Introduction
Keloids, which are fibroproliferative, arise due to
a disruption in the wound-healing process (1). Keloids exhibit cancer-like
characteristics, and are raised complex lesions that grow rapidly
and uncontrollably, infiltrate surrounding tissues, do not regress
spontaneously and often recur after excision (2). These lesions can extend beyond the
original incision site, leading to pain, functional impairment and
physical distress (3). Despite
significant progress in the prevention and treatment of keloids,
the precise mechanism underlying their formation remains unknown.
Thus, a comprehensive understanding of keloid pathogenesis is
essential for the development of efficacious therapeutic
interventions. Fructus arctii, a popular vegetable in China
and Japan, is widely appreciated for its various health-promoting
properties (4). Studies have
demonstrated that β-sitosterol (SIT) possesses diverse
pharmacological properties, including anti-tumor (5), anti-angiogenic (6) and anti-inflammatory effects (7). However, the exact pharmacological
effects of SIT on keloids are not yet fully understood. The
PI3K/AKT pathway is an intrinsic signal transduction pathway in
mammalian cells that regulates angiogenesis, cell growth,
proliferation and metabolism and plays a crucial role in keloid
formation (8–10). PTEN serves as a negative regulator
of the PI3K/AKT/mTOR pathway (11). Consequently, it governs a broad
spectrum of cellular processes related to cell viability,
proliferation and growth (12).
Emerging evidence indicates the downregulation of PTEN in keloids
(13). The hypothesis of the
present study was that SIT exerts a therapeutic effect on keloids
by inhibiting their growth through the PTEN/PI3K/AKT signaling
pathway. The present study comprehensively investigated the
potential mechanism underlying the therapeutic effects of SIT in
keloid treatment through network pharmacology, molecular docking
and in vitro and in vivo experiments. Additionally,
it explored the potential association between SIT and the
PTEN/PI3K/AKT signaling pathway. These findings may offer a
potential remedy for keloids.
Materials and methods
F. arctii active ingredient
acquisition and screening for F. arctii targets and keloids
targets
The present study used The Chinese Medicine Systems
Pharmacology Database and Analysis Platform (TCMSP; http://tcmsp-e.com/) for data analysis (14). The bioactive compounds isolated
from F. arctii were identified via ‘Fructus Arctii’
as a keyword and the drug-likeness screening criterion had to have
a minimum value of 0.18, while oral bioavailability screening had
to have a minimum value of 30%. The number of targets for the
active ingredients were recorded. The UniProt ID was acquired from
the UniProt database for the target (https://www.uniprot.org/) (15). Using ‘keloid’ as a keyword, targets
related to keloid is searched and obtained from the Therapeutic
Target Database (TTD; http://db.idrblab.org/ttd/) (16), DrugBank (https://www.drugbank.ca/) (17), GeneCards (https://www.genecards.org/) (18) and Online Mendelian Inheritance in
Man (OMIM; http://www.omim.org/) (19). Duplicates were removed from the
merged data sets originating from each database. Afterward, a Venn
diagram was created to illustrate the overlap between the targets
of F. arctii and keloids (Fig.
1).
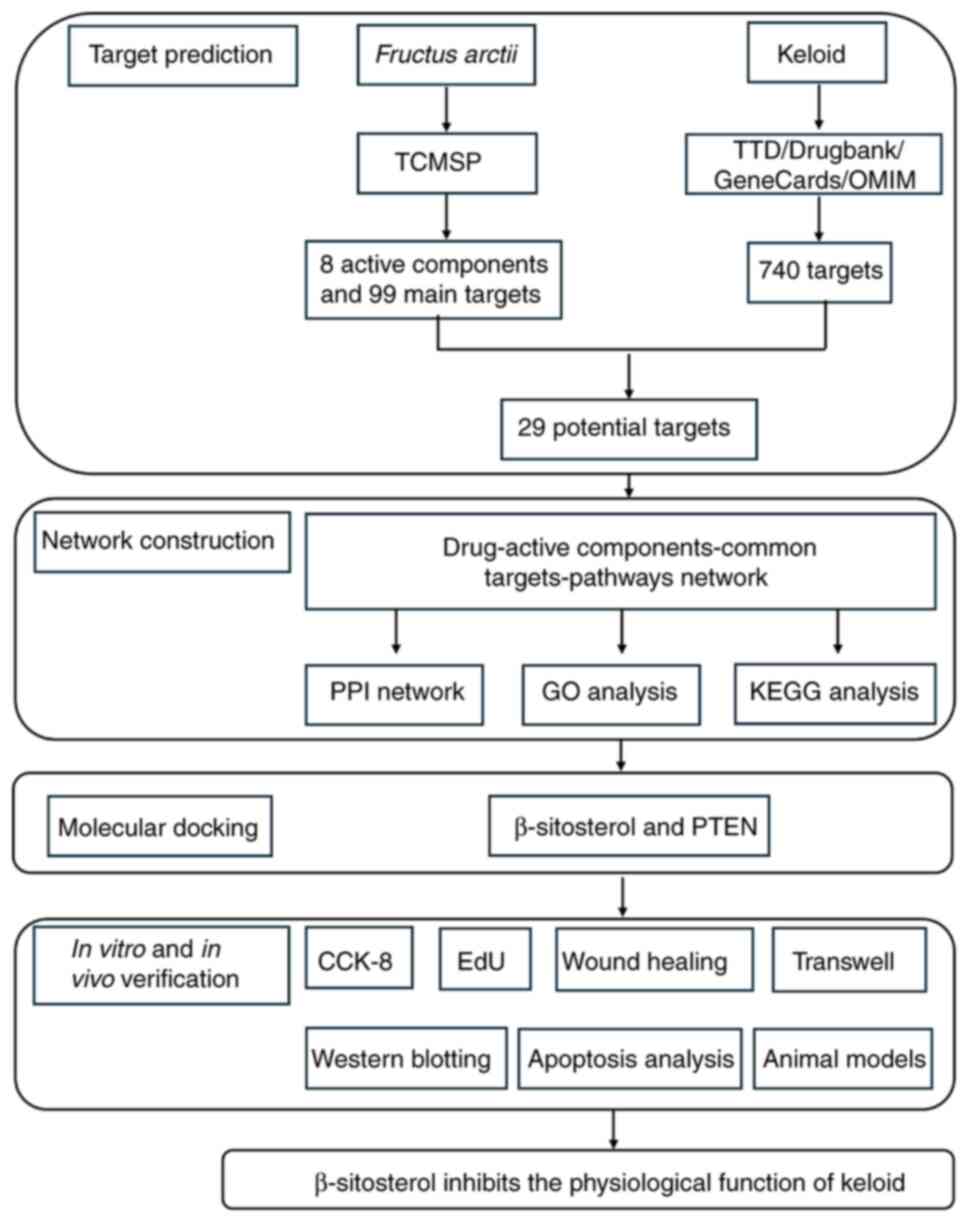 | Figure 1.Flow chart of the present study. It
was divided into four parts: Target prediction, network
construction, molecular docking and in vitro verification.
TCMSP, The Chinese Medicine Systems Pharmacology Database and
Analysis Platform; TTD, Therapeutic Target Database; OMIM, Online
Mendelian Inheritance in Man; PPI, protein-protein interaction;
F. arctii, Fructus arctii; GO, Gene Ontology; KEGG, Kyoto
Encyclopedia of Genes and Genomes; EdU,
5-Ethynyl-2′-deoxyuridine. |
Construction of a drug-active
ingredient-target-pathway network
Following the data entry requirements of Cytoscape
software (version 3.7.0; http://cytoscape.org), the data from the previous
steps were organized and imported into Cytoscape to generate a
network depicting the relationships among active ingredients,
targets and pathways (20).
Following this, the outcomes of the analysis pertaining to F.
arctii and common targets were saved from the network.
Following this, visual representations of the connections between
drug-component-targets were achieved by modifying the shape and
color of each node.
Protein-protein interaction (PPI)
network construction and analysis
The common targets of F. arctii and keloid
were uploaded to the STRING database (21). Homo sapiens was designated
as the species, a high confidence level of 0.700 was established as
the minimum interaction threshold and all other parameters were
maintained at their default values. This allowed the construction
of a PPI core network of keloids, with F. arctii acting upon
it (22). The Network Analyzer and
cytoHubba plug-ins in Cytoscape 3.7.0 software were used to
calculate the topological properties of the PPI network and the top
10 Hub genes were selected.
Pathway analysis of the target
genes
The aforementioned predicted primary targets were
entered into the Metascape database (http://metascape.org), with the species defined as
Homo sapiens. Following database retrieval and
transformation operations, enrichment analyses via Kyoto
Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO)
were conducted. The significant GO functions and KEGG signaling
pathways were screened on the basis of the P-value. This allowed
for the prediction of the mechanism of action of F. arctii
in the treatment of keloids.
Molecular docking verification
The structures of SIT and PTEN (PDB ID:7JVX) were
obtained via the PubChem database (https://pubchem.ncbi.nlm.nih.gov) (23) and RCSB Protein Data Bank (RCSB PDB;
http://www.rcsb.org/structure) (24) for molecular docking studies. They
were then reconstructed using PyMOL software (25). The AutoDock Tools (version 1.5.6;
http://autodock.scripps.edu) were used
for protein-ligand docking analysis. The 3D interactions were
generated using PyMOL software. The results of successful molecular
docking were visualized with PyMOL 2.5.4 software. The binding
affinity energy (kcal/mol) was computed via CB-Dock2 (https://cadd.labshare.cn/cb-dock2/php/index.php) to
determine the optimal docking model with the lowest energy.
Molecular visualization plays a crucial role in analyzing and
communicating modeling studies. Therefore, the details of the
ligand-receptor interactions were revealed via PyMol2 software
(Free version; http://pymol.org) and BIOVIA Discovery
Studio Visualizer software (version 2021; Dassault Systèmes)
(26).
Cells and therapies
The KEL FIB (CRL-1762) human keloid fibroblast (KF)
cell line was obtained from ATCC. The cells were cultured in
Dulbecco's modified Eagle's medium (DMEM; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Beyotime Institute of Biotechnology) at
37°C in a humidified atmosphere with 5% CO2. To
investigate the effects of SIT, KF cells (KFs) were treated with
various concentrations (15, 30, 60 and 120 µM) of SIT
(MedChemExpress) prior to functional analyses. KFs were treated
with YS-49 (10 µM) (cat. no. HY-15477; MedChemExpress), a PI3K/AKT
pathway activator, for 24 h to assess cell replication (27). SIT and YS-49 were dissolved in
dimethyl sulfoxide (DMSO) and the cells in the corresponding
control groups were incubated with an equal volume of DMSO.
Cell counting kit-8 (CCK-8)
assays
To establish an initial concentration range for SIT,
5×103 cells per well were seeded in a 96-well plate. The
cells were subsequently exposed to a gradient of SIT concentrations
(15, 30, 60 and 120 µM) for 12, 24 and 48 h. Cell viability was
subsequently assessed via a CCK-8 assay. After the addition of the
CCK-8 reagent, the cells were incubated for 2 h, after which the
absorbance (OD value) at 450 nm was measured via a microplate
reader (Molecular Devices, LLC). The obtained absorbance values
were normalized to those of the control group.
5-Ethynyl-2′-deoxyuridine (EdU)
assays
A Cell-Light EdU Apollo567 In Vitro Kit
(C10310-1, RiboBio, Guangzhou, Guangdong, China) was used for EdU
staining. Cell proliferation imaging experiments were conducted
using 96-well cell culture plates. The manufacturer's guidelines
were followed to detect EdU-labeled positive cells. Cells were
incubated with Hoechst 33342 dye (10 µg/ml) for 30 min at room
temperature in the dark. After staining, they were washed twice
with PBS. EdU assays were used to assess the activity of DNA
replication as an indicator of changes in the proliferative
capacity of KFs.
Wound-healing assay
KFs were seeded in a six-well plate at a density of
1×105 cells per well and grown to 90% confluence.
Subsequently, the cells were scraped with a 1-ml sterile pipette
and the resulting cell pellets were rinsed with PBS three times.
After incubation in serum-free medium, the cells were treated with
different concentrations of SIT. The samples were observed under a
microscope at 0 and 24 h after drug treatment. Cell migration was
assessed by quantifying the percentage of the wound closure area in
three independent experiments via ImageJ version 1.53t software
(National Institutes of Health).
Transwell assay
In 24-well Transwell chambers, cell invasion and
migration were evaluated. To analyze invasion, the plate was
precoated with Matrigel (Beijing Solarbio Science & Technology
Co., Ltd.) for 4 h at 37°C before cell seeding; for migration,
chambers devoid of Matrigel were used. A total of 1×104
cells were seeded into the upper compartment of each Transwell
chamber, which contained 100 µl of DMEM (Corning, Inc.). The lower
chamber was filled with 500 µl of complete medium containing 10%
FBS as a chemoattractant. The cells were fixed and incubated for 20
min with 0.1% crystal violet after 24 h of SIT treatment. A cotton
swab was subsequently used to delicately remove the cells that were
still present on the upper surface of the membrane insert. The
quantity of cells that had successfully migrated to the lower
surface was then determined.
Western blot analysis
Western blot analysis was used to determine protein
expression in KFs in accordance with previously described
procedures (28). Protein lysate
was extracted using RIPA buffer (Beijing Solarbio Science &
Technology Co., Ltd.) and protein concentration was determined
using the BCA Protein Assay Kit (Beyotime Institute of
Biotechnology). Proteins (30 µg/lane) were separated by SDS-PAGE on
8–10% gels, transferred onto PVDF membranes, blocked with 5%
non-fat dry milk (MilliporeSigma) at room temperature for 1 h and
were then incubated with primary antibodies overnight at 4°C. The
primary antibodies used were as follows: anti-PI3K (4249T; 1:1,000;
CST Biological Reagents Co., Ltd.), anti-phosphorylated (p-)PI3K
(17366; 1:1,000; CST Biological Reagents Co., Ltd.), anti-PTEN
(AF5447; 1:1,000; Affinity), anti-AKT (4691; 1:1,000; CST
Biological Reagents Co., Ltd.), anti-p-AKT (4060; 1:1,000; CST
Biological Reagents Co., Ltd.), anti-E-cadherin (3195; 1:1,000; CST
Biological Reagents Co., Ltd.), anti-Vimentin (5741; 1:1,000; CST
Biological Reagents Co., Ltd.), anti-Snail (3879; 1:1,000; CST
Biological Reagents Co., Ltd.), anti-zonula occludens-1 (ZO-1;
8193; 1:1,000; CST Biological Reagents Co., Ltd.) and anti-β-actin
(4967; 1:1,000; CST Biological Reagents Co., Ltd.). Subsequently,
the blots were probed with HRP-conjugated anti-rabbit and
anti-mouse secondary antibodies (1:5,000; cat. nos. RGAR001 and
RGAM001; Proteintech Group, Inc.), which were diluted in TBS-0.1%
Tween 20 solution, at room temperature for 2 h. The blots were
observed with an enhanced chemiluminescence Western blot detection
reagent (MilliporeSigma). ImageJ version 1.53t software (National
Institutes of Health) was used to conduct a statistical analysis of
the grayscale values of the proteins. After normalization to
β-actin, the internal control, the relative expression of the
target protein in each group of cells was computed.
Animal models
A total of 10 male SPF-grade BALB/c nude mice, aged
six weeks and weighing 20±2 g, were procured from the Experimental
Animal Center of Yanbian University (Yanji, China) and were
randomly assigned to two groups (n=5 mice/group). Nude mice were
housed in a specific pathogen-free environment with a temperature
of 25°C and humidity of 30%. The mice were exposed to a 12-h
light/dark cycle, and were given free access to adult mouse feed
sterilized with cobalt-60 and water sterilized by autoclaving.
Following a 1-week acclimatization period with standard feeding, a
nude mouse keloid model was established. This was achieved by
injecting a keloid fibroblast suspension with a density of
5.0×107 cells/ml diluted in Matrigel (Corning, Inc.)
under aseptic conditions. The mixture was then kept on ice.
Subsequently, 0.1 ml of the prepared keloid fibroblast suspension
was subcutaneously injected into the axilla of the right forelimb
of each nude mouse. The keloid volume, weight and growth of the
mice were monitored every three days. The average tumor volume was
calculated via the following formula: tumor volume=(length ×
width2)/2. After 7 days, keloid formation became visible
under the skin in the right axilla of the nude mice. Upon reaching
a size of approximately 50 mm3, the SIT-treated group of
nude mice received intraperitoneal injections of SIT at a safe
therapeutic dose of 50 mg/kg, as determined in a previous study
(29). The treatments were
repeated every two days for a total of 10 cycles. The control group
received equivalent saline injections. Throughout the course of the
experiment, the mice were closely monitored for any signs of poor
health, including weight loss, abnormal posture, changes in
activity levels, labored breathing and signs of distress. No signs
of severe distress were observed in the animals prior to sacrifice.
At the end of the experiment, all mice were sacrificed by cervical
dislocation following anesthesia with an injection of sodium
pentobarbital at a dose of 70 mg/kg body weight and death was
confirmed by the absence of respiration and heartbeat for more than
5 min. The fibroproliferative tissues were collected for
measurement and further experiments.
Hematoxylin and eosin (H&E)
staining and immunohistochemistry (IHC)
The subcutaneous tumors were fixed in 10% formalin
at room temperature for 24 h, dehydrated in graded ethanol and
embedded in paraffin. The paraffin-embedded tumors were then
sectioned into 4-µm slices, which were oven-baked at 56°C overnight
and stained using the H&E Stain kit (Beijing Solarbio Science
& Technology Co., Ltd.), according to the manufacturer's
protocol. For IHC, the slides were placed in 10 mM sodium citrate
buffer (pH 6.0) and boiled, then simmered for 10 min, for antigen
retrieval, before being cooled for 30 min. To remove endogenous
peroxidase activity, slides were incubated with 3% hydrogen
peroxide aqueous solution for 10 min at room temperature (20–25°C).
Furthermore, blocking was performed using TBS-0.1% Tween with 5%
normal goat serum for 1 h at room temperature (Cell Signaling
Technology, Inc.). The sections were then incubated overnight at
4°C with primary antibodies against PTEN (cat. no. AF5447; 1:200
dilution; Affinity Biosciences). The next day, a biotinylated Goat
Anti-Rabbit IgG H&L (Biotin) secondary antibody (1:100; cat.
no. ab207995; Abcam) was added, and the slides were incubated at
room temperature for 30 min. Streptavidin-horseradish peroxidase
(cat. no. SA10001; Invitrogen; Thermo Fisher Scientific, Inc.) was
then used to incubate the slides at room temperature for 30 min and
200 µl DAB was added to each section as the chromogen. Hematoxylin
was used to counterstain the sections at room temperature for 1
min. Finally, images of the slides were captured using a Nikon
Eclipse Ti-S/L100 inverted phase contrast fluorescence microscope
with a 20× objective.
In vivo safety
Blood biochemical analysis was conducted to evaluate
biosafety parameters, including creatinine (CREA), alanine
aminotransferase (ALT), serum aspartate aminotransferase (AST) and
blood urea nitrogen (BUN) (27).
The serum was separated by centrifugation at 4,000 × g for 10 min
at 4°C. ALT, AST, BUN and CREA levels were determined following the
instructions provided with the kit (Nanjing Jiancheng
Bioengineering Institute).
Statistical analysis
Data analysis was performed via GraphPad Prism 7.0
software (Dotmatics). Group variables were characterized by the
number of cases, scores, measurements and percentages. Data that
followed a normal distribution are presented as the means ±
standard deviations. Multiple group comparisons were conducted via
one-way ANOVA with either Tukey's or Dunnett's post hoc tests to
determine significant differences between groups. P<0.05 was
considered to indicate a statistically significant difference. Each
experiment was performed in triplicate.
Results
Identification of active components in
F. arctii and screening of shared targets between drugs and
diseases
The present study used the TCMSP screening criteria
to identify eight chemical components (Table I). Among these components were
neoarctin A, arctiin, SIT, kaempferol, aupraene, β-carotene,
(3R,4R)-3,4-bis[(3,4-dimethoxyphenyl)methyl]oxolan-2-one and
cynarine. By setting the species to Homo sapiens, the genes
of each target pair were cross-referenced with the UniProt protein
database to obtain the standardized symbols. A total of 99 target
genes associated with the active ingredients and 740 targets
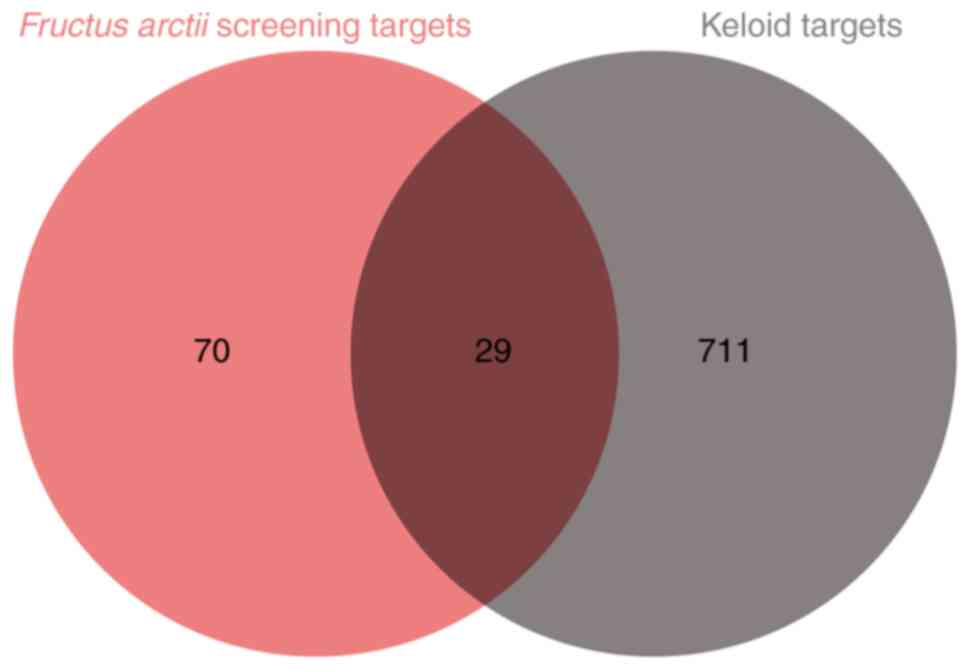
related to keloid formation were identified (Table SI). A Venn diagram depicting the
overlapping targets between F. arctii and keloid formation
showed 29 shared targets (Fig. 2).
The details of these targets are provided in Table II.
 | Table I.A total of eight Fructus
arctii active ingredients from the TCMSP database were screened
according to oral bioavailability (OB) and drug-likeness (DL). |
Table I.
A total of eight Fructus
arctii active ingredients from the TCMSP database were screened
according to oral bioavailability (OB) and drug-likeness (DL).
| Mol ID | Molecule Name | MW | AlogP | Hdon | Hacc | OB, % | Caco-2 | BBB | DL | FASA- | HL | Targets |
|---|
| MOL010868 | Neoarctin A | 742.88 | 7.44 | 1 | 12 | 39.99 | 0.32 | −0.36 | 0.27 | 0.21 | 5.82 | 0 |
| MOL000522 | Arctiin | 534.61 | 1.81 | 4 | 11 | 34.45 | −0.73 | −1.5 | 0.84 | 0.24 | 1.41 | 10 |
| MOL000358 | β-sitosterol | 414.79 | 8.08 | 1 | 1 | 36.91 | 1.32 | 0.99 | 0.75 | 0.23 | 5.36 | 38 |
| MOL000422 | kaempferol | 286.25 | 1.77 | 4 | 6 | 41.88 | 0.26 | −0.55 | 0.24 | 0 | 14.74 | 63 |
| MOL001506 | Supraene | 410.8 | 11.33 | 0 | 0 | 33.55 | 2.08 | 1.73 | 0.42 | 0.27 | 2.72 | 0 |
| MOL002773 | β-Carotene | 536.96 | 12 | 0 | 0 | 37.18 | 2.25 | 1.52 | 0.58 | 0.33 | 4.36 | 22 |
| MOL003290 |
(3R,4R)-3,4-bis[(3,4-dimethoxy-phenyl)
methyl] oxolan-2-one | 386.48 | 3.97 | 0 | 6 | 52.3 | 0.78 | 0.17 | 0.48 | 0.23 | 3.07 | 16 |
| MOL007326 | Cynarin(e) | 516.49 | 1.56 | 7 | 12 | 31.76 | −0.95 | −2.37 | 0.68 | 0.37 | 3.13 | 0 |
 | Table II.Gene names overlapping targets
between Fructus arctii and keloid formation. |
Table II.
Gene names overlapping targets
between Fructus arctii and keloid formation.
| Gene number | Target
intersection |
|---|
| 1 | PTGS2 |
| 2 | PTGS1 |
| 3 | PIK3CG |
| 4 | ADRA1A |
| 5 | ADRA1B |
| 6 | BAX |
| 7 | CASP9 |
| 8 | JUN |
| 9 | CASP3 |
| 10 | CASP8 |
| 11 | TGFB1 |
| 12 | NOS2 |
| 13 | PTEN |
| 14 | DPP4 |
| 15 | F2R |
| 16 | AKT1 |
| 17 | MAP3K7 |
| 18 | XDH |
| 19 | MMP1 |
| 20 | CYP3A4 |
| 21 | AKR1C3 |
| 22 | SLPI |
| 23 | MMP2 |
| 24 | CAV1 |
| 25 | CTNNB1 |
| 26 | MYC |
| 27 | GJA1 |
| 28 | MMP10 |
| 29 | ADRA1D |
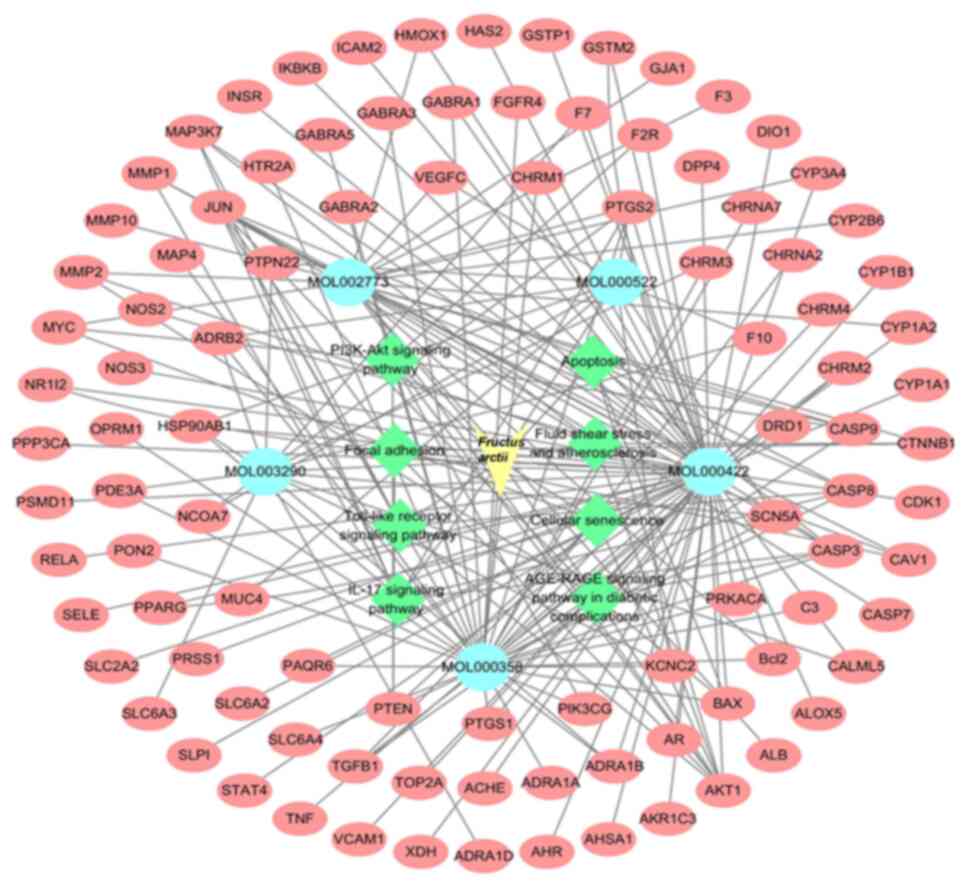
Network of drug-active
ingredient-target-pathway interactions
The present study used Cytoscape 3.7.0 software to
analyze the drug-active ingredient-target-pathway network for F.
arctii and keloids. Fig. 3
shows the exported analytical data. A total of eight KEGG pathways
linked to keloid development were selected to highlight the ones
with targets from F. arctii. It was found that five primary
active ingredients potentially interact with 99 targets across
eight signaling pathways, suggesting that F. arctii may
inhibit keloid development through multiple chemical components,
targets and pathways. In the network, the light blue ovals denoted
active ingredients, the green diamonds indicated pathways and the
red circles represented common targets among components and keloids
and between the pathways and keloids.
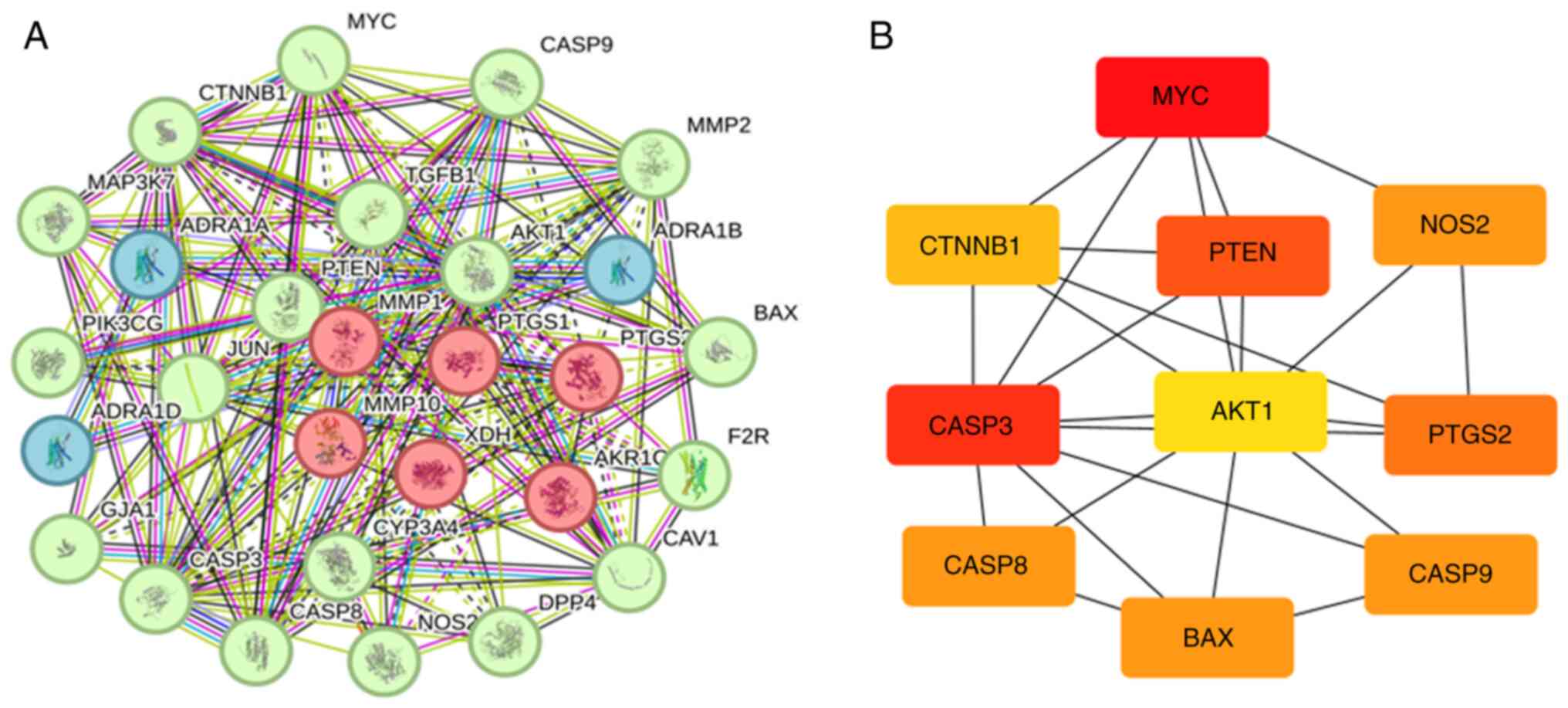
Creation of a PPI network and
screening of core genes
A PPI network for common genes was constructed using
the STRING database. The network comprised 29 nodes and 151 edges,
with an average node degree of 10.4 (Fig. 4A; Table SII). Fig. 4B and Table III show the nodes, including MYC,
CASP3, PTEN, PTGS2, NOS2, CASP9, CASP8, BAX, CTNNB1 and AKT1,
arranged in descending order of their degrees. Research shows that
SIT significantly inhibits the progression of various malignant
tumors by regulating PTEN expression (30,31).
However, investigations on the combined mechanism of action of SIT
and PTEN in keloids have yet to be conducted. The authors aim to
explore the potential roles and mechanisms of action of SIT and
PTEN in keloids. More network connections indicate stronger
associations between the proteins. The PPI network suggested that
F. arctii treatment affected keloids via multiple pathways,
components and targets.
 | Table III.The top 10 genes with highest
confidence scores. |
Table III.
The top 10 genes with highest
confidence scores.
| Rank | Name | Score |
|---|
| 1 | MYC | 0.57059 |
| 2 | CASP3 | 0.477329 |
| 3 | PTEN | 0.475638 |
| 4 | PTGS2 | 0.475492 |
| 5 | NOS2 | 0.473661 |
| 5 | CASP9 | 0.473661 |
| 5 | CASP8 | 0.473661 |
| 5 | BAX | 0.473661 |
| 9 | CTNNB1 | 0.453462 |
| 10 | AKT1 | 0.395144 |
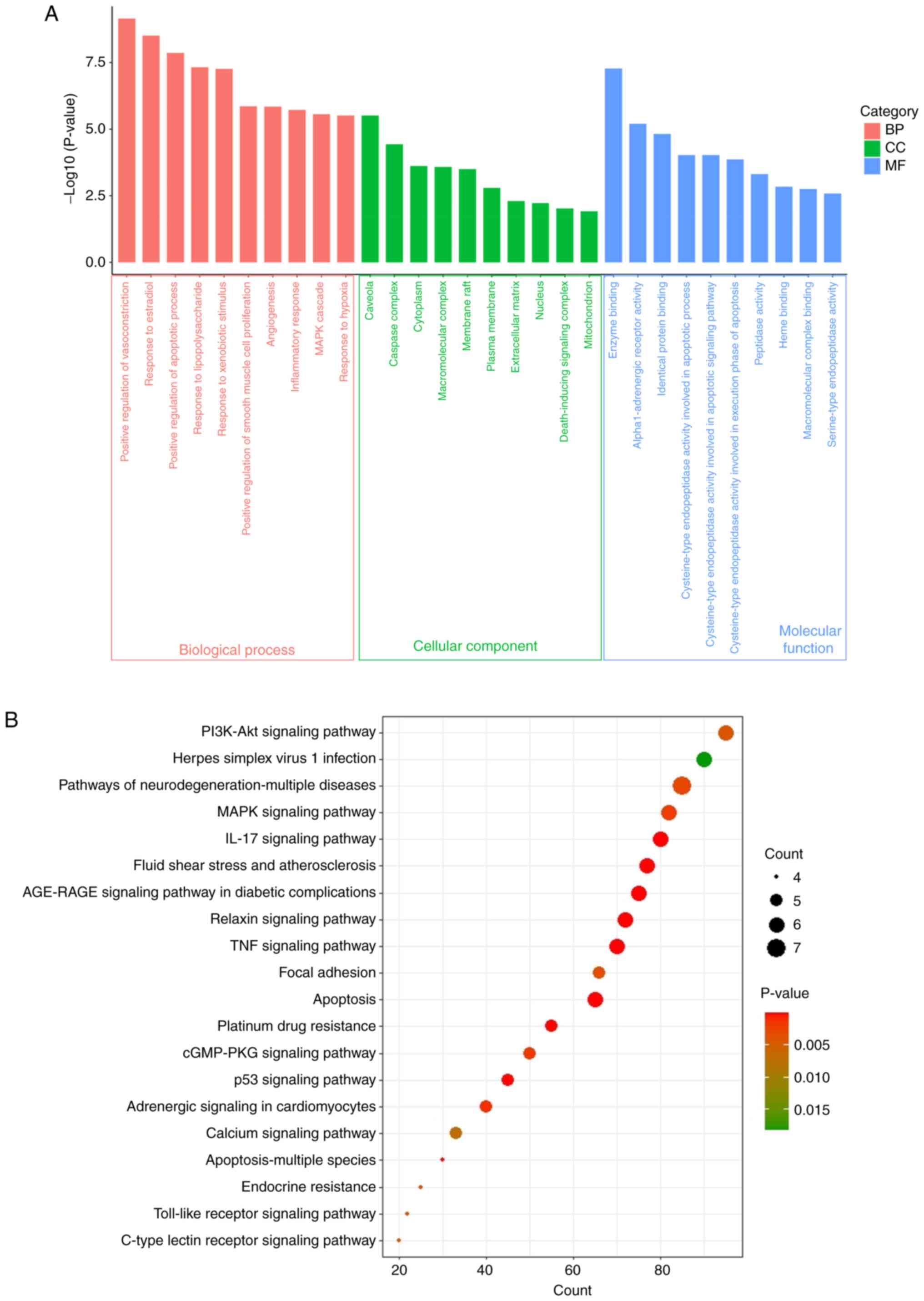
GO and KEGG functional analyses
The results of the GO functional enrichment analysis
for molecular functions (MFs), cellular components (CCs) and
biological processes (BPs) are illustrated in Fig. 5A and Table SIII. The interaction targets were
associated with 198 BPs, 27 CCs and 27 MFs. A histogram depicting
the first 10 GO terms with P<0.01 was prepared. The primary BP
terms included ‘positive regulation of vasoconstriction’, ‘response
to estradiol and the apoptotic process’, ‘response to
lipopolysaccharide’, ‘response to xenobiotic stimuli’, ‘response to
hypoxia’, ‘positive regulation of smooth muscle cell
proliferation’, ‘angiogenesis’, ‘inflammatory response’ and ‘MAPK
cascade’. The primary CC terms were ‘caveolae’, ‘caspase complex’,
‘cytoplasm’, ‘macromolecular complex’, ‘membrane raft’, ‘plasma
membrane’, ‘extracellular matrix’, ‘nucleus’, ‘death-inducing
signaling complex’ and ‘mitochondrion’. The enriched MF terms
included ‘enzyme binding’, ‘alpha1-adrenergic receptor activity’,
‘identical protein binding’, ‘peptidase activity’, ‘heme binding’,
‘macromolecular complex binding’ and ‘serine-type endopeptidase
activity’.
The findings of pathway enrichment analysis
indicated significant enrichment of the 28 core targets in 99
pathways (Table SIV). A bubble
plot was prepared displaying the top 20 pathways with P<0.01
(Fig. 5B). The top 10 significant
pathways included the PI3K-AKT signaling pathway, neurodegeneration
pathways, the MAPK signaling pathway, the IL-17 signaling pathway,
fluid shear stress and atherosclerosis, AGE-RAGE signaling in
diabetic complications, the relaxin signaling pathway, the TNF
signaling pathway, focal adhesion and apoptosis. Among these
pathways, the PI3K-AKT signaling pathway may exhibit the closest
association with keloids.
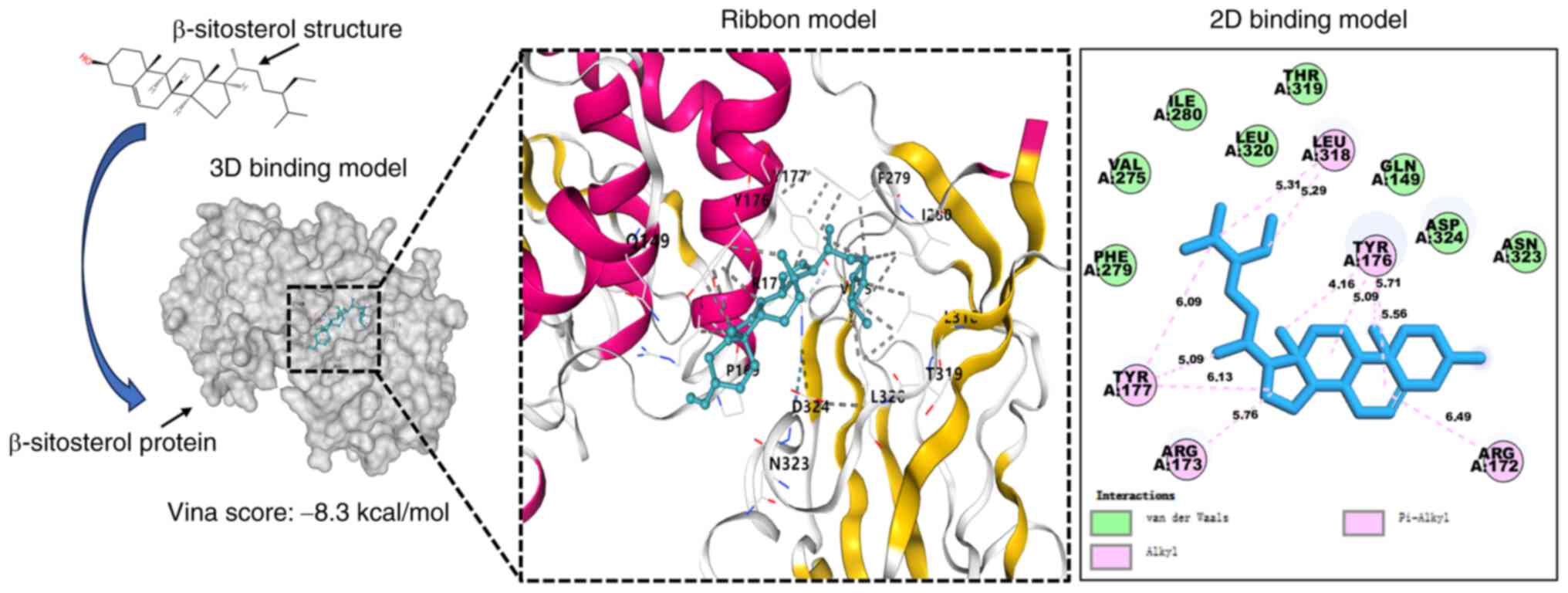
Molecular docking of the active
ingredients of SIT target genes
The molecular model revealed that SIT strongly
interacts with PTEN, as depicted in Fig. 6. The space-filling model clearly
demonstrated the perfect embedding of SIT within the crystal pocket
of PTEN. Similarly, both the ribbon model image and local 2D
binding model revealed the specific interactions between SIT and
the associated protein residues. After simulating the components
using AutoDock tools, a high affinity of −8.3 kcal/mol was
computed. The receptor-ligand docking hypothesis suggests an
inverse relationship between the docking energy and binding
affinity. A lower docking energy indicates stronger binding
affinity. SIT engages in van der Waals interactions with the PTEN
residues PHE279, VAL275, ILE280, LEU320, THR319, GLN149, ASP324 and
ASN323. The stable binding of SIT to PTEN involves the formation of
alkyl and pi-alkyl interaction bonds with residues LEU318, TYR176,
TYR177, ARG172 and ARG173.
SIT inhibits the physiological
function of KFs
Fig. 7A displays
the chemical structure of SIT. To assess the effect of SIT on KFs
proliferation and determine the optimal drug concentration for
cellular experiments, the changes in cellular activity at 12, 24
and 48 h post-SIT treatment were compared to that in the control
group using the CCK-8 assay. Fig.
7B shows that SIT significantly reduced the viability of KFs in
a concentration- and time-dependent manner. The treatment of KFs
with SIT of various concentrations for 48 h decreased the cell
viability from 70 to 5%. The IC50 values for KFs at 12,
24 and 48 h were 70.09, 33.78 and 33.75 µM, respectively. KFs
treated with 30 µM SIT for 48 h showed a drug concentration of
33.75 µM, which was the 48 h IC50 value. The inhibitory
effects observed at other drug concentrations and time points
deviated significantly from the corresponding IC50
values. Subsequently, a drug gradient study on keloid cell cultures
was conducted using 15 and 30 µM SIT as well as a drug reversion
analysis using 30 µM SIT, both for 48 h.
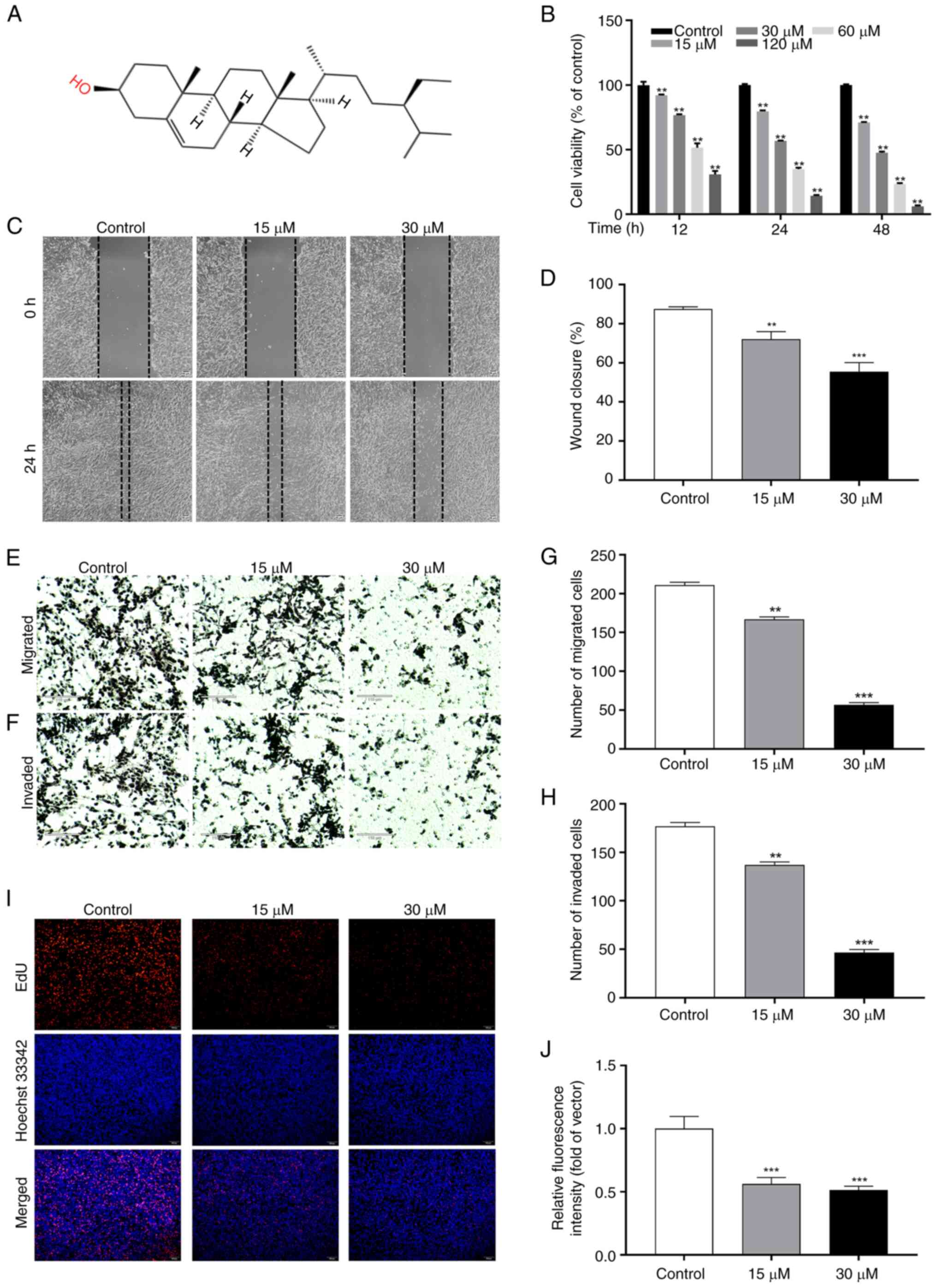 | Figure 7.SIT inhibits the physiological
function of KFs. (A) Chemical structure of SIT. (B) The relative
cell viability of KFs following treatment with different
concentrations of SIT for 12, 24 and 48 h. (C and D) The migration
was evaluated by wound healing assay (magnificaion, ×40). (E-H) The
number of migrated and the number of invaded cells was evaluated by
Transwell assay (scale bar, 110 µm; magnification, ×40). (I and J)
The result of EdU staining was presented as the percentage of
EdU-positive cells whose nuclei were stained in red (scale bar, 100
µm; magnification, ×100). The results are the mean ± SD (n=3).
**P<0.01, ***P<0.001 vs. control. SIT, β-sitosterol; KFs,
keloid fibroblast cells; EdU, 5-Ethynyl-2′-deoxyuridine. |
SIT inhibits the migration and
invasion of KFs
Evidence from previous studies has shown that SIT
inhibits the migration of various cancer cells (32). To assess the effect of SIT on the
behavior of KFs, scratch and Transwell assays were performed and
cell migration and invasion evaluated. The results revealed that in
the control group, the scratch gap gradually closed over 24 h. SIT
effectively impeded KFs migration in a concentration- and
time-dependent manner (Fig. 7C and
D). Migration was significantly reduced in cells treated with
15 and 30 µM SIT compared with that in control cells. Furthermore,
to assess the migratory and invasive potential of cells, they were
treated with SIT at various concentrations over a 24 h treatment
period (Fig. 7E and F). The
Transwell assay results revealed that treatment with SIT at various
concentrations effectively inhibited keloid cell migration and
invasion (Fig. 7G and H).
SIT modulates DNA synthesis and
proliferation in KFs
The present study assessed DNA synthesis by
measuring the incorporation of EdU, a thymidine nucleotide analog,
during the S phase of the cell cycle. The EdU incorporation rate
indicates the DNA synthesis rate. Fig.
7I and J show the effective inhibition of KFs growth by SIT.
Compared with the control treatment, treatment with 15 and 30 µM
SIT significantly reduced the number of EdU-labeled cells
(P<0.01).
SIT modulates the expression of
EMT-associated proteins in a dose-dependent manner
E-cadherin, Vimentin and Snail are established key
biomarkers of EMT (33). To
determine the importance of EMT in cell migration and invasion
(34), the present study evaluated
the effect of SIT on EMT regulation in KFs. It was found that the
inhibitory effect of SIT on migration is correlated with the
attenuation of EMT. The morphological changes in KFs after 48 h of
SIT treatment were assessed. Post treatment, the KFs transitioned
from a multi-protruding spindle shape to a more regular triangular
or short fusiform shape (Fig. 8A).
The expression of EMT markers, including E-cadherin, ZO-1 and
Vimentin, in SIT-treated KFs were analyzed using western blotting.
The expression levels of E-cadherin and ZO-1 increased in a
concentration-dependent manner in the SIT-treated groups, whereas
the Snail and Vimentin levels decreased in a dose-dependent manner
(Fig. 8B and C).
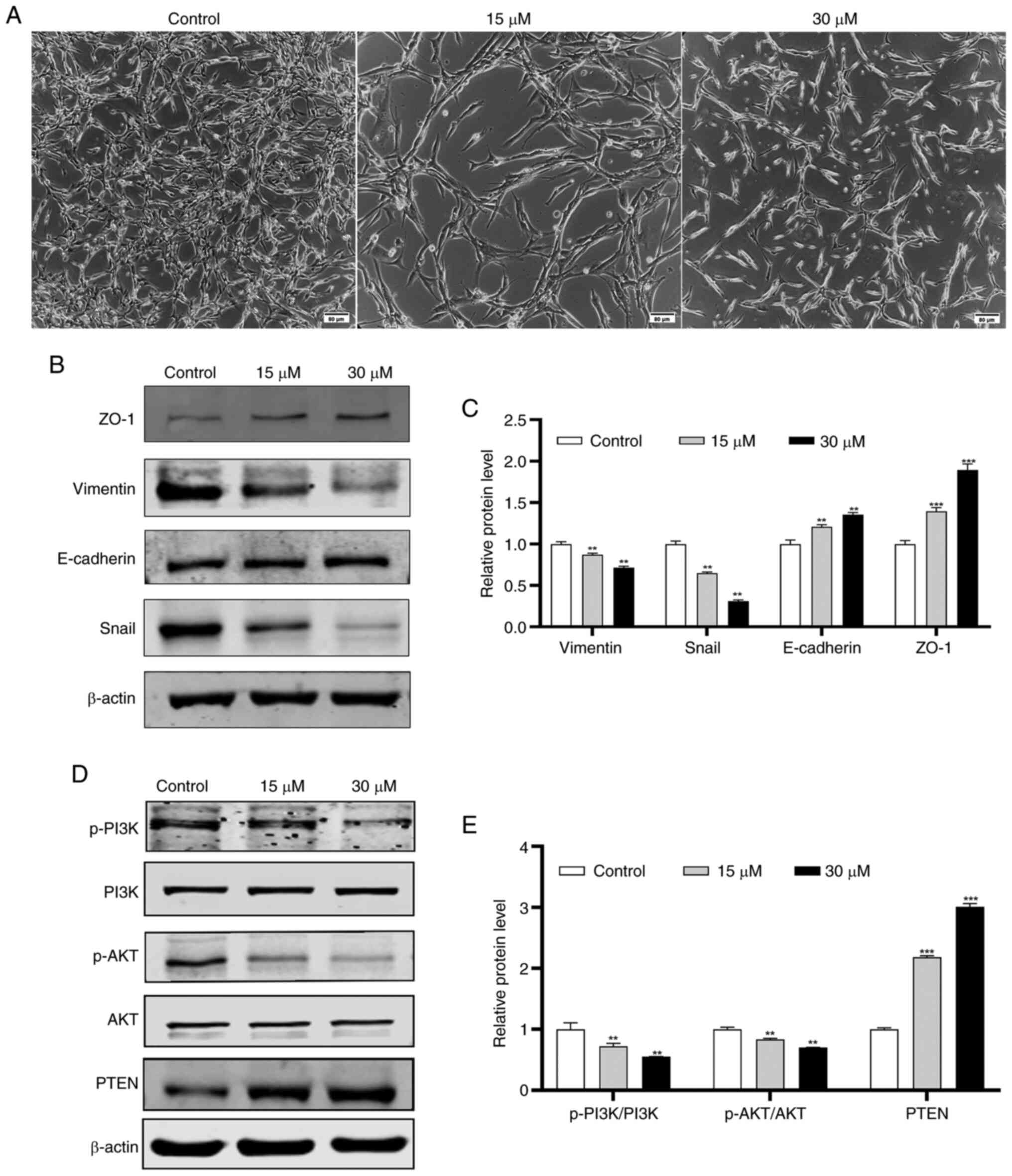 | Figure 8.Effect of SIT on PTEN/Akt signaling
pathway and EMT in KFs. KFs were untreated or treated with 15 and
30 µM SIT for 48 h. (A) Representative images of treated cells were
shown (scale bar, 80 µm; magnification, ×100). (B and C) the
expression of the EMT-related proteins in KFs following treatment
with SIT by western blotting. (D and E) the expression of
PI3K,p-PI3K, Akt, p-Akt and PTEN in KFs following treatment with
SIT by western blotting. The results are the mean ± SD (n=3).
**P<0.01, ***P<0.001. SIT, β-sitosterol; EMT,
epithelial-mesenchymal transition; KFs, keloid fibroblast cells;
p-, phosphorylated; ZO-1, zonula occludens-1. |
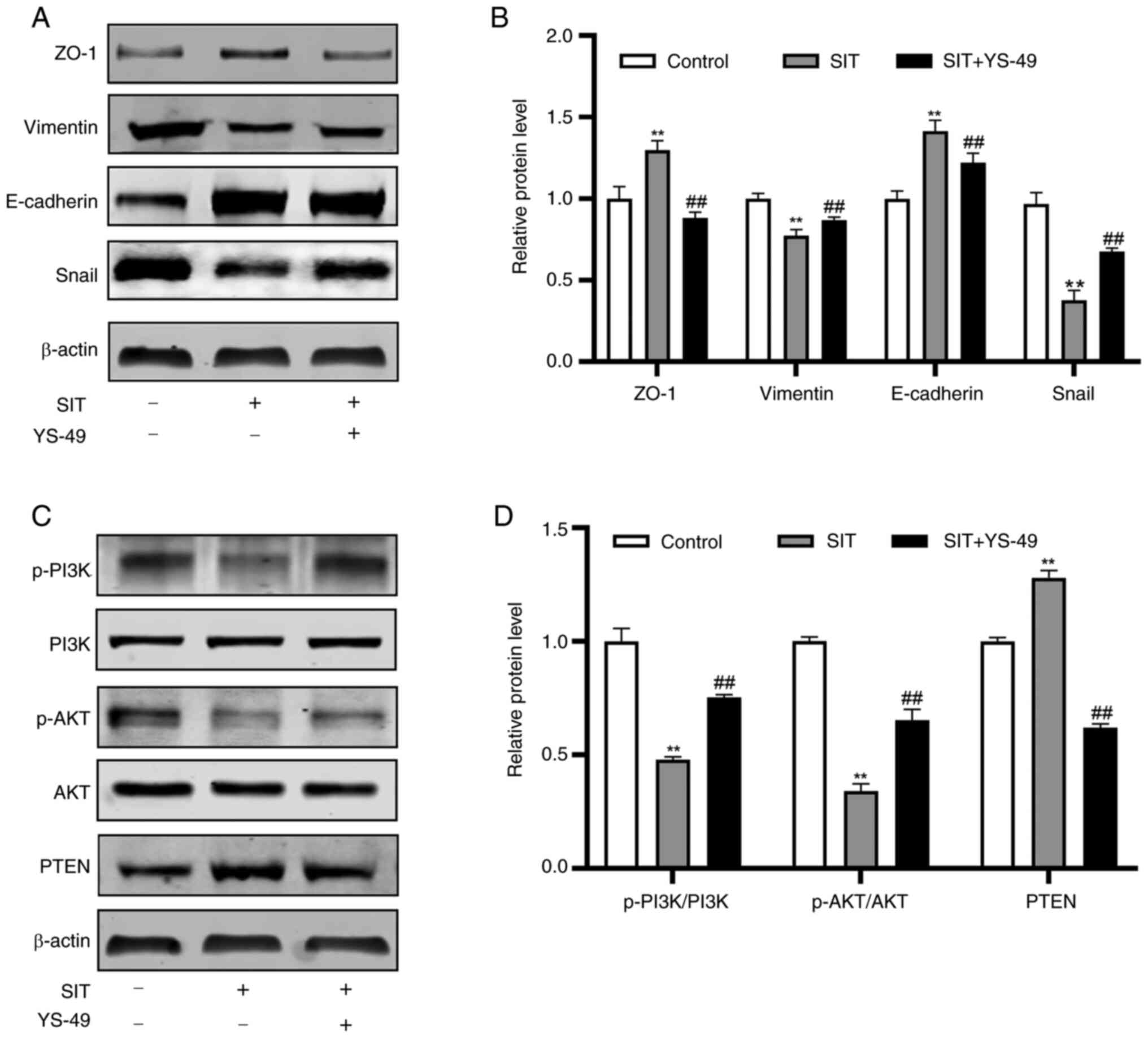
SIT regulates KFs via the
PTEN/PI3K/AKT pathway
After it was established that SIT regulates EMT and
the expression of associated markers, the present study
investigated its inhibitory mechanisms in KFs. SIT treatment
significantly increased PTEN protein expression (Fig. 8D and E). Given that PI3K and AKT
are crucial downstream targets of PTEN (35), the expression levels of PI3K,
p-PI3K, p-AKT and AKT were assessed concurrently. Total cellular
proteins were extracted from cells treated with 15 and 30 µM SIT
and control cells. SIT treatment reduced p-PI3K and p-AKT
expression; however, it did not alter the total PI3K and AKT
protein levels. Moreover, the changes in the p-PI3K and p-AKT
levels were inversely correlated with the PTEN levels. These
findings suggest that SIT may suppress growth of KFs via the
PTEN/PI3K/AKT pathway.
Reversing the SIT-mediated inhibition
of KFs proliferation, migration, invasion and EMT through PI3K/AKT
pathway activation
SIT may suppress KFs growth via the PTEN/PI3K/AKT
pathway. To confirm this, rescue experiments were conducted with
YS-49, a PI3K/AKT activator (27).
Findings from CCK-8 assays indicated that YS-49 treatment
significantly reversed the inhibition of proliferation in
SIT-treated KFs (Fig. 9A).
Additionally, the results of the EdU, scratch and Transwell assays
indicated that YS-49 reversed the inhibitory effects on the
proliferation, migration and invasion of cells caused by SIT
(Fig. 9B-H). Following YS-49
treatment, the PTEN, E-cadherin and ZO-1 levels decreased in
SIT-treated KFs, whereas the Snail, Vimentin, p-PI3K and p-AKT
levels increased (Fig. 10A-D).
Collectively, these findings confirmed that SIT effectively
suppresses the proliferation, migration and invasion and EMT
progression of KFs by inhibiting the PTEN/PI3K/AKT signaling
pathway.
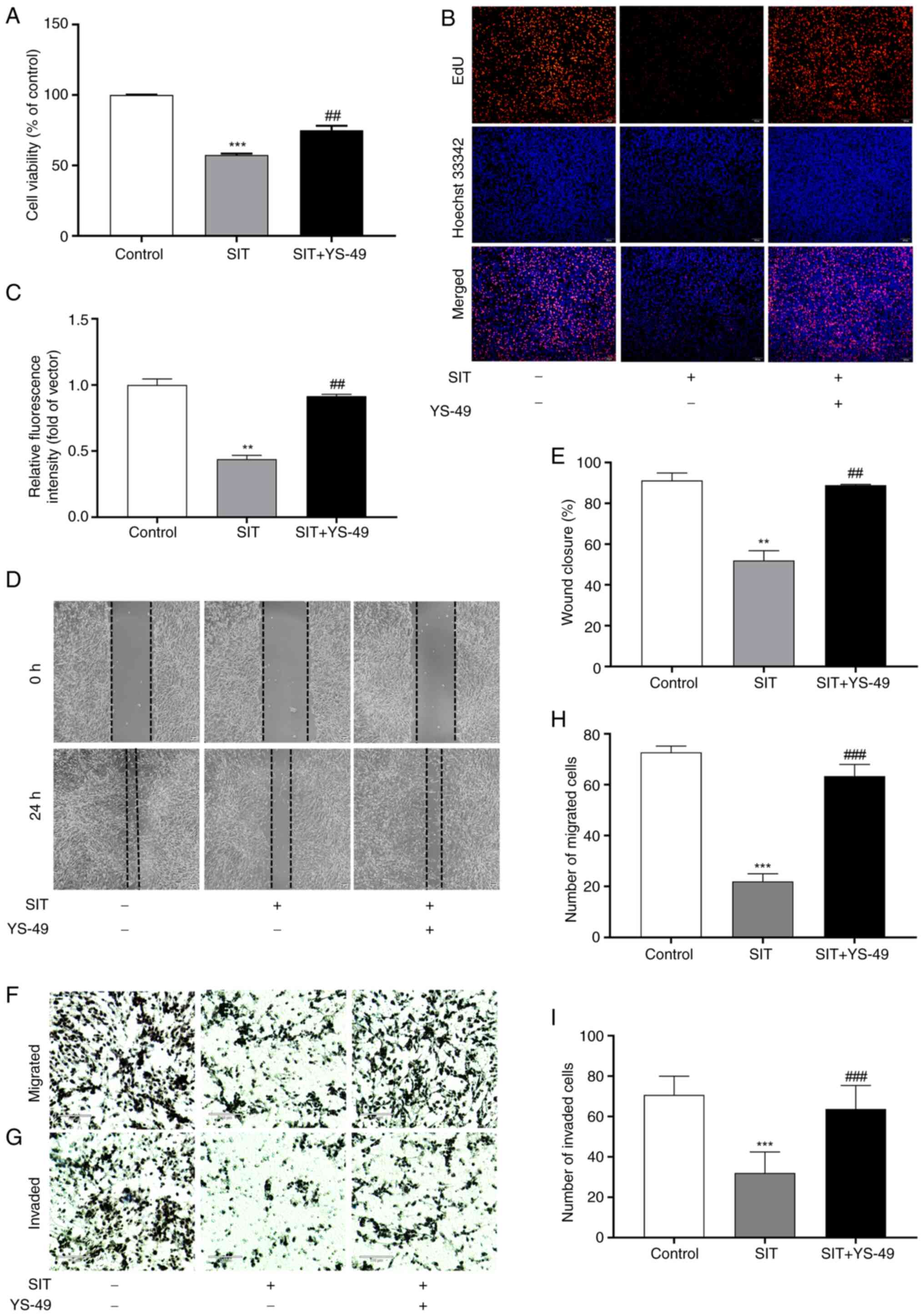 | Figure 9.Counteraction of SIT's inhibitory
effects on KFs cell proliferation, migration and invasion by
PI3K/AKT pathway activation. (A) The effect of SIT on KFs viability
was detected following treatment with YS-49 by CCK-8 assay. (B and
C) The effect of SIT on KFs proliferation was evaluated following
treatment with YS-49 by EdU staining (magnification, ×100). (D and
E) The effect of SIT on the migration of KFs following treatment
with YS-49 was observed by wound healing assay (scale bar, 50 µm;
magnification, ×40). (F-I) The effect of SIT on the invasion and
migration of KFs was evaluated following treatment with YS-49 by
Transwell assay (scale bar, 110 µm; magnification, ×40).
Statistical analyses were performed using one-way ANOVA with
Tukey's post hoc test. **P<0.01, ***P<0.001 vs. Control group
(without treatment with SIT). ##P<0.01,
###P<0.001 vs. SIT treatment group. SIT,
β-sitosterol; KFs, keloid fibroblast cells; EdU,
5-Ethynyl-2′-deoxyuridine. |
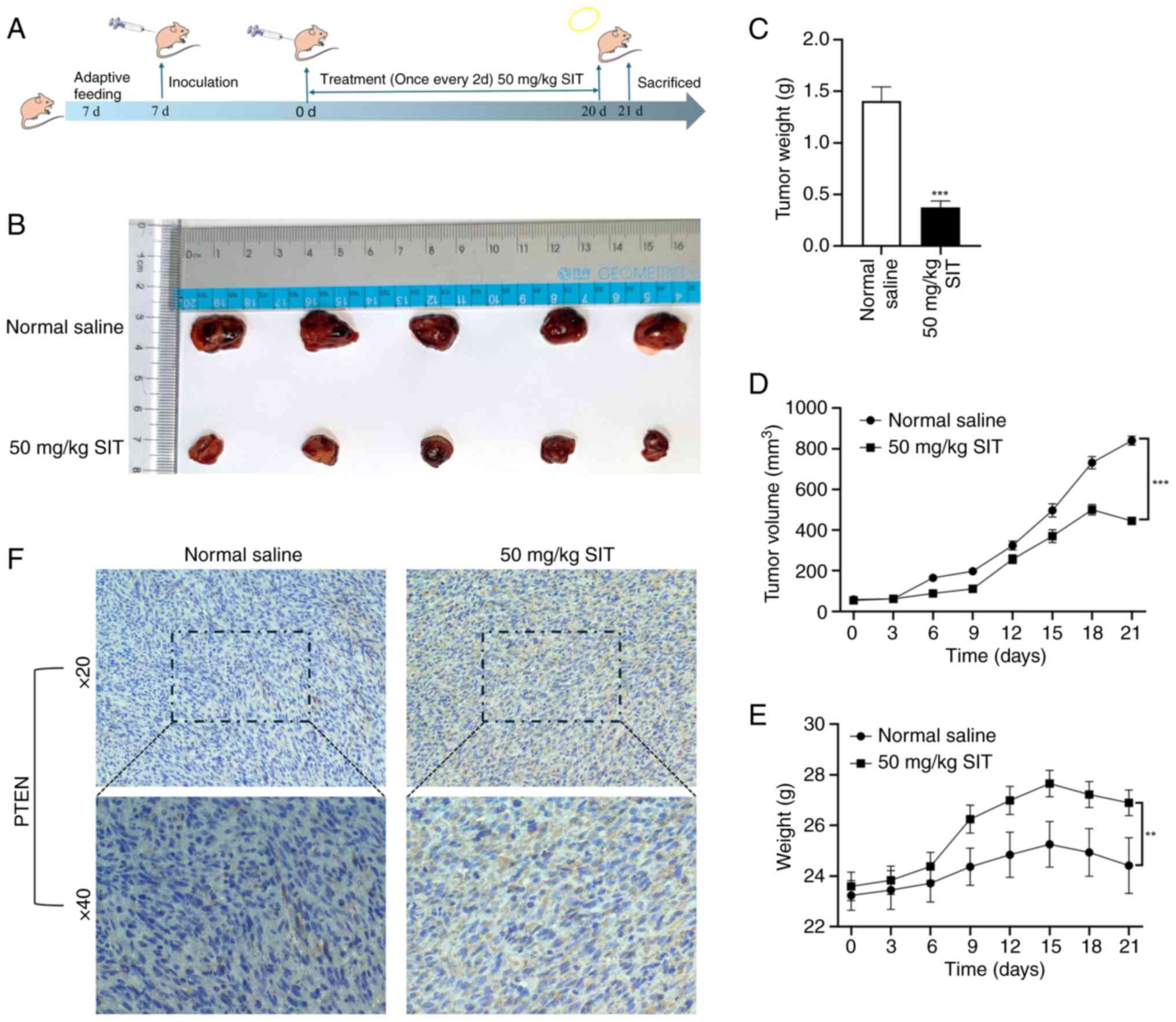
SIT suppresses KFs xenograft growth by
inhibiting PTEN expression
After confirming the in vitro effects of SIT
on cell proliferation, migration and invasion, its effect on keloid
formation in vivo was assessed using a nude mice model with
KFs xenografts. Fig. 11A shows
the flowchart of the animal experiments, detailing the model
establishment and treatment procedures. Keloid growth was markedly
slower in the SIT group than in the saline group, which indicated
the inhibitory potential of SIT (Fig.
11D). The results for keloid measurements (Fig. 11B), tumor weight (Fig. 11C) and nude mice weight were
consistent (Fig. 11E). The saline
group showed the lowest weight, whereas the treatment group showed
normal growth trends. Nude mice in the SIT group began losing
weight after the 15th day of treatment, probably due to keloid
reduction. This indicated effective in vivo anti-keloid
effects. To further elucidate the mechanism underlying the
inhibition of keloid growth by SIT in vivo, PTEN expression
in fibroproliferative tissues was evaluated using
immunohistochemistry. SIT treatment markedly increased the PTEN
protein levels (Fig. 11F).
Notably, H&E staining revealed no significant effects on the
heart, liver, kidney, or spleen, indicating the absence of adverse
effects of SIT treatment on these vital organs (Fig. S1). No evidence was observed of
myocardial fiber disarray, necrosis, or inflammatory infiltration.
Cardiac muscle structure and cellular integrity remained intact. No
signs of hepatocyte degeneration, necrosis, or inflammatory cell
infiltration were observed. There was no indication of glomerular
damage, tubular necrosis, or interstitial inflammation. No evidence
of hemorrhage or structural disorganization was detected. These
results, which are consistent with the in vitro findings,
indicated that SIT inhibits keloid growth by modulating PTEN
expression in vivo.
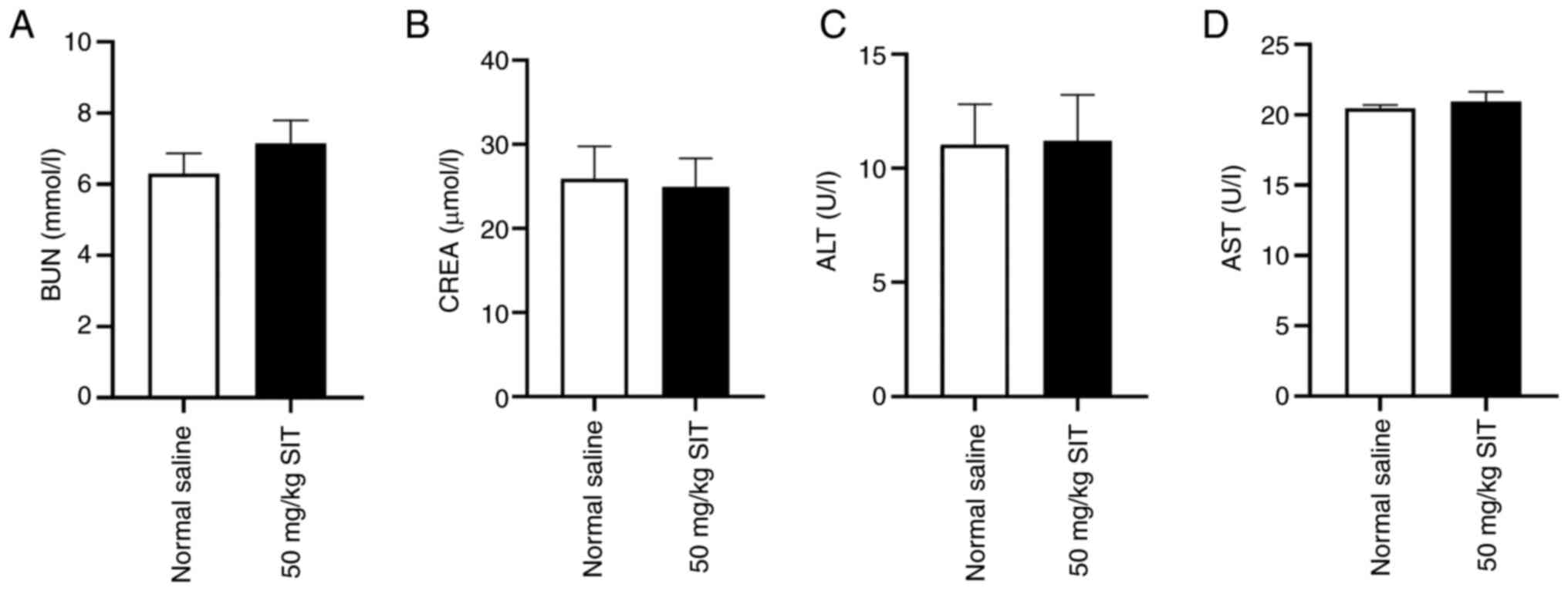
In vivo cytotoxicity
The present study assessed formulation safety via
blood biochemical analyses, including those for BUN, CREA, ALT and
AST. The levels of BUN, CREA, ALT and AST remained consistently low
across groups (Fig. 12). The
findings indicate that SIT has a favorable safety profile for
keloid treatment in nude mouse.
Discussion
Keloids extend beyond the initial dermal lesion,
where they proliferate (2).
Keloids and tumors share key characteristics, including epigenetic
methylation profiles, disease-related biological behaviors and
cellular bioenergetics (36). EMT
activation increases invasiveness by inducing microenvironmental
and phenotypic changes. Transformation involves the downregulation
of epithelial genes, such as E-cadherin and ZO-1 genes and the
upregulation of mesenchymal genes, such as Vimentin and Snail genes
(37). The current treatments
available for keloids are unclear, partly due to ineffective
outcomes and unavailability of sufficient evidence.
HIF-1α activates the TGF-β/Smad signaling pathway in
keloids (38), increasing collagen
production by dermal fibroblasts (39). The inhibition of this pathway is a
potential strategy for keloid treatment. TGF-β induces abnormal
VEGF expression, leading to unusual blood vessel proliferation in
the dermis (40). Inflammation and
angiogenesis are essential for keloid progression. Additionally,
these processes have been linked to the PI3K/AKT signaling pathway
(41). Oral CDK12/13 degraders,
which target this pathway, have been shown to be promising for
therapy (42). TGF-β has been
shown to activate the Wnt/β-catenin pathway, which subsequently
leads to abnormal fibroblast proliferation and differentiation and
impedes scar tissue repair (43).
The present study used network pharmacology to determine whether
SIT affects keloid progression through the PTEN/PI3K/AKT pathway.
The findings suggested that exploring the interactions among the
PTEN/PI3K/AKT, TGF-β/Smad and Wnt/β-catenin pathways can provide a
novel research direction for SIT-mediated keloid treatment. Keloid
pathogenesis is complex, with current treatments primarily
including surgery and localized drug injections. These strategies
are often associated with high recurrence rates, limited treatment
options and adverse effects (44).
SIT, a compound found in lipid-rich plants, effectively blocks cell
migration and invasion in several cancers by modulating the
PI3K/AKT pathway (45). SIT has
been shown to regulate EMT in various tumors and fibrotic
conditions. STRING analysis identified Caspases 3, 8 and 9 as well
as Bax as key apoptotic factors in keloid formation (46,47).
Activating these factors is crucial for inducing apoptosis and can
potentially affect tissue repair and remodeling. MYC mediates
keloid fibroblast proliferation and collagen deposition (48). These findings suggest that the
effect of SIT on apoptotic factors could be crucial in treating
keloids and represents a promising research direction.
Additionally, pathway enrichment analysis revealed a strong link
between keloid and the PTEN/PI3K/AKT signaling pathway (Fig. 5B). Findings from molecular docking
studies also confirmed that SIT interacts with PTEN to form a
stable complex.
Experiments with a PI3K/AKT activator revealed the
role of SIT in keloid pathology. SIT treatment increases E-cadherin
and ZO-1 levels while decreasing Snail and Vimentin levels, thus
affecting EMT in keloids. The present study assessed the safety of
the experimental dose by analyzing the levels of serum biochemical
markers for liver and kidney function and performing H&E
staining on vital organs in nude mice. The selected dose was deemed
safe in the nude mouse model, which is consistent with findings
from previous studies (49,50).
The present study revealed that SIT suppresses keloid
proliferation, migration, invasion and EMT via the PTEN/PI3K/AKT
pathway. However, the present study had several limitations. First,
it focused on the effects of SIT on keloid characteristics such as
proliferation, migration, invasion and EMT mediated via the
PTEN/PI3K/AKT pathway. A comprehensive understanding requires
further investigation into additional pathways and molecules to
elucidate fully the mechanism of action of SIT in keloid treatment.
Second, it only used a subcutaneous keloid proliferation model in
nude mouse for validation. Although this model is standard in
keloid research and widely used across various biological
disciplines, further studies with alternative models, such as pig
skin and in vitro 3D models, are essential for establishing
the minimum effective human dose of SIT. Pig skin and in
vitro 3D keloid models might yield further information in the
future. Finally, SIT may exert a synergistic effect by targeting
multiple pathways. Addressing off-target effects, a significant
challenge in drug discovery, will be a key focus of future research
on this drug (51). Addressing
these limitations will help provide crucial experimental evidence
for the clinical applications of SIT. The present study aimed to
introduce a new treatment for keloids in clinical practice, which
could eventually improve the quality of life of patients with this
condition. This significant finding has important implications for
future innovative treatment strategies for keloids.
The present study was based on the results of
network pharmacology research. The findings provided initial
evidence that SIT suppresses keloid function by inhibiting the
PTEN/PI3K/AKT signaling pathway, potentially impeding keloid growth
and invasion. These conclusions were further supported by evidence
from experiments conducted with PI3K/AKT activators. Overall, these
findings highlighted the promising anti-keloid effects of SIT,
indicating its strong safety profile and potential as a novel
therapeutic agent for keloids. In the future, the authors aim to
delve deeper into the anti-keloid mechanisms of SIT, which could
help pave the way for its clinical application in keloid
treatment.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural Science
Foundation of China (grant no. 82260617).
Availability of data and materials
The data generated in the present study are included
in the figures and/or tables of this article.
Authors' contributions
PPH and ZHJ designed the project. PPH, ZNL and SJ
performed the experiments. ZNL and SJW made substantial
contributions to data analysis. PPH wrote this article. LHZ, SJW
and YLL edited the manuscript and made substantial contributions to
the bioinformatics analysis. SJ and LHZ supervised the project. ZHJ
and LHZ confirm the authenticity of all the raw data. All authors
read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was reviewed and approved by the
Yanbian University Experimental Animal Welfare Ethics Committee
(approval no. YD20230911009). All procedures adhered to the ethical
guidelines for animal use set forth by the National Institutes of
Health.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
SIT
|
β-sitosterol
|
|
TCMSP
|
The Chinese Medicine Systems
Pharmacology Database and Analysis Platform
|
|
TTD
|
Therapeutic Target Database
|
|
OMIM
|
Online Mendelian Inheritance in
Man
|
|
PPI
|
protein-protein interaction
|
|
F. arctii
|
Fructus arctii
|
|
GO
|
Gene Ontology
|
|
KEGG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
EMT
|
epithelial-mesenchymal transition
|
|
KF
|
keloid fibroblast
|
|
MF
|
molecular functions
|
|
CC
|
cellular components
|
|
BP
|
biological processes
|
|
CCK-8
|
Cell Counting Kit-8
|
|
EdU
|
5-Ethynyl-2′-deoxyuridine
|
References
|
1
|
Limandjaja GC, Niessen FB, Scheper RJ and
Gibbs S: The keloid disorder: Heterogeneity, histopathology,
mechanisms and models. Front Cell Dev Biol. 8:3602020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tan S, Khumalo N and Bayat A:
Understanding keloid pathobiology from a quasi-neoplastic
perspective: Less of a scar and more of a chronic inflammatory
disease with cancer-like tendencies. Front Immunol. 10:18102019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hsu CK, Lin HH, Harn HI, Ogawa R, Wang YK,
Ho YT, Chen WR, Lee YC, Lee JY, Shieh SJ, et al: Caveolin-1
controls hyperresponsiveness to mechanical stimuli and
fibrogenesis-associated RUNX2 activation in keloid fibroblasts. J
Invest Dermatol. 138:208–218. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jin X, Liu S, Chen S, Wang L, Cui Y, He J,
Fang S, Li J and Chang Y: A systematic review on botany,
ethnopharmacology, quality control, phytochemistry, pharmacology
and toxicity of Arctium lappa L. fruit. J Ethnopharmacol.
308:1162232023. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Vo TK, Ta QTH, Chu QT, Nguyen TT and Vo
VG: Anti-hepatocellular-cancer activity exerted by β-sitosterol and
β-sitosterol-glucoside from Indigofera zollingeriana Miq.
Molecules. 25:30212020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nandi S, Nag A, Khatua S, Sen S,
Chakraborty N, Naskar A, Acharya K, Calina D and Sharifi-Rad J:
Anticancer activity and other biomedical properties of
β-sitosterol: Bridging phytochemistry and current pharmacological
evidence for future translational approaches. Phytother Res.
38:592–619. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liao PC, Lai MH, Hsu KP, Kuo YH, Chen J,
Tsai MC, Li CX, Yin XJ, Jeyashoke N and Chao LK: Identification of
β-sitosterol as in vitro anti-inflammatory constituent in moringa
oleifera. J Agric Food Chem. 66:10748–10759. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nunnery SE and Mayer IA: Targeting the
PI3K/AKT/mTOR pathway in hormone-positive breast cancer. Drugs.
80:1685–1697. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bathina S, Gundala NKV, Rhenghachar P,
Polavarapu S, Hari AD, Sadananda M and Das UN: Resolvin D1
ameliorates nicotinamide-streptozotocin-induced type 2 diabetes
mellitus by its anti-inflammatory action and modulating
PI3K/Akt/mTOR pathway in the brain. Arch Med Res. 51:492–503. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xin Y, Min P, Xu H, Zhang Z and Zhang Y
and Zhang Y: CD26 upregulates proliferation and invasion in keloid
fibroblasts through an IGF-1-induced PI3K/AKT/mTOR pathway. Burns
Trauma. 8:tkaa0252020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee YR, Chen M and Pandolfi PP: The
functions and regulation of the PTEN tumour suppressor: New modes
and prospects. Nat Rev Mol Cell Biol. 19:547–562. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mukherjee R, Vanaja KG, Boyer JA, Gadal S,
Solomon H, Chandarlapaty S, Levchenko A and Rosen N: Regulation of
PTEN translation by PI3K signaling maintains pathway homeostasis.
Mol Cell. 81:708–723.e5. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yan L, Wang LZ, Xiao R, Cao R, Pan B, Lv
XY, Jiao H, Zhuang Q, Sun XJ and Liu YB: Inhibition of
microRNA-21-5p reduces keloid fibroblast autophagy and migration by
targeting PTEN after electron beam irradiation. Lab Invest.
100:387–399. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ru J, Li P, Wang J, Zhou W, Li B, Huang C,
Li P, Guo Z, Tao W, Yang Y, et al: TCMSP: A database of systems
pharmacology for drug discovery from herbal medicines. J
Cheminform. 6:132014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
UniProt Consortium: UniProt: The universal
protein knowledgebase in 2023. Nucleic Acids Res. 51(D1):
D523–D531. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu F, Shi Z, Qin C, Tao L, Liu X, Xu F,
Zhang L, Song Y, Liu X, Zhang J, et al: Therapeutic target database
update 2012: A resource for facilitating target-oriented drug
discovery. Nucleic Acids Res. 40((Database Issue)): D1128–D1136.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wishart DS, Knox C, Guo AC, Cheng D,
Shrivastava S, Tzur D, Gautam B and Hassanali M: DrugBank: A
knowledgebase for drugs, drug actions and drug targets. Nucleic
Acids Res. 36((Database Issue)): D901–D906. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fishilevich S, Zimmerman S, Kohn A, Iny
Stein T, Olender T, Kolker E, Safran M and Lancet D: Genic insights
from integrated human proteomics in GeneCards. Database (Oxford).
2016:baw0302016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shao C, Wang H, Sang F and Xu L: Study on
the mechanism of improving HIV/AIDS immune function with jian
aikang concentrated pill based on network pharmacology combined
with experimental validation. Drug Des Devel Ther. 16:2731–2753.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szklarczyk D, Gable AL, Lyon D, Junge A,
Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork
P, et al: STRING v11: protein-protein association networks with
increased coverage, supporting functional discovery in genome-wide
experimental datasets. Nucleic Acids Res. 47(D1): D607–D613. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41((Database Issue)): D808–D815. 2013.PubMed/NCBI
|
|
22
|
Scott DE, Bayly AR, Abell C and Skidmore
J: Small molecules, big targets: Drug discovery faces the
protein-protein interaction challenge. Nat Rev Drug Discov.
15:533–550. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim S, Thiessen PA, Bolton EE, Chen J, Fu
G, Gindulyte A, Han L, He J, He S, Shoemaker BA, et al: PubChem
substance and compound databases. Nucleic Acids Res. 44(D1):
D1202–D1213. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rose PW, Prlić A, Altunkaya A, Bi C,
Bradley AR, Christie CH, Costanzo LD, Duarte JM, Dutta S, Feng Z,
et al: The RCSB protein data bank: Integrative view of protein,
gene and 3D structural information. Nucleic Acids Res. 45(D1):
D271–D281. 2017.PubMed/NCBI
|
|
25
|
Ji L, Song T, Ge C, Wu Q, Ma L, Chen X,
Chen T, Chen Q, Chen Z and Chen W: Identification of bioactive
compounds and potential mechanisms of scutellariae radix-coptidis
rhizoma in the treatment of atherosclerosis by integrating network
pharmacology and experimental validation. Biomed Pharmacother.
165:1152102023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Khanh Nguyen NP, Kwon JH, Kim MK, Tran KN,
Huong Nguyen LT and Yang IJ: Antidepressant and anxiolytic
potential of Citrus reticulata Blanco essential oil: A network
pharmacology and animal model study. Front Pharmacol.
15:13594272024. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang X, Liu J, Sun Y, Zhou Q, Ding X and
Chen X: Chinese herbal compound Huangqin Qingrechubi capsule
reduces lipid metabolism disorder and inflammatory response in
gouty arthritis via the LncRNA H19/APN/PI3K/AKT cascade. Pharm
Biol. 61:541–555. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui J, Jin S, Jin C and Jin Z: Syndecan-1
regulates extracellular matrix expression in keloid fibroblasts via
TGF-β1/Smad and MAPK signaling pathways. Life Sci. 254:1173262020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen Y, Yang Y, Wang N, Liu R, Wu Q, Pei H
and Li W: β-Sitosterol suppresses hepatocellular carcinoma growth
and metastasis via FOXM1-regulated Wnt/β-catenin pathway. J Cell
Mol Med. 28:e180722024. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang S, Meng S, Zhou X, Gao Z and Piao MG:
pH-Responsive and mucoadhesive nanoparticles for enhanced oral
insulin delivery: The effect of hyaluronic acid with different
molecular weights. Pharmaceutics. 15:8202023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang H, Liu J, Zhang Z, Peng J, Wang Z,
Yang L, Wang X, Hu S and Hong L: β-Sitosterol targets ASS1 for Nrf2
ubiquitin-dependent degradation, inducing ROS-mediated apoptosis
via the PTEN/PI3K/AKT signaling pathway in ovarian cancer. Free
Radic Biol Med. 214:137–157. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang F, Zhou K, Yuan W and Sun K: Radix
bupleuri-radix paeoniae alba inhibits the development of
hepatocellular carcinoma through activation of the PTEN/PD-L1 axis
within the immune microenvironment. Nutr Cancer. 76:63–79. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chaw SY, Abdul Majeed A, Dalley AJ, Chan
A, Stein S and Farah CS: Epithelial to mesenchymal transition (EMT)
biomarkers-E-cadherin, beta-catenin, APC and Vimentin-in oral
squamous cell carcinogenesis and transformation. Oral Oncol.
48:997–1006. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lee YI, Shim JE, Kim J, Lee WJ, Kim JW,
Nam KH and Lee JH: WNT5A drives interleukin-6-dependent
epithelial-mesenchymal transition via the JAK/STAT pathway in
keloid pathogenesis. Burns Trauma. 10:tkac0232022. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lv D, Xu Z, Cheng P, Hu Z, Dong Y, Rong Y,
Xu H, Wang Z, Cao X, Deng W and Tang B: S-Nitrosylation-mediated
coupling of DJ-1 with PTEN induces PI3K/AKT/mTOR pathway-dependent
keloid formation. Burns Trauma. 11:tkad0242023. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Vincent AS, Phan TT, Mukhopadhyay A, Lim
HY, Halliwell B and Wong KP: Human skin keloid fibroblasts display
bioenergetics of cancer cells. J Invest Dermatol. 128:702–709.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang Y, Hong W and Wei X: The molecular
mechanisms and therapeutic strategies of EMT in tumor progression
and metastasis. J Hematol Oncol. 15:1292022. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Mingyuan X, Qianqian P, Shengquan X,
Chenyi Y, Rui L, Yichen S and Jinghong X: Hypoxia-inducible
factor-1α activates transforming growth factor-β1/Smad signaling
and increases collagen deposition in dermal fibroblasts.
Oncotarget. 9:3188–3197. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang T, Wang XF, Wang ZC, Lou D, Fang QQ,
Hu YY, Zhao WY, Zhang LY, Wu LH and Tan WQ: Current potential
therapeutic strategies targeting the TGF-β/Smad signaling pathway
to attenuate keloid and hypertrophic scar formation. Biomed
Pharmacother. 129:1102872020. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang XM, Liu XM, Wang Y and Chen ZY:
Activating transcription factor 3 (ATF3) regulates cell growth,
apoptosis, invasion and collagen synthesis in keloid fibroblast
through transforming growth factor beta (TGF-beta)/SMAD signaling
pathway. Bioengineered. 12:117–126. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Qin W, Gao J, Yan J, Han X, Lu W, Ma Z,
Niu L and Jiao K: Microarray analysis of signalling interactions
between inflammation and angiogenesis in subchondral bone in
temporomandibular joint osteoarthritis. Biomater Transl. 5:175–184.
2024.PubMed/NCBI
|
|
42
|
Chang Y, Wang X, Yang J, Tien JC, Mannan
R, Cruz G, Zhang Y, Vo JN, Magnuson B, Mahapatra S, et al:
Development of an orally bioavailable CDK12/13 degrader and
induction of synthetic lethality with AKT pathway inhibition. Cell
Rep Med. 5:1017522024. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chaudet KM, Goyal A, Veprauskas KR and
Nazarian RM: Wnt signaling pathway proteins in scar, hypertrophic
scar, and keloid: Evidence for a continuum? Am J Dermatopathol.
42:842–847. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Juckett G and Hartman-Adams H: Management
of keloids and hypertrophic scars. Am Fam Physician. 80:253–260.
2009.PubMed/NCBI
|
|
45
|
Bao X, Zhang Y, Zhang H and Xia L:
Molecular mechanism of β-sitosterol and its derivatives in tumor
progression. Front Oncol. 12:9269752022. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Park YJ, Bang IJ, Jeong MH, Kim HR, Lee
DE, Kwak JH and Chung KH: Effects of β-sitosterol from corn silk on
TGF-β1-induced epithelial-mesenchymal transition in lung alveolar
epithelial cells. J Agric Food Chem. 67:9789–9795. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xie Y, Chen Z, Li S, Yan M, He W, Li L, Si
J, Wang Y, Li X and Ma K: A network pharmacology- and
transcriptomics-based investigation reveals an inhibitory role of
β-sitosterol in glioma via the EGFR/MAPK signaling pathway. Acta
Biochim Biophys Sin (Shanghai). 56:223–238. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Feng G, Sun H and Piao M: FBXL6 is
dysregulated in keloids and promotes keloid fibroblast growth by
inducing c-Myc expression. Int Wound J. 20:131–139. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang KN, Hu Y, Han LL, Zhao SS, Song C,
Sun SW, Lv HY, Jiang NN, Xv LZ, Zhao ZW and Li M: Salvia chinensis
benth inhibits triple-negative breast cancer progression by
inducing the DNA damage pathway. Front Oncol. 12:8827842022.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li J, Meng ZY, Wen H, Lu CH, Qin Y, Xie
YM, Chen Q, Lv JH, Huang F and Zeng ZY: β-sitosterol alleviates
pulmonary arterial hypertension by altering smooth muscle cell
phenotype and DNA damage/cGAS/STING signaling. Phytomedicine.
135:1560302024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Olorundare O, Adeneye A, Akinsola A, Kolo
P, Agede O, Soyemi S, Mgbehoma A, Okoye I, Albrecht R and Mukhtar
H: Irvingia gabonensis seed extract: An effective attenuator of
doxorubicin-mediated cardiotoxicity in wistar rats. Oxid Med Cell
Longev. 2020:16028162020. View Article : Google Scholar : PubMed/NCBI
|