). The membranes were blocked
for 2 h at room temperature in TBST with 5% non-fat dry milk
(Bio-Rad Laboratories, Inc.) and incubated overnight at 4°C with
antibodies against HOXA5 (1:1,000 dilution; cat. no. sc-81289;
Santa Cruz Biotechnology, Inc.), p53 (1:1,000 dilution; cat. no.
sc-126; Santa Cruz Biotechnology, Inc.) and GAPDH (1:1,000
dilution; cat. no. sc-47724; Santa Cruz Biotechnology, Inc.).
Subsequently, the membranes were incubated with a horseradish
peroxidase-conjugated secondary antibody (1:10,000 dilution; cat.
no. 31430; Thermo Fisher Scientific, Inc.) for 1 h at room
temperature. The specific protein signals were detected using an
enhanced chemiluminescence kit (cat. no. WBKLS0500;
MilliporeSigma). Quantification of HOXA5 and p53 band intensities
was determined using ImageJ software v1.49 (National Institutes of
Health) and normalized to that of GAPDH.
Plasmid construction and lentivirus
vector production
The full-length human HOXA5 cDNA was
amplified from the immortalized ovarian surface epithelia cell line
IOSE (26) by RT-PCR. The amplicon
was cloned into a 2nd generation lentiviral vector plasmid
pSin-IRES-GFP (IG) (27) at the
XbaI and EcoRI sites to generate pSin-HOXA5-IRES-GFP
(HIG). The plasmids pIG or pHIG (12 µg) were cotransfected with
lentiviral packaging plasmids pCMVdeltaR8.91 (10.8 µg) and pMD.G
(1.2 µg) (obtained from the National RNAi Core Facility, Taipei,
Taiwan) into 293T cells. At 48 h post-transfection the
lentivirus-containing supernatants were collected and filtered
through 0.45-µm pore size filters (MilliporeSigma) prior to use in
transduction assays. Lentiviral vector stock at a multiplicity of
infection (MOI) of 5 was used to infect OSCC cells. Successful
infection of lentiviral vector IG or HIG was monitored by GFP
expression 3 days later and the infected cells were sorted by
FACSAria III Cell Sorter (BD Bioscience) for use in the RT-qPCR
assay. Lentiviral vector expressing short hairpin RNA (shRNA) was
produced by transfection of pCMVdeltaR8.91 (10.8 µg), pMD.G (1.2
µg) and pLKO.1-puro plasmid carrying an shRNA (12 µg) (obtained
from the National RNAi Core Facility, Taipei, Taiwan) into 293T
cells (28). The shRNA sequences
specific for HOXA5 and GFP were 5′-CCGCAGAAGGAGGATTGAAAT-3′
and 5′-CAACAGCCACAACGTCTATAT-3′, respectively. OSCC cells infected
with shHOXA5-expressing lentivirus vector at an MOI of 5 were
selected by incubating with 2 µg/ml puromycin (MilliporeSigma) for
2 days. Then cells were maintained in the presence of 1 µg/ml
puromycin. Cells infected with shGFP-expressing lentiviral vector
were used as a control.
Luciferase reporter assay
Two pGL2b plasmids (Promega Corporation) containing
a wild-type and an AP-1 motif mutated p53 promoters inserted into
polycloning sites upstream of the firefly luciferase (FL) reporter
gene (29) were used in the assay.
Each of the pGL2b plasmids were cotransfected with pRL-TK
Renilla luciferase (RL) control reporter vector (Promega
Corporation) into cells by using Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.). At 48 h post-transfection, cells were washed
twice with phosphate buffered saline (PBS), lysed in the Luciferase
assay lysis buffer (Promega Corporation), scraped from the culture
dish and transferred to a tube. After vortex and centrifugation at
10,000 × g for 1 min at room temperature, the supernatant was
collected for quantification of luciferase activity using
Dual-Luciferase Reporter Assay System (Promega Corporation)
according to the manufacturer's protocol. The relative
transcriptional activity was determined as the ratio of FL to
RL.
Cell viability analysis by MTS
assay
Cells were seeded onto replicate 96-well plates at a
density of 2,000 cells/well on day 0. The cell viability was
determined on day 4 by the MTS assay using the CellTiter Aqueous
One Solution Cell Proliferation Assay kit (Promega
Corporation).
Cell necrosis assay
Cells were seeded onto replicate 6-well plate at a
density of 6×104 cells/well on day 0. On day 1, the
cells were washed with PBS and cultured in the medium containing 2
µg/ml cisplatin. Three days later, the cells were harvested with
trypsin, fixed with 70% ethanol for 24 h, stained with propidium
iodide (MilliporeSigma) for 30 min at 37°C and analyzed by
FACSCalibur (BD Biosciences). The proportion of necrotic cells in
sub-G1 area was quantified using CellQuest Pro software (BD
Biosciences).
In vivo experiments
The in vivo experiments were conducted in
accordance with the principles of laboratory animal care at the
National Institutes of Health and with the approval of the
Institutional Animal Care and Use Committee at National Chung Cheng
University, Taiwan, R.O.C. (IACUC no. 1080401). A total of 29
female athymic BALB/c nude mice (5–6 weeks old; ~25 g) were
supplied by National Laboratory Animal Center (Taipei, Taiwan). The
housing conditions of the mice were as follows: 12-h light/dark
cycle; temperatures of 18–23°C with 40–60% humidity; and water and
food accessible at all times. Following the induction of anesthesia
using 2% isoflurane for four minutes and 1.5% maintenance, the mice
were inoculated with 5×106 OC2 cells in the right dorsal
flank. When the tumor volumes reached ~100 mm3 (16 days
after tumor implantation), the mice received intraperitoneal
injections of PBS or cisplatin at a weekly dose of 3 mg/kg for a
total of six treatments. Tumor length (L) and width (W) were
measured daily with calipers and tumor volumes were calculated
using the formula (LxWxW/2). At the experimental endpoint (60 days
after tumor implantation), all mice were humanely sacrificed under
a 2% isoflurane gas anesthesia by cervical dislocation. Mortality
was verified through a physical examination for the absence of
cardiac activity and respiration.
Statistical analyses
Wilcoxon signed rank test was used to test the
difference in fold-change of RNA expression between paired normal
and tumor samples. One-way ANOVA was conducted for the comparion of
methylation level and relative protein expression among three dose
levels of 5-Aza-dC followed by Dunnett's test for the multiple
comparison against the control group. Student's t-tests were
performed for the comparion of relative RNA expression, cell
viability, and cell necrosis between two treatment groups. For the
comparison of tumor growth rates, generalized linear models with
generalized estimated equations were used to account for the
correlation between the repeated measurements. The interaction
terms between time and group in the generalized linear models were
used to evaluate the difference in tumor growth rates. Analyses
were performed using SAS version 9.4 (SAS Institute).
Results
HOXA5 hypermethylation as a biomarker
for OSCC detection
We have previously investigated the methylation
levels of three CpG sites (−1324, −479 and +187 related to TSS) in
the promoter region of HOXA5 using the Illumina GoldenGate
Methylation Array (Illumina, Inc). The methylation levels
(presented as β values) at the three query sites (−1324, −479 and
+187) were all significantly higher in the OSCC tissues than in
normal oral tissues and the −1324 site was effective for detecting
OSCC with an AUC of 0.83 (specificity=87%, sensitivity=80%)
(13). To validate the array data,
the present study analyzed the methylation status of HOXA5
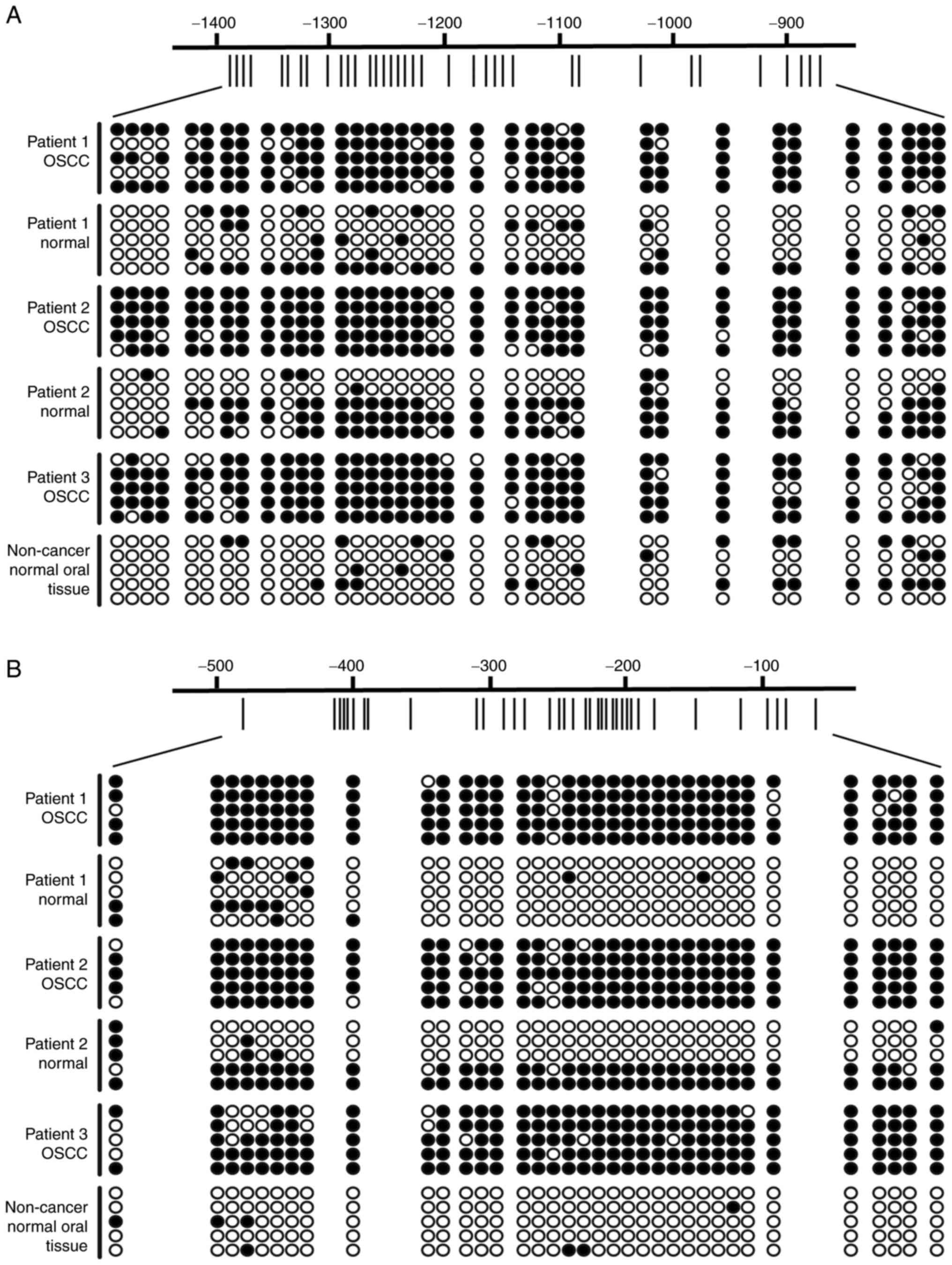
promoter regions from −1423 to −841 and from −505 to −31 in several
oral tissue specimens by using bisulfite sequencing assay. The
comparison of three normal tissues with low β values (β=0.06, 0.08
and 0.03 for the −1324 site) indicated much higher methylation
density in the promoter region of three OSCC tissues with high β
values (β=0.31, 0.32 and 0.43 for the −1324 site) (Fig. 1).
To obtain more-complete information on the
methylation status of HOXA5, the present study applied the
Infinium MethylationEPIC BeadChip assay (Illumina, Inc.) to 25
paired OSCC and adjacent normal tissue specimens. Among the 44 CpG
sites of HOXA5 analyzed using the BeadChip assay, 43
presented significantly higher methylation in OSCC tissues than in
the normal oral tissues, with 25 CpG sites exhibiting AUCs of
>0.80 (Table I). Comparison of
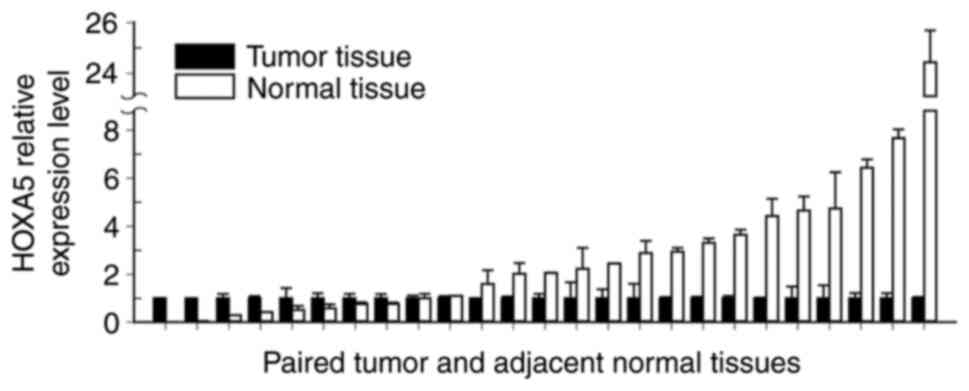
HOXA5 RNA expression between these paired normal oral and
OSCC tissue specimens yielded results consistent with those
obtained by the BeadChip assay. The median HOXA5 expression
level was 2.06-fold higher in normal tissues than in OSCC tissues
(interquartile range=2.89; P<0.005; Fig. 2). These results indicated that
HOXA5 is hypermethylated in OSCC tissues and that
HOXA5 hypermethylation might hold great potential as a
biomarker for detecting OSCC.
 | Table I.Performance of 44 CpG sites of HOXA5
for detecting OSCC. |
Table I.
Performance of 44 CpG sites of HOXA5
for detecting OSCC.
|
|
|
| Discrimination
statistics |
|---|
|
|
|
|
|
|---|
| Probe | UCSC RefGene
Group | Mean ∆β | AUC | (95% CI) | P-value |
|---|
| cg00809969 | 3′-UTR | 0.10 | 0.84 | (0.72, 0.96) | <0.0001 |
| cg19701577 | 3′-UTR | 0.15 | 0.91 | (0.83, 0.99) | <0.0001 |
| cg14974749 | Body | 0.28 | 0.93 | (0.87, 1.00) | <0.0001 |
| cg20974609 | Body | 0.13 | 0.77 | (0.62, 0.91) | 0.0003 |
| cg10592111 | Body | 0.09 | 0.75 | (0.60, 0.89) | 0.0011 |
| cg16139219 | Body | 0.09 | 0.76 | (0.61, 0.91) | 0.0007 |
| cg11724970 | Body | 0.13 | 0.77 | (0.63, 0.91) | 0.0002 |
| cg05076221 | Body | 0.10 | 0.79 | (0.65, 0.93) | <0.0001 |
| cg23936031 | 1st Exon | 0.10 | 0.69 | (0.53, 0.85) | 0.0178 |
| cg09549073 | 5′-UTR | 0.08 | 0.7 | (0.54, 0.86) | 0.0124 |
| cg04863892 | TSS200 | 0.07 | 0.66 | (0.49, 0.83) | 0.0615 |
| cg09207400 | TSS200 | 0.07 | 0.7 | (0.54, 0.85) | 0.0134 |
| cg19759481 | TSS200 | 0.09 | 0.69 | (0.53, 0.86) | 0.0188 |
| cg02916332 | TSS1500 | 0.09 | 0.74 | (0.59, 0.89) | 0.0021 |
| cg17569124 | TSS1500 | 0.08 | 0.72 | (0.56, 0.88) | 0.0062 |
| cg02005600 | TSS1500 | 0.09 | 0.74 | (0.59, 0.90) | 0.0018 |
| cg25307665 | TSS1500 | 0.15 | 0.8 | (0.67, 0.93) | <0.0001 |
| cg14014955 | TSS1500 | 0.09 | 0.74 | (0.58, 0.89) | 0.0033 |
| cg02646423 | TSS1500 | 0.10 | 0.7 | (0.54, 0.86) | 0.0151 |
| cg20517050 | TSS1500 | 0.15 | 0.8 | (0.66, 0.93) | <0.0001 |
| cg23204968 | TSS1500 | 0.12 | 0.75 | (0.60, 0.91) | 0.0013 |
| cg14058329 | TSS1500 | 0.20 | 0.9 | (0.81, 0.99) | <0.0001 |
| cg03207666 | TSS1500 | 0.17 | 0.9 | (0.81, 0.99) | <0.0001 |
| cg23454797 | TSS1500 | 0.13 | 0.8 | (0.67, 0.93) | <0.0001 |
| cg12015737 | TSS1500 | 0.28 | 0.91 | (0.82, 1.00) | <0.0001 |
| cg08070327 | TSS1500 | 0.17 | 0.87 | (0.76, 0.98) | <0.0001 |
| cg25506432 | TSS1500 | 0.12 | 0.87 | (0.76, 0.98) | <0.0001 |
| cg16997642 | TSS1500 | 0.20 | 0.89 | (0.80, 0.99) | <0.0001 |
| cg20817131 | TSS1500 | 0.19 | 0.91 | (0.81, 1.00) | <0.0001 |
| cg14013695 | TSS1500 | 0.22 | 0.91 | (0.83, 1.00) | <0.0001 |
| cg25390165 | TSS1500 | 0.15 | 0.9 | (0.81, 1.00) | <0.0001 |
| cg01323381 | TSS1500 | 0.26 | 0.92 | (0.85, 1.00) | <0.0001 |
| cg19643053 | TSS1500 | 0.27 | 0.94 | (0.87, 1.00) | <0.0001 |
| cg05774699 | TSS1500 | 0.20 | 0.91 | (0.84, 0.99) | <0.0001 |
| cg26023912 | TSS1500 | 0.18 | 0.91 | (0.83, 0.99) | <0.0001 |
| cg14882265 | TSS1500 | 0.16 | 0.89 | (0.80, 0.99) | <0.0001 |
| cg17432857 | TSS1500 | 0.25 | 0.94 | (0.87, 1.00) | <0.0001 |
| cg00969405 | TSS1500 | 0.25 | 0.93 | (0.87, 1.00) | <0.0001 |
| cg07049592 | TSS1500 | 0.19 | 0.93 | (0.85, 1.00) | <0.0001 |
| cg02106682 | TSS1500 | 0.19 | 0.93 | (0.85, 1.00) | <0.0001 |
| cg03368099 | TSS1500 | 0.21 | 0.92 | (0.82, 1.00) | <0.0001 |
| cg01748892 | TSS1500 | 0.16 | 0.88 | (0.78, 0.98) | <0.0001 |
| cg13694927 | TSS1500 | 0.17 | 0.88 | (0.77, 0.98) | <0.0001 |
| cg03744763 | TSS1500 | 0.18 | 0.86 | (0.75, 0.98) | <0.0001 |
HOXA5 expression regulated by DNA
methylation
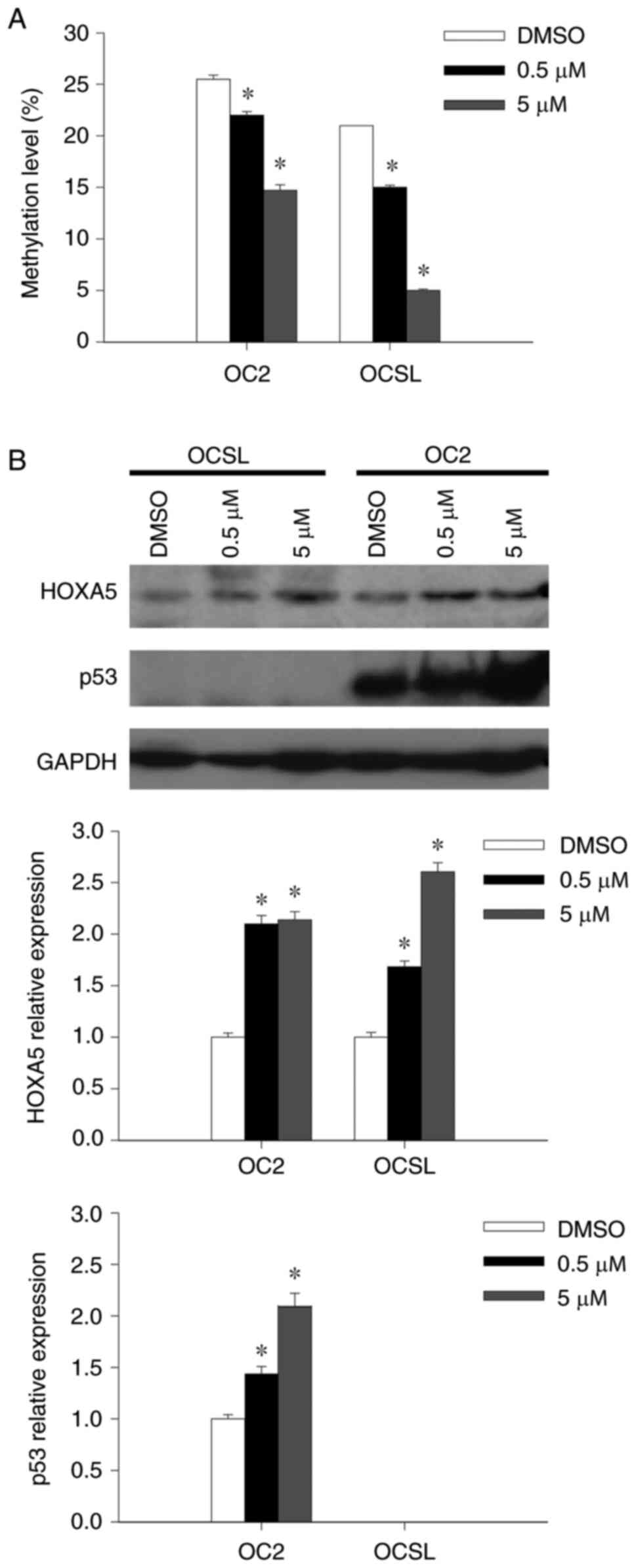
The present study treated the OSCC cell lines OCSL
and OC2 using the demethylating agent 5-Aza-dC to determine the
correlation between HOXA5 methylation status and its gene
expression and to observe whether HOXA5 expression could be
augmented. Quantitative MSP analysis revealed that HOXA5
methylation levels decreased in a dose-dependent manner after
treatment using 5-Aza-dC (Fig.
3A). HOXA5 expression was observed to increase in the
protein in both cell lines, which was consistent with the reduced
DNA methylation (Fig. 3B).
Increased p53 expression was also observed in OC2 after 5-Aza-dC
treatment, however, p53 expression was negligible in OCSL before
and after 5-Aza-dC treatment (Fig.
3B). The results suggested that the methylation level and gene
expression of HOXA5 were negatively correlated and that DNA
methylation plays an important role in HOXA5 downregulation
in OSCC.
HOXA5 upregulates p53 transcriptional
activity in OSCC
A previous study found that HOXA5 is a p53
transcription factor and induces the apoptosis of breast cancer
cells (16). To determine if HOXA5
plays a role in regulating p53 expression and inducing OSCC cell
death, the present study either overexpressed or knocked down
HOXA5 in the OC2 cell line by employing lentiviral
vector-mediated HOXA5 transfer or RNA interference. When
HOXA5 was overexpressed, the expression of p53 was also
upregulated in OC2 (Fig. 4A). By
contrast, HOXA5 knockdown using the
lentiviral-vector-delivered shRNA targeting HOXA5
downregulates the p53 expression (Fig.
4B). These results indicated a positive correlation between the
expression levels of HOXA5 and p53. To further determine if p53
expression was transcriptionally regulated by HOXA5, we transfected
OC2 cells using the wild-type (pGL2b-WTp53-FL) or mutant-type
(pGL2b-Mutp53-FL) p53-responsive luciferase expression plasmid
followed by performing a luciferase reporter assay. A greater
transcriptional activity of luciferase in
HOXA5-overexpressed cells was observed compared with control
cells (Fig. 4C). These results
indicated that HOXA5 may act as a transcription factor in the
upregulation of p53 expression in OC2 cells.
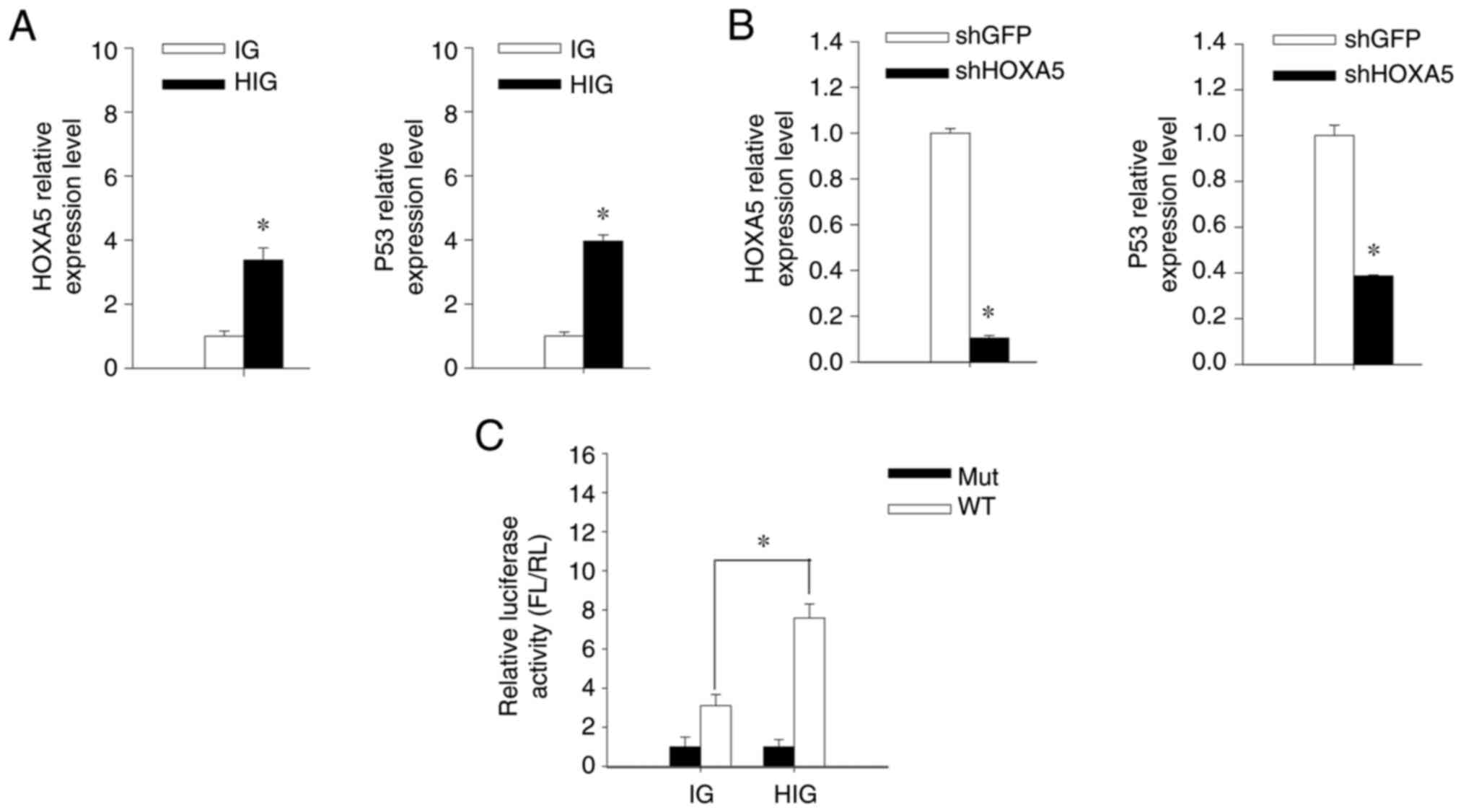 | Figure 4.HOXA5 upregulates p53 expression in
OSCC. (A) OC2 cells were transduced using IG or HIG followed by
fluorescence-activated cell sorting and the mRNA levels of HOXA5
and p53 in OC2 were quantified using RT-qPCR. The expression level
of each IG-transduced cell was assigned a value of 1. (B) OC2 cells
were transduced using lentiviral vector carrying shHOXA5 or shGFP
and selected using puromycin. The mRNA expression of HOXA5 and p53
in OC2 were quantified using RT-qPCR. The expression level of each
shGFP-transduced cell was assigned a value of 1. (C) The IG- and
HIG-transduced OC2 cells were transfected with pGL2b-WTp53-FL or
pGL2b-Mutp53-FL and cotransfected with pRL-TK to act as internal
control reporters. Bioluminescence intensities of FL and RL were
measured using a luminometer and the luciferase activity was
calculated from the FL-to-RL ratio. The IG-transduced OC2 cells
transfected with pGL2b-Mutp53-FL were assigned a value of 1. Error
bars indicate standard deviations. *P<0.05. WT, pGL2b-WTp53-FL;
Mut, pGL2b-Mutp53-FL. OSCC, oral squamous cell carcinoma; IG,
control lentiviral vector; HIG, HOXA5-expressing lentiviral vector;
RT-qPCR, reverse transcription-quantitative PCR; sh, short hairpin;
FL, firefly luciferase; RL, Renilla luciferase; Mut, mutant;
WT, wild-type. |
Restoration of HOXA5 reduces OSCC cell
viability
Previous studies have demonstrated that p53 affects
the viability of various cancer cell types via mechanisms such as
apoptosis, cell-cycle arrest and senescence (30,31);
the present study also demonstrated that HOXA5 can upregulate p53
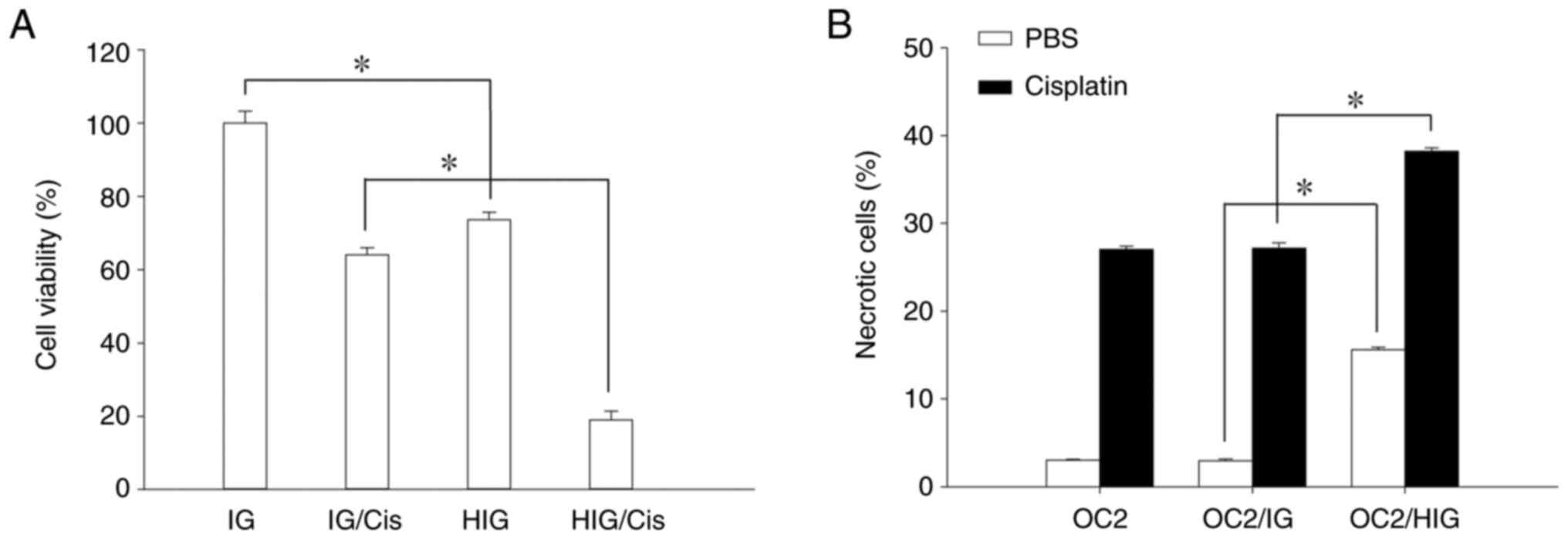
expression in OC2 cells. An MTS assay was performed to measure the
viability of HOXA5-overexpressed OC2 cells and determine if
HOXA5-upregulated p53 expression influences the viability of OSCC
cells. The results in Fig. 5A
revealed that the viability of HOXA5-overexpressed cells was
inhibited relative to the IGs. Cisplatin is a common chemotherapy
drug for treating various cancers, including OSCC (32). Nevertheless, the therapeutic effect
of cisplatin can be attenuated by the loss of p53 function in
cancer cells, which also induces resistance to cisplatin (33). HOXA5-overexpressed cells
were exposed to cisplatin and then cell viability was analyzed
using the MTS assay to determine whether HOXA5-upregulated p53
expression enhanced the sensitivity of cisplatin. The results in
Fig. 5A revealed that the
viability of HOXA5-overexpressed cells was inhibited
relative to the IGs after exposure to cisplatin. Furthermore, a
cell necrosis assay confirmed that HOXA5 overexpression
induces necrosis in OSCC cells and hence reduces their viability
(Fig. 5B).
HOXA5 enhances the therapeutic effect
of cisplatin in OSCC in vivo
The in vitro experiments indicated that
HOXA5 can reduce cell viability and enhance the therapeutic
effect of cisplatin in OC2 cells. Cisplatin was administered to
mice bearing subcutaneous OC2 ×enografts to confirm the role of
HOXA5 in enhancing the efficiency of chemotherapy against
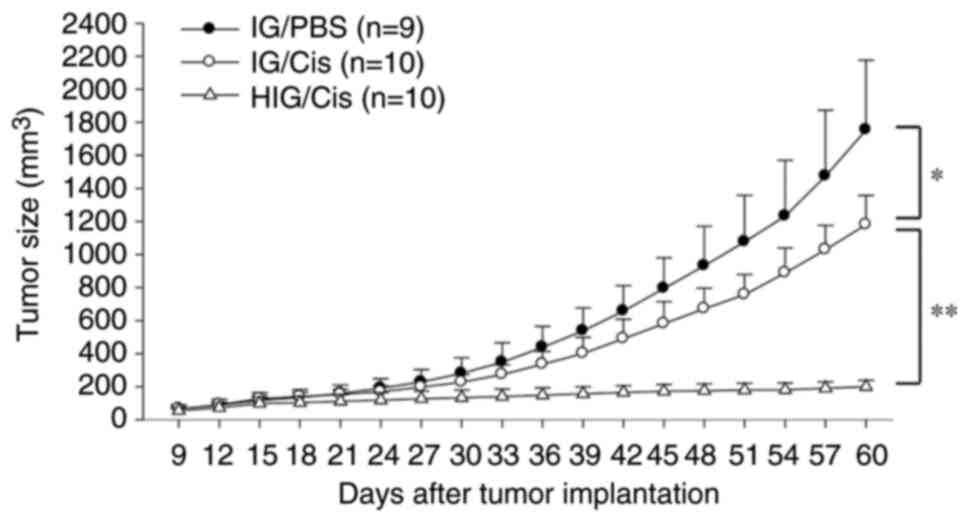
OSCC in vivo. In comparison with the IG/PBS group, cisplatin
treatment slightly suppressed tumor growth in the IG/cisplatin
group. HOXA5 overexpression further enhanced the therapeutic
effect of cisplatin in the HIG/cisplatin group, indicating a strong
antitumor effect from combining HOXA5 and cisplatin
(Fig. 6).
Discussion
The high-throughput methylation array applied in the
present study revealed that HOXA5 was hypermethylated in
OSCC tissues, an observation that was also confirmed by the
bisulfite sequencing analysis (Fig.
1). Hypermethylation of HOXA5 has been identified as a
mechanism that suppresses its expression in breast and skin
tumorigenesis (16,34). Consistent with RT-qPCR data that
revealed the HOXA5 expression level to be lower in OSCC
tissues than in normal oral tissues (Fig. 2), HOXA5 expression can be
promoted in OSCC cells after treating the cells using the
demethylating agent 5-Aza-dC (Fig.
3).
While it is clear that DNA methylation plays an
important role in inhibiting HOXA5 expression in OSCC, other
epigenetic regulations including histone hypoacetylation were also
considered as contributing events in the process of oral
carcinogenesis (11). HOXA5
expression in OSCC cells increased following 5-Aza-dC treatment,
while no such effects occurred following treatment with histone
deacetylase inhibitor trichostatin A (TSA) (data not shown).
Consistent with the previous study (35) finding that DNA methylation was more
dominant than histone hypoacetylation in regulating HOXA5
expression in breast cancer cells, in which TSA treatment did not
reactivate the silenced HOXA5, HOXA5 hypermethylation is the
main mechanism underlying the inhibition of HOXA5 expression
in OSCC cells.
The present study demonstrated that restoring
HOXA5 expression not only inhibited OSCC growth in
vitro but also enhanced the therapeutic effect of cisplatin
both in vitro and in vivo. These effects were in part
achieved by HOXA5 upregulating p53 expression, which has
also been demonstrated to activate an apoptotic pathway in breast
cancer cells (16). p53
upregulation has also been demonstrated to subsequently increase
the expression of its downstream target genes, p21 and
Bax, in OSCC cells treated with natural compounds and
aspirin (36–38). However, p53 mutations have been
reported to occur in ~50% of human cancers (39) and it is also the gene that most
frequently mutates in OSCC and causes failure of cisplatin
treatments and poor disease outcomes (40). A previous study on the p53-mutated
breast cancer cell line Hs578T found that HOXA5 could
alternatively induce cell apoptosis via a p53-independent pathway
through the activation of caspase-2 and caspase-8 (41). HOXA5 restoration therefore
might still be able to activate caspases in the case of p53-mutated
OSCC, causing p53-independent apoptosis.
Through monitoring the growth curves of tumors, it
was found that HOXA5 greatly enhanced the therapeutic effect
of cisplatin in vivo, achieving a major reduction in tumor
size. However, at the experimental endpoint, all mice were humanely
sacrificed without surgical excision of tumors. As a result, the
present study was unable to show the appearance and the actual size
of excised tumors.
In summary, the present study found a differential
pattern of HOXA5 methylation between normal oral and OSCC
tissues, indicating that HOXA5 hypermethylation is a
reliable biomarker for detecting OSCC. The data suggested that
HOXA5 is a downregulated proapoptotic gene in OSCC and that
restoring expression can help to treat OSCC regardless of the
presence of chemotherapy.
Acknowledgements
Not applicable.
Funding
The present study was supported in part by the Ministry of
Science and Technology of Taiwan (grant no. MOST105-2320-B-194-003)
and Ditmanson Medical Foundation Chia-Yi Christian Hospital (grant
no. R108-018).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YC and SL performed the laboratory experiments and
drafted the manuscript. YeL contributed to the laboratory work. YuL
performed analysis and interpretation of data. YiL and CT conceived
and coordinated the overall study and revised the manuscript. YiL
and CT confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Tissue samples were collected from patients after
obtaining written informed consent in accordance with a protocol
approved by the Institutional Review Board of China Medical
University Hospital, Taiwan, R.O.C. (IRB no. CMUH102-REC1-054). All
animal experiments were conducted in accordance with the principles
of laboratory animal care at the National Institutes of Health and
with the approval of the Institutional Animal Care and Use
Committee at National Chung Cheng University, Taiwan, R.O.C. (IACUC
no. 1080401).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–e386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ministry of Health and Welfare, . 2021
Cancer Registry Annual Report. https://twcr.tw/wp-content/uploads/2024/02/Top-10-cancers-in-Taiwan-2021.pdfDecember
18–2024
|
|
4
|
Health Promotion Administration and
Ministry of Health Welfare, . 2023 Health Promotion Administration
Annual Report. https://www.hpa.gov.tw/EngPages/Detail.aspx?nodeid=1070&pid=18165December
18–2024
|
|
5
|
Warnakulasuriya S: Global epidemiology of
oral and oropharyngeal cancer. Oral Oncol. 45:309–316. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pedruzzi PAG, Kowalski LP, Nishimoto IN,
Oliveira BV, Tironi F and Ramos GHA: Analysis of prognostic factors
in patients with oropharyngeal squamous cell carcinoma treated with
radiotherapy alone or in combination with systemic chemotherapy.
Arch Otolaryngol Head Neck Surg. 134:1196–1204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ishida K, Tomita H, Nakashima T, Hirata A,
Tanaka T, Shibata T and Hara A: Current mouse models of oral
squamous cell carcinoma: Genetic and chemically induced models.
Oral Oncol. 73:16–20. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin CS, Lin YC, Adebayo BO, Wu A, Chen JH,
Peng YJ, Cheng MF, Lee WH, Hsiao M, Chao TY and Yeh CT: Silencing
JARID1B suppresses oncogenicity, stemness and increases radiation
sensitivity in human oral carcinoma. Cancer Lett. 368:36–45. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Koch FP, Kunkel M, Biesterfeld S and
Wagner W: Diagnostic efficiency of differentiating small cancerous
and precancerous lesions using mucosal brush smears of the oral
cavity-a prospective and blinded study. Clin Oral Investig.
15:763–769. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hadzic S, Gojkov-Vukelic M, Pasic E and
Dervisevic A: Importance of early detection of potentially
malignant lesions in the prevention of oral cancer. Mater Sociomed.
29:129–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
D'Souza W and Saranath D: Clinical
implications of epigenetic regulation in oral cancer. Oral Oncol.
51:1061–1068. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hema KN, Smitha T, Sheethal HS and
Mirnalini SA: Epigenetics in oral squamous cell carcinoma. J Oral
Maxillofac Pathol. 21:252–259. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li YF, Hsiao YH, Lai YH, Chen YC, Chen YJ,
Chou JL, Chan MW, Lin YH, Tsou YA, Tsai MH and Tai CK: DNA
methylation profiles and biomarkers of oral squamous cell
carcinoma. Epigenetics. 10:229–236. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lizen B, Moens C, Mouheiche J, Sacré T,
Ahn MT, Jeannotte L, Salti A and Gofflot F: Conditional loss of
Hoxa5 function early after birth impacts on expression of genes
with synaptic function. Front Mol Neurosci. 10:3692017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Landry-Truchon K, Houde N, Boucherat O,
Joncas FH, Dasen JS, Philippidou P, Mansfield JH and Jeannotte L:
HOXA5 plays tissue-specific roles in the developing respiratory
system. Development. 144:3547–3561. 2017.PubMed/NCBI
|
|
16
|
Raman V, Martensen SA, Reisman D, Evron E,
Odenwald WF, Jaffee E, Marks J and Sukumar S: Compromised HOXA5
function can limit p53 expression in human breast tumours. Nature.
405:974–978. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kikuyama M, Takeshima H, Kinoshita T,
Okochi-Takada E, Wakabayashi M, Akashi-Tanaka S, Ogawa T, Seto Y
and Ushijima T: Development of a novel approach, the
epigenome-based outlier approach, to identify tumor-suppressor
genes silenced by aberrant DNA methylation. Cancer Lett.
322:204–212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Conway K, Edmiston SN, May R, Kuan PF, Chu
H, Bryant C, Tse CK, Swift-Scanlan T, Geradts J, Troester MA and
Millikan RC: DNA methylation profiling in the Carolina breast
cancer study defines cancer subclasses differing in
clinicopathologic characteristics and survival. Breast Cancer Res.
16:4502014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Daugaard I, Dominguez D, Kjeldsen TE,
Kristensen LS, Hager H, Wojdacz TK and Hansen LL: Identification
and validation of candidate epigenetic biomarkers in lung
adenocarcinoma. Sci Rep. 6:358072016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Martinez R, Carmona FJ, Vizoso M, Rohde V,
Kirsch M, Schackert G, Ropero S, Paulus W, Barrantes A, Gomez A and
Esteller M: DNA methylation alterations in grade II- and anaplastic
pleomorphic xanthoastrocytoma. BMC Cancer. 14:2132014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanai M, Hamada J, Takada M, Asano T,
Murakawa K, Takahashi Y, Murai T, Tada M, Miyamoto M, Kondo S and
Moriuchi T: Aberrant expressions of HOX genes in colorectal and
hepatocellular carcinomas. Oncol Rep. 23:843–851. 2010.PubMed/NCBI
|
|
22
|
Rodini CO, Xavier FCA, Paiva KBS, De Souza
Setúbal Destro MF, Moyses RA, Michaluarte P, Carvalho MB, Fukuyama
EE; Head and Neck Genome Project Gencapo, ; Tajara EH, et al:
Homeobox gene expression profile indicates HOXA5 as a candidate
prognostic marker in oral squamous cell carcinoma. Int J Oncol.
40:1180–1188. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wong DY, Chang KW, Chen CF and Chang RC:
Characterization of two new cell lines derived from oral cavity
human squamous cell carcinomas-OC1 and OC2. J Oral Maxillofac Surg.
48:385–390. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lai YH, He RY, Chou JL, Chan MWY, Li YF
and Tai CK: Promoter hypermethylation and silencing of tissue
factor pathway inhibitor-2 in oral squamous cell carcinoma. J
Transl Med. 12:2372014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gillan L, Matei D, Fishman DA, Gerbin CS,
Karlan BY and Chang DD: Periostin secreted by epithelial ovarian
carcinoma is a ligand for alpha(V)beta(3) and alpha(V)beta(5)
integrins and promotes cell motility. Cancer Res. 62:5358–5364.
2002.PubMed/NCBI
|
|
27
|
Shichinohe T, Bochner BH, Mizutani K,
Nishida M, Hegerich-Gilliam S, Naldini L and Kasahara N:
Development of lentiviral vectors for antiangiogenic gene delivery.
Cancer Gene Ther. 8:879–889. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen YJ, Chen SY, Lovel R, Ku YC, Lai YH,
Hung CL, Li YF, Lu YC and Tai CK: Enhancing chemosensitivity in
oral squamous cell carcinoma by lentivirus vector-mediated RNA
interference targeting EGFR and MRP2. Oncol Lett. 12:2107–2114.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kirch HC, Flaswinkel S, Rumpf H, Brockmann
D and Esche H: Expression of human p53 requires synergistic
activation of transcription from the p53 promoter by AP-1,
NF-kappaB and Myc/Max. Oncogene. 18:2728–2738. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kaiser AM and Attardi LD: Deconstructing
networks of p53-mediated tumor suppression in vivo. Cell Death
Differ. 25:93–103. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mello SS and Attardi LD: Deciphering p53
signaling in tumor suppression. Curr Opin Cell Biol. 51:65–72.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Andreadis C, Vahtsevanos K, Sidiras T,
Thomaidis I, Antoniadis K and Mouratidou D: 5-Fluorouracil and
cisplatin in the treatment of advanced oral cancer. Oral Oncol.
39:380–385. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jo DW, Kim YK and Yun PY: The influence of
p53 mutation status on the anti-cancer effect of cisplatin in oral
squamous cell carcinoma cell lines. J Korean Assoc Oral Maxillofac
Surg. 42:337–344. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Watson RE, Curtin GM, Hellmann GM,
Doolittle DJ and Goodman JI: Increased DNA methylation in the HoxA5
promoter region correlates with decreased expression of the gene
during tumor promotion. Mol Carcinog. 41:54–66. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Novak P, Jensen T, Oshiro MM, Wozniak RJ,
Nouzova M, Watts GS, Klimecki WT, Kim C and Futscher BW: Epigenetic
inactivation of the HOXA gene cluster in breast cancer. Cancer Res.
66:10664–10670. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang SH, Liao PH, Pan YF, Chen SL, Chou SS
and Chou MY: The novel p53-dependent metastatic and apoptotic
pathway induced by vitexin in human oral cancer OC2 cells.
Phytother Res. 27:1154–1161. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yeh YT, Yeh H, Su SH, Lin JS, Lee KJ, Shyu
HW, Chen ZF, Huang SY and Su SJ: Phenethyl isothiocyanate induces
DNA damage-associated G2/M arrest and subsequent apoptosis in oral
cancer cells with varying p53 mutations. Free Radic Biol Med.
74:1–13. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ho CC, Yang XW, Lee TL, Liao PH, Yang SH,
Tsai CH and Chou MY: Activation of p53 signalling in
acetylsalicylic acid-induced apoptosis in OC2 human oral cancer
cells. Eur J Clin Invest. 33:875–882. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Olivier M, Hollstein M and Hainaut P: TP53
mutations in human cancers: Origins, consequences, and clinical
use. Cold Spring Harb Perspect Biol. 2:a0010082010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lindemann A, Takahashi H, Patel AA, Osman
AA and Myers JN: Targeting the DNA damage response in OSCC with
TP53 mutations. J Dent Res. 97:635–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen H, Chung S and Sukumar S:
HOXA5-induced apoptosis in breast cancer cells is mediated by
caspases 2 and 8. Mol Cell Biol. 24:924–935. 2004. View Article : Google Scholar : PubMed/NCBI
|