|
1
|
Gilbert W: Why genes in pieces? Nature.
271:5011978. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jurica MS and Moore MJ: Pre-mRNA splicing:
Awash in a sea of proteins. Mol Cell. 12:5–14. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Will CL and Luhrmann R: Spliceosome
structure and function. Cold Spring Harb Perspect Biol.
3:a0037072011. View Article : Google Scholar : PubMed/NCBI
|
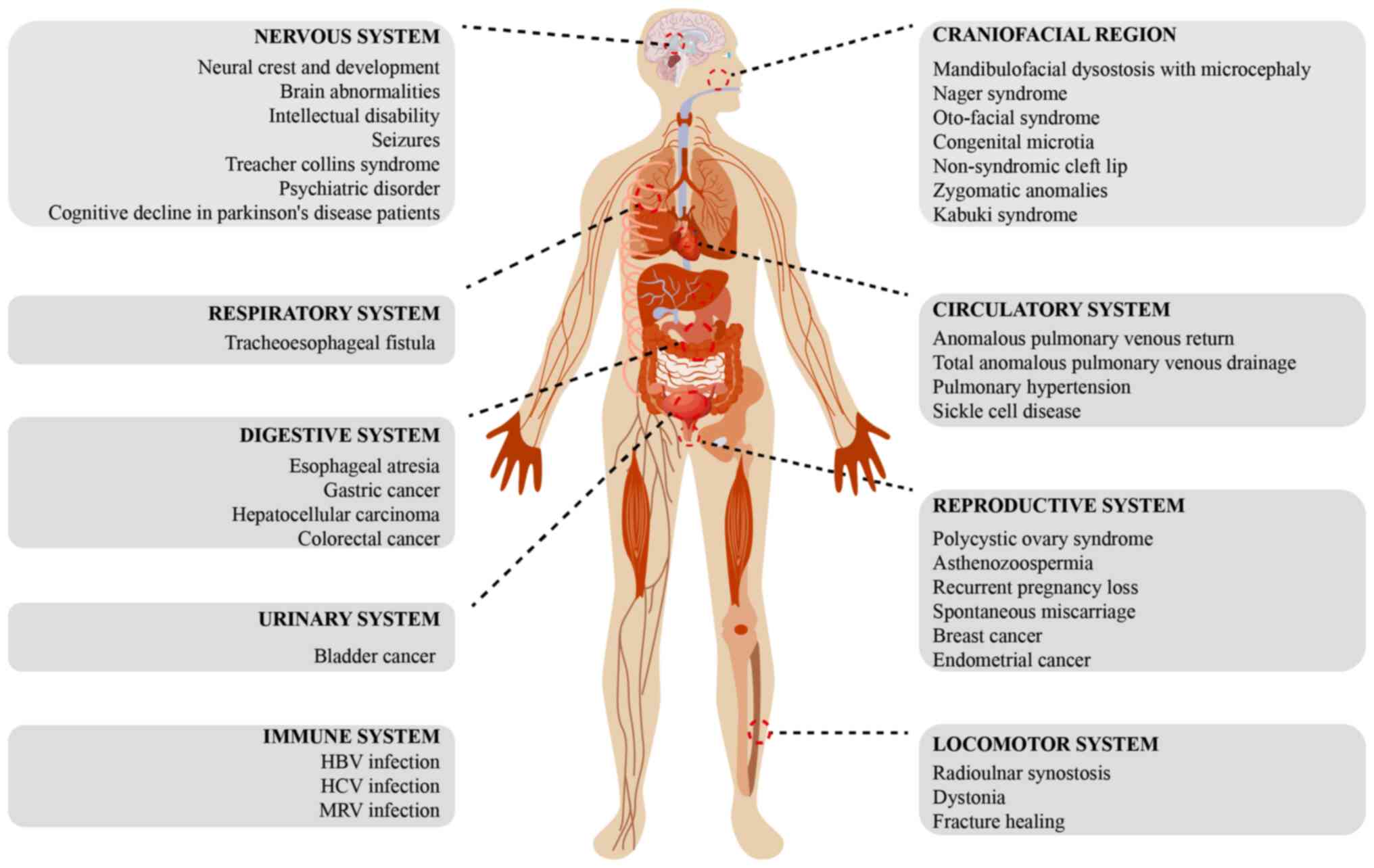
|
4
|
Boesler C, Rigo N, Anokhina MM, Tauchert
MJ, Agafonov DE, Kastner B, Urlaub H, Ficner R, Will CL and
Lührmann R: A spliceosome intermediate with loosely associated
tri-snRNP accumulates in the absence of Prp28 ATPase activity. Nat
Commun. 7:119972016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saez B, Walter MJ and Graubert TA:
Splicing factor gene mutations in hematologic malignancies. Blood.
129:1260–1269. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo R, Zheng L, Park JW, Lv R, Chen H,
Jiao F, Xu W, Mu S, Wen H, Qiu J, et al: BS69/ZMYND11 reads and
connects histone H3.3 lysine 36 trimethylation-decorated chromatin
to regulated pre-mRNA processing. Mol Cell. 56:298–310. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
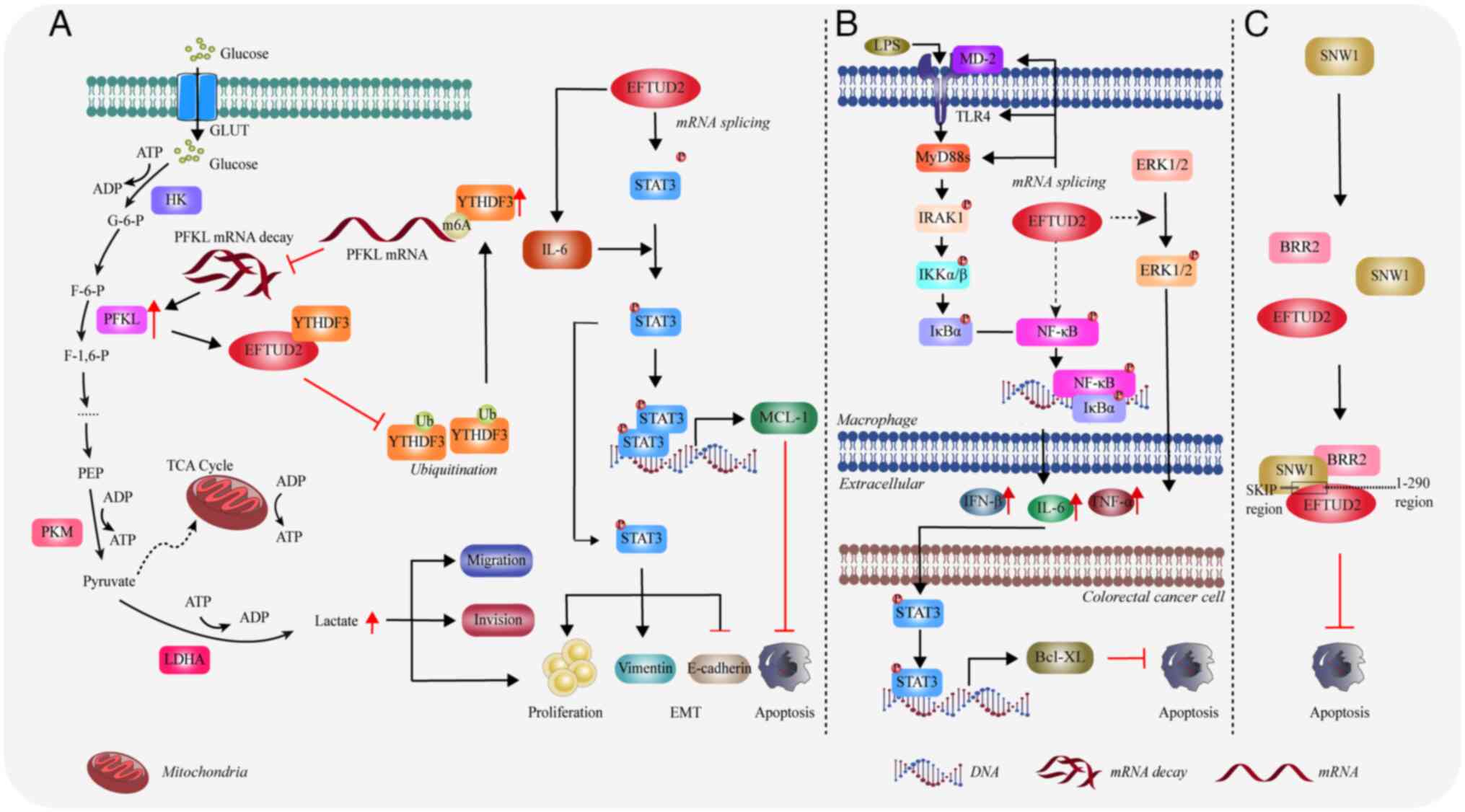
Zody MC, Garber M, Adams DJ, Sharpe T,
Harrow J, Lupski JR, Nicholson C, Searle SM, Wilming L, Young SK,
et al: DNA sequence of human chromosome 17 and analysis of
rearrangement in the human lineage. Nature. 440:1045–1049. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zahn-Zabal M, Michel PA, Gateau A, Nikitin
F, Schaeffer M, Audot E, Gaudet P, Duek PD, Teixeira D, Rech de
Laval V, et al: The neXtProt knowledgebase in 2020: Data, tools and
usability improvements. Nucleic Acids Res. 48(D1): D328–D334.
2020.PubMed/NCBI
|
|
9
|
Fagerberg L, Hallstrom BM, Oksvold P,
Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S,
Danielsson A, Edlund K, et al: Analysis of the human
tissue-specific expression by genome-wide integration of
transcriptomics and antibody-based proteomics. Mol Cell Proteomics.
13:397–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Plaschka C, Newman AJ and Nagai K:
Structural basis of nuclear pre-mRNA splicing: Lessons from yeast.
Cold Spring Harb Perspect Biol. 11:a0323912019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Papasaikas P and Valcarcel J: The
spliceosome: The ultimate RNA chaperone and sculptor. Trends
Biochem Sci. 41:33–45. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
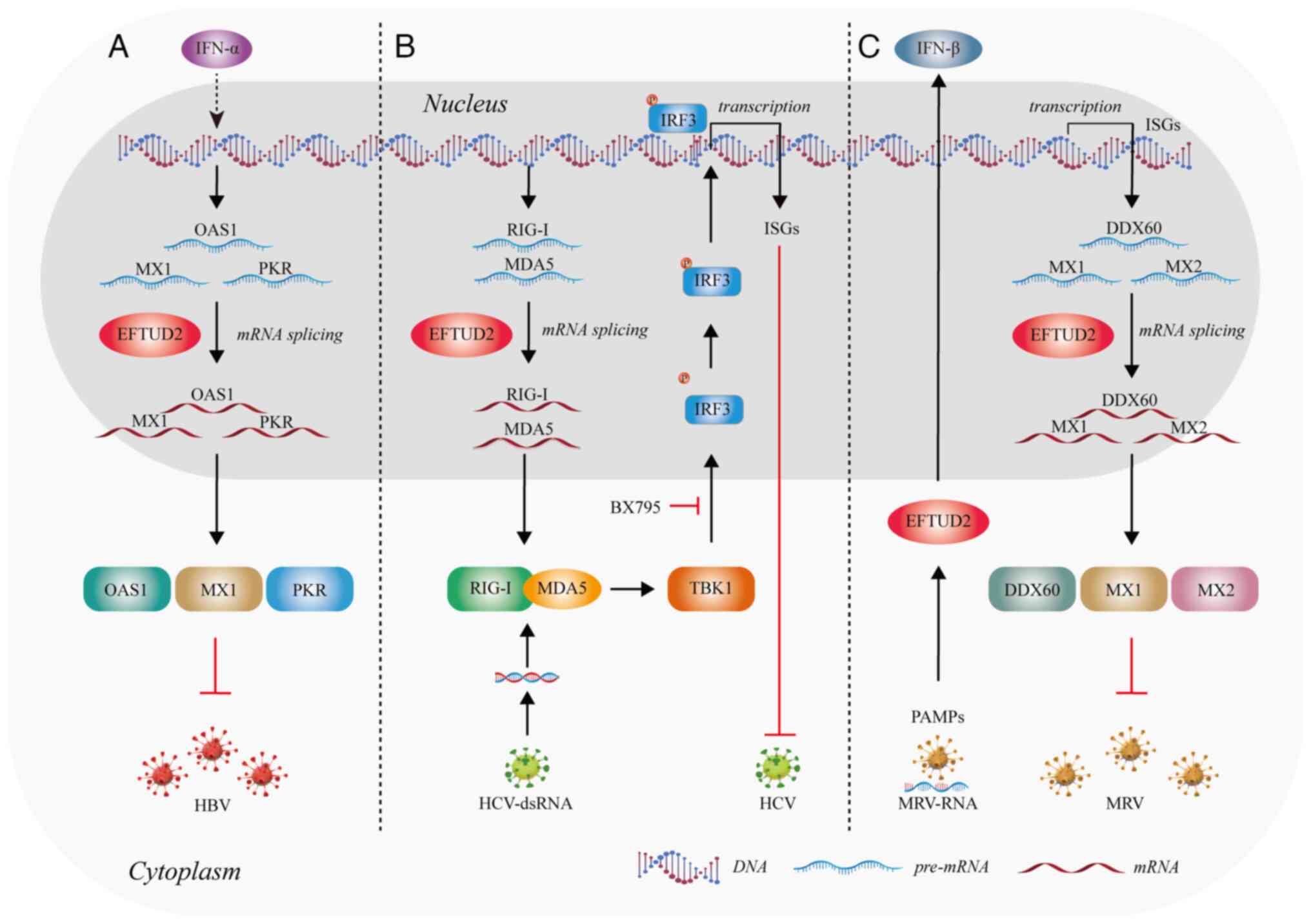
12
|
Gozani O, Feld R and Reed R: Evidence that
sequence-independent binding of highly conserved U2 snRNP proteins
upstream of the branch site is required for assembly of
spliceosomal complex A. Genes Dev. 10:233–243. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Misra B, Wagner R and Boneval H: Injuries
of hepatic veins and retrohepatic vena cava. Am Surg. 49:55–60.
1983.PubMed/NCBI
|
|
14
|
Agafonov DE, Kastner B, Dybkov O, Hofele
RV, Liu WT, Urlaub H, Lührmann R and Stark H: Molecular
architecture of the human U4/U6.U5 tri-snRNP. Science.
351:1416–1420. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Laggerbauer B, Achsel T and Luhrmann R:
The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in
vitro. Proc Natl Acad Sci USA. 95:4188–4192. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Maeder C, Kutach AK and Guthrie C:
ATP-dependent unwinding of U4/U6 snRNAs by the Brr2 helicase
requires the C terminus of Prp8. Nat Struct Mol Biol. 16:42–48.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Boehm V and Gehring NH: Exon junction
complexes: Supervising the gene expression assembly line. Trends
Genet. 32:724–735. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Yan C, Hang J, Finci LI, Lei J
and Shi Y: An atomic structure of the human spliceosome. Cell.
169:918–929. e142017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Park BY, Tachi-Duprat M, Ihewulezi C,
Devotta A and Saint-Jeannet JP: The Core splicing factors EFTUD2,
SNRPB and TXNL4A are essential for neural crest and craniofacial
development. J Dev Biol. 10:292022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Beauchamp MC, Djedid A, Bareke E, Merkuri
F, Aber R, Tam AS, Lines MA, Boycott KM, Stirling PC, Fish JL, et
al: Mutation in Eftud2 causes craniofacial defects in mice via
mis-splicing of Mdm2 and increased P53. Hum Mol Genet. 30:739–757.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Beauchamp MC, Djedid A, Daupin K, Clokie
K, Kumar S, Majewski J and Jerome-Majewska LA: Loss of function
mutation of Eftud2, the gene responsible for mandibulofacial
dysostosis with microcephaly (MFDM), leads to pre-implantation
arrest in mouse. PLoS One. 14:e02192802019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Janeway CA Jr and Medzhitov R: Innate
immune recognition. Annu Rev Immunol. 20:197–216. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
De Arras L, Laws R, Leach SM, Pontis K,
Freedman JH, Schwartz DA and Alper S: Comparative genomics RNAi
screen identifies Eftud2 as a novel regulator of innate immunity.
Genetics. 197:485–496. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
White CR, Dungan M and Carrithers MD:
Activation of human macrophage sodium channels regulates RNA
processing to increase expression of the DNA repair protein
PPP1R10. Immunobiology. 224:80–93. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun J, Li L, Hu J, Gao Y, Song J, Zhang X
and Hu H: Time-course RNA-Seq profiling reveals isoform-level gene
expression dynamics of the cGAS-STING pathway. Comput Struct
Biotechnol J. 20:6490–6500. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang S, Zhao M and Jia S: Macrophage: Key
player in the pathogenesis of autoimmune diseases. Front Immunol.
14:10803102023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kawai T and Akira S: The role of
pattern-recognition receptors in innate immunity: Update on
Toll-like receptors. Nat Immunol. 11:373–384. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mendoza-Barbera E, Corral-Rodriguez MA,
Soares-Schanoski A, Velarde M, Macieira S, Messerschmidt A,
López-Collazo E and Fuentes-Prior P: Contribution of globular death
domains and unstructured linkers to MyD88.IRAK-4 heterodimer
formation: An explanation for the antagonistic activity of MyD88s.
Biochem Biophys Res Commun. 380:183–187. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu MM and Shu HB: Innate immune response
to cytoplasmic DNA: Mechanisms and diseases. Annu Rev Immunol.
38:79–98. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Maelfait J, Bridgeman A, Benlahrech A,
Cursi C and Rehwinkel J: Restriction by SAMHD1 Limits
cGAS/STING-dependent innate and adaptive immune responses to HIV-1.
Cell Rep. 16:1492–1501. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sato S, Li K, Sakurai N, Hashizume M,
Baidya S, Nonaka H, Noguchi K, Ishikawa K, Obuse C and Takaoka A:
Regulation of an adaptor protein STING by Hsp90β to enhance innate
immune responses against microbial infections. Cell Immunol.
356:1041882020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sarkar A, Emrick LT, Smith EM, Austin EG,
Yang Y, Hunter JV, Scaglia F and Lalani SR: Novel de novo mutations
in EFTUD2 detected by exome sequencing in mandibulofacial
dysostosis with Microcephaly syndrome. Am J Med Genet A.
167A:914–918. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Smigiel R, Bezniakow N, Jakubiak A, Błoch
M, Patkowski D, Obersztyn E and Sasiadek MM: Phenotype analysis of
Polish patients with mandibulofacial dysostosis type Guion-Almeida
associated with esophageal atresia and choanal atresia caused by
EFTUD2 gene mutations. J Appl Genet. 56:199–204. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matsuo M, Yamauchi A, Ito Y, Sakauchi M,
Yamamoto T, Okamoto N, Tsurusaki Y, Miyake N, Matsumoto N and Saito
K: Mandibulofacial dysostosis with microcephaly: A case presenting
with seizures. Brain Dev. 39:177–181. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Narumi-Kishimoto Y, Ozawa H, Yanagi K,
Kawai T, Okamura K, Hata K, Kaname T and Matsubara Y: A novel
EFTUD2 mutation identified an adult male with mandibulofacial
dysostosis Guion-Almeida type. Clin Dysmorphol. 29:186–188. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
McDermott JH, Study DD and Clayton-Smith
J: Sibling recurrence of total anomalous pulmonary venous drainage.
Eur J Med Genet. 60:265–267. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang J, Ahimaz PR, Hashemifar S, Khlevner
J, Picoraro JA, Middlesworth W, Elfiky MM, Que J, Shen Y and Chung
WK: Novel candidate genes in esophageal atresia/tracheoesophageal
fistula identified by exome sequencing. Eur J Hum Genet.
29:122–130. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Khattar D and Suhrie KR: Esophageal
atresia with or without tracheoesophageal fistula: Comorbidities,
genetic evaluations and neonatal outcomes. Cureus.
15:e347792023.PubMed/NCBI
|
|
39
|
Bukowska-Olech E, Materna-Kiryluk A,
Walczak-Sztulpa J, Popiel D, Badura-Stronka M, Koczyk G, Dawidziuk
A and Jamsheer A: Targeted Next-generation sequencing in the
diagnosis of facial dysostoses. Front Genet. 11:5804772020.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lines MA, Huang L, Schwartzentruber J,
Douglas SL, Lynch DC, Beaulieu C, Guion-Almeida ML, Zechi-Ceide RM,
Gener B, Gillessen-Kaesbach G, et al: Haploinsufficiency of a
spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial
dysostosis with microcephaly. Am J Hum Genet. 90:369–377. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lacour JC, McBride L, St Hilaire H,
Mundinger GS, Moses M, Koon J, Torres JI and Lacassie Y: Novel de
novo EFTUD2 Mutations in 2 Cases With MFDM, initially suspected to
have alternative craniofacial diagnoses. Cleft Palate Craniofac J.
56:674–678. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Luquetti DV, Hing AV, Rieder MJ, Nickerson
DA, Turner EH, Smith J, Park S and Cunningham ML: ‘Mandibulofacial
dysostosis with microcephaly’ caused by EFTUD2 mutations: Expanding
the phenotype. Am J Med Genet A. 161A:108–113. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kim SY, Lee DH, Han JH and Choi BY: Novel
splice site pathogenic variant of EFTUD2 is associated with
mandibulofacial dysostosis with microcephaly and extracranial
symptoms in Korea. Diagnostics (Basel). 10:2962020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Voigt C, Megarbane A, Neveling K, Czeschik
JC, Albrecht B, Callewaert B, von Deimling F, Hehr A, Falkenberg
Smeland M, König R, et al: Oto-facial syndrome and esophageal
atresia, intellectual disability and zygomatic anomalies-expanding
the phenotypes associated with EFTUD2 mutations. Orphanet J Rare
Dis. 8:1102013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rengasamy Venugopalan S, Farrow EG and
Lypka M: Whole-exome sequencing identified a variant in EFTUD2 gene
in establishing a genetic diagnosis. Orthod Craniofac Res. 20
(Suppl 1):S50–S56. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang M, Sun H, Liu Y and Hu T: Whole exome
sequencing revealed a heterozygous elongation factor Tu GTP-binding
domain containing 2 (EFTUD2) mutation in a couple experiencing
recurrent pregnancy loss. Chin Med J (Engl). 135:1108–1110. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tu M, He L, You Y, Li J, Yao N, Qu C,
Huang W, Xu L, Luo R and Hong J: EFTUD2 maintains the survival of
tumor cells and promotes hepatocellular carcinoma progression via
the activation of STAT3. Cell Death Dis. 11:8302020. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lv C, Li XJ, Hao LX, Zhang S, Song Z, Ji
XD and Gong B: Over-activation of EFTUD2 correlates with tumor
propagation and poor survival outcomes in hepatocellular carcinoma.
Clin Transl Oncol. 24:93–103. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou R, Ni W, Qin C, Zhou Y, Li Y, Huo J,
Bian L, Zhou A and Li J: A functional loop between YTH domain
family protein YTHDF3 mediated m(6)A modification and
phosphofructokinase PFKL in glycolysis of hepatocellular carcinoma.
J Exp Clin Cancer Res. 41:3342022. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Johnson DE, O'Keefe RA and Grandis JR:
Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev
Clin Oncol. 15:234–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhou W, Chen Y, Luo R, Li Z, Jiang G and
Ou X: Identification of biomarkers related to immune cell
infiltration in hepatocellular carcinoma using gene co-expression
network. Pathol Oncol Res. 27:6016932021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lv Z, Wang Z, Luo L, Chen Y, Han G, Wang
R, Xiao H, Li X, Hou C, Feng J, et al: Spliceosome protein Eftud2
promotes colitis-associated tumorigenesis by modulating
inflammatory response of macrophage. Mucosal Immunol. 12:1164–1173.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fukata M, Chen A, Vamadevan AS, Cohen J,
Breglio K, Krishnareddy S, Hsu D, Xu R, Harpaz N, Dannenberg AJ, et
al: Toll-like receptor-4 promotes the development of
colitis-associated colorectal tumors. Gastroenterology.
133:1869–1881. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Grivennikov S, Karin E, Terzic J, Mucida
D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H,
Eckmann L and Karin M: IL-6 and Stat3 are required for survival of
intestinal epithelial cells and development of colitis-associated
cancer. Cancer Cell. 15:103–113. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Popivanova BK, Kitamura K, Wu Y, Kondo T,
Kagaya T, Kaneko S, Oshima M, Fujii C and Mukaida N: Blocking
TNF-alpha in mice reduces colorectal carcinogenesis associated with
chronic colitis. J Clin Invest. 118:560–570. 2008.PubMed/NCBI
|
|
56
|
Matsumoto S, Hara T, Mitsuyama K, Yamamoto
M, Tsuruta O, Sata M, Scheller J, Rose-John S, Kado S and Takada T:
Essential roles of IL-6 trans-signaling in colonic epithelial
cells, induced by the IL-6/soluble-IL-6 receptor derived from
lamina propria macrophages, on the development of
colitis-associated premalignant cancer in a murine model. J
Immunol. 184:1543–1551. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ramesh P, Lannagan TRM, Jackstadt R,
Atencia Taboada L, Lansu N, Wirapati P, van Hooff SR, Dekker D,
Pritchard J, Kirov AB, et al: BCL-XL is crucial for progression
through the adenoma-to-carcinoma sequence of colorectal cancer.
Cell Death Differ. 28:3282–3296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hashimoto K, Nishimura S, Shinyashiki Y,
Ito T and Akagi M: Characterizing inflammatory markers in highly
aggressive soft tissue sarcomas. Medicine (Baltimore).
101:e306882022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Guo G, Wang Y, Zhou Y, Quan Q, Zhang Y,
Wang H, Zhang B and Xia L: Immune cell concentrations among the
primary tumor microenvironment in colorectal cancer patients
predicted by clinicopathologic characteristics and blood indexes. J
Immunother Cancer. 7:1792019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Yu L, Jiang R, Chen W, Liu Y, Wang G, Gong
X and Wang Y: Novel prognostic indicator combining inflammatory
indicators and tumor markers for gastric cancer. World J Surg
Oncol. 21:502023. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sato N, Maeda M, Sugiyama M, Ito S, Hyodo
T, Masuda A, Tsunoda N, Kokuryo T, Hamaguchi M, Nagino M and Senga
T: Inhibition of SNW1 association with spliceosomal proteins
promotes apoptosis in breast cancer cells. Cancer Med. 4:268–277.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Albers M, Diment A, Muraru M, Russell CS
and Beggs JD: Identification and characterization of Prp45p and
Prp46p, essential pre-mRNA splicing factors. RNA. 9:138–150. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chen F, Wang Q and Zhou Y: The
construction and validation of an RNA binding protein-related
prognostic model for bladder cancer. BMC Cancer. 21:2442021.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Beyer S, Muller L, Mitter S, Keilmann L,
Meister S, Buschmann C, Kraus F, Topalov NE, Czogalla B, Trillsch
F, et al: High RIG-I and EFTUD2 expression predicts poor survival
in endometrial cancer. J Cancer Res Clin Oncol. 149:4293–4303.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Wieczorek D: Human facial dysostoses. Clin
Genet. 83:499–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wood KA, Eadsforth MA, Newman WG and
O'Keefe RT: The Role of the U5 snRNP in genetic disorders and
cancer. Front Genet. 12:6366202021. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Griffin C and Saint-Jeannet JP:
Spliceosomopathies: Diseases and mechanisms. Dev Dyn.
249:1038–1046. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lehalle D, Wieczorek D, Zechi-Ceide RM,
Passos-Bueno MR, Lyonnet S, Amiel J and Gordon CT: A review of
craniofacial disorders caused by spliceosomal defects. Clin Genet.
88:405–415. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Guion-Almeida ML, Zechi-Ceide RM,
Vendramini S and Ju Nior AT: A new syndrome with growth and mental
retardation, mandibulofacial dysostosis, microcephaly and cleft
palate. Clin Dysmorphol. 15:171–174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Abell K, Hopkin RJ, Bender PL, Jackson F,
Smallwood K, Sullivan B, Stottmann RW, Saal HM and Weaver KN:
Mandibulofacial dysostosis with microcephaly: An expansion of the
phenotype via parental survey. Am J Med Genet A. 185:413–423. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Silva JB, Soares D, Leao M and Santos H:
Mandibulofacial dysostosis with microcephaly: A syndrome to
remember. BMJ Case Rep. 12:e2298312019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yu KPT, Luk HM, Gordon CT, Fung G, Oufadem
M, Garcia-Barcelo MM, Amiel J, Chung BHY, Lo IFM and Tiong YT:
Mandibulofacial dysostosis Guion-Almeida type caused by novel
EFTUD2 splice site variants in two Asian children. Clin Dysmorphol.
27:31–35. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Huang L, Vanstone MR, Hartley T, Osmond M,
Barrowman N, Allanson J, Baker L, Dabir TA, Dipple KM, Dobyns WB,
et al: Mandibulofacial dysostosis with microcephaly: Mutation and
database update. Hum Mutat. 37:148–154. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Vincent M, Genevieve D, Ostertag A, Marlin
S, Lacombe D, Martin-Coignard D, Coubes C, David A, Lyonnet S,
Vilain C, et al: Treacher collins syndrome: A clinical and
molecular study based on a large series of patients. Genet Med.
18:49–56. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Need AC, Shashi V, Hitomi Y, Schoch K,
Shianna KV, McDonald MT, Meisler MH and Goldstein DB: Clinical
application of exome sequencing in undiagnosed genetic conditions.
J Med Genet. 49:353–361. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Park CY, Zhou J, Wong AK, Chen KM,
Theesfeld CL, Darnell RB and Troyanskaya OG: Genome-wide landscape
of RNA-binding protein target site dysregulation reveals a major
impact on psychiatric disorder risk. Nat Genet. 53:166–173. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Santiago JA and Potashkin JA: Blood
biomarkers associated with cognitive decline in early stage and
drug-naive Parkinson's disease patients. PLoS One. 10:e01425822015.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang S, Sun D, Liu C, Guo Y, Ma J, Ge RL
and Cui S: Weighted gene co-expression network analysis reveals the
hub genes associated with pulmonary hypertension. Exp Biol Med
(Maywood). 248:217–231. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Liu L, Pertsemlidis A, Ding LH, Story MD,
Steinberg MH, Sebastiani P, Hoppe C, Ballas SK and Pace BS:
Original research: A case-control genome-wide association study
identifies genetic modifiers of fetal hemoglobin in sickle cell
disease. Exp Biol Med (Maywood). 241:706–718. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Gordon CT, Petit F, Oufadem M,
Decaestecker C, Jourdain AS, Andrieux J, Malan V, Alessandri JL,
Baujat G, Baumann C, et al: EFTUD2 haploinsufficiency leads to
syndromic oesophageal atresia. J Med Genet. 49:737–746. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Heidarzadehpilehrood R, Pirhoushiaran M,
Binti Osman M, Abdul Hamid H and Ling KH: Weighted gene
co-expression network analysis (WGCNA) Discovered novel long
non-coding RNAs for polycystic ovary syndrome. Biomedicines.
11:5182023. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Hou Y, Wang Y, Xu S, Qi G and Wu X:
Bioinformatics identification of microRNAs involved in polycystic
ovary syndrome based on microarray data. Mol Med Rep. 20:281–291.
2019.PubMed/NCBI
|
|
83
|
Li L and Chen S: Screening, identification
and interaction analysis of key MicroRNAs and genes in
Asthenozoospermia. Int J Med Sci. 18:1670–1679. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Czeschik JC, Voigt C, Alanay Y, Albrecht
B, Avci S, Fitzpatrick D, Goudie DR, Hehr U, Hoogeboom AJ,
Kayserili H, et al: Clinical and mutation data in 12 patients with
the clinical diagnosis of Nager syndrome. Hum Genet. 132:885–898.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Zarate YA, Bell C and Schaefer GB:
Radioulnar synostosis and brain abnormalities in a patient with
17q21.31 microdeletion involving EFTUD2. Cleft Palate Craniofac J.
52:237–239. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
86
|
Zech M, Boesch S, Skorvanek M, Necpál J,
Švantnerová J, Wagner M, Dincer Y, Sadr-Nabavi A, Serranová T,
Rektorová I, et al: Clinically relevant copy-number variants in
exome sequencing data of patients with dystonia. Parkinsonism Relat
Disord. 84:129–134. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Tian A, Li Y, Fan H, Hu P, Xu R, Yuan H,
Cai J, Zhang W, Yue M, Li J, et al: Association of elongation
factor Tu GTP-binding Domain-containing 2 Gene (EFTUD2)
polymorphism with the risk of hepatitis B virus infection. Immunol
Invest. 51:1485–1497. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hu P, Li Y, Zhang W, Liu R, Peng L, Xu R,
Cai J, Yuan H, Feng T, Tian A, et al: The spliceosome factor EFTUD2
promotes IFN Anti-HBV effect through mRNA splicing. Mediators
Inflamm. 2023:25462782023. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Sumpter R Jr, Loo YM, Foy E, Li K,
Yoneyama M, Fujita T, Lemon SM and Gale M Jr: Regulating
intracellular antiviral defense and permissiveness to hepatitis C
virus RNA replication through a cellular RNA helicase, RIG-I. J
Virol. 79:2689–2699. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Wang N, Liang Y, Devaraj S, Wang J, Lemon
SM and Li K: Toll-like receptor 3 mediates establishment of an
antiviral state against hepatitis C virus in hepatoma cells. J
Virol. 83:9824–9834. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Zhu C, Xiao F, Hong J, Wang K, Liu X, Cai
D, Fusco DN, Zhao L, Jeong SW, Brisac C, et al: EFTUD2 is a novel
innate immune regulator restricting hepatitis C virus infection
through the RIG-I/MDA5 pathway. J Virol. 89:6608–6618. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Metz P, Reuter A, Bender S and
Bartenschlager R: Interferon-stimulated genes and their role in
controlling hepatitis C virus. J Hepatol. 59:1331–1341. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Boudreault S, Lemay G and Bisaillon M: U5
snRNP core proteins are key components of the defense response
against viral infection through their roles in programmed cell
death and interferon induction. Viruses. 14:27102022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Kawai T, Takahashi K, Sato S, Coban C,
Kumar H, Kato H, Ishii KJ, Takeuchi O and Akira S: IPS-1, an
adaptor triggering RIG-I- and Mda5-mediated type I interferon
induction. Nat Immunol. 6:981–988. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lau DT, Fish PM, Sinha M, Owen DM, Lemon
SM and Gale M Jr: Interferon regulatory factor-3 activation,
hepatic interferon-stimulated gene expression and immune cell
infiltration in hepatitis C virus patients. Hepatology. 47:799–809.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Schoggins JW, Wilson SJ, Panis M, Murphy
MY, Jones CT, Bieniasz P and Rice CM: A diverse range of gene
products are effectors of the type I interferon antiviral response.
Nature. 472:481–485. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chi Q, Geng X, Xu K, Wang C and Zhao H:
Potential targets and molecular mechanism of miR-331-3p in
hepatocellular carcinoma identified by weighted gene coexpression
network analysis. Biosci Rep. 40:BSR202001242020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhang ZG, Shi ZD, Dong JJ, Chen YA, Cao
MY, Li YT, Ma WM, Hao L, Pang K, Zhou JH, et al: Novel potential
urinary biomarkers for effective diagnosis and prognostic
evaluation of high-grade bladder cancer. Transl Cancer Res.
12:1992–2007. 2023. View Article : Google Scholar : PubMed/NCBI
|