Introduction
MicroRNAs (miRNAs) are a broad class of small,
non-coding RNAs that negatively regulate protein expression. miRNAs
can post-transcriptionally regulate the expression of hundreds of
their target genes, thereby controlling a wide range of biological
functions such as cellular proliferation, differentiation and
apoptosis (1). The expression of
miRNAs was shown to be temporally and spatially regulated, whereas
the disruption of miRNA physiological expression patterns was
associated with a number of examples of human tumorigenesis,
suggesting that they play a role as a novel class of oncogenes or
tumor suppressor genes (2).
miRNA expression profiles were shown to be potential
tools for cancer diagnosis and prediction of prognosis. Various
miRNAs were reported to be associated with the clinical outcome of
chronic lymphocytic leukemia (2),
lung adenocarcinoma (3,4), breast cancer (5) and pancreatic cancers (6,7).
However, whether a miRNA signature can predict the clinical
outcomes of gastric cancer has yet to be determined.
Formalin-fixed, paraffin-embedded (FFPE) tissue
samples are an invaluable source for the study of human disease. A
large number of the tissue blocks are archived worldwide with
corresponding well-documented clinical histories and
histopathological reports. The potential value of these archives
for retrospective molecular studies has been well-recognized
(8).
In this study, miRNA expression profiles from FFPE
samples in gastric cancer were examined and compared with
clinicopathological factors.
Materials and methods
Patients and tissue specimens
FFPE specimens of gastric cancer and associated
patient information were collected from gastrointestinal surgery at
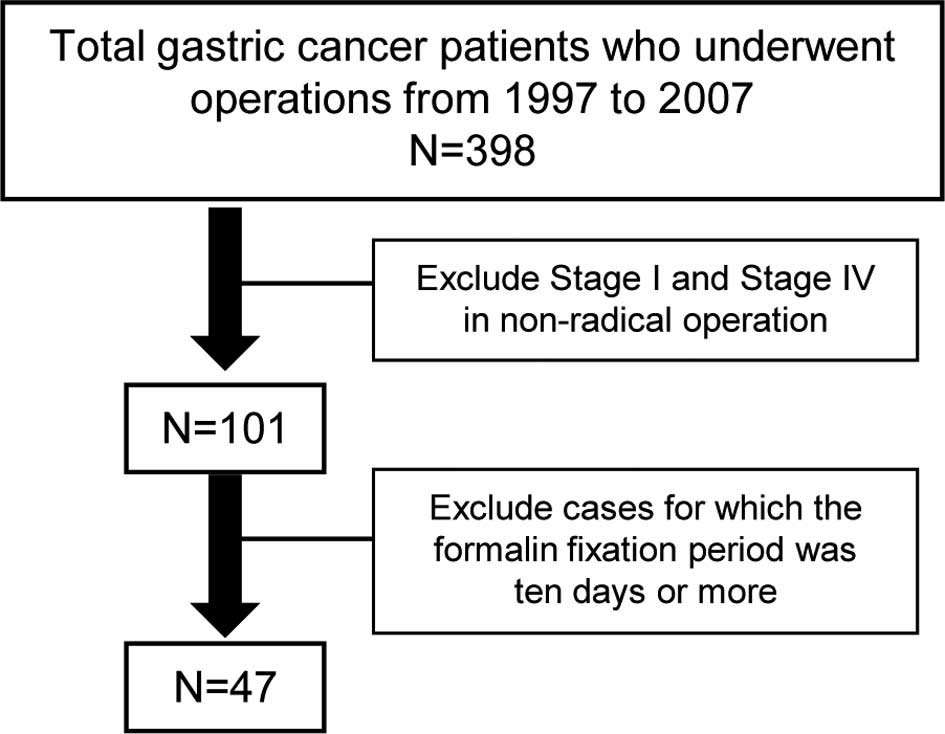
Toyama University Hospital, Japan. A total of 47 specimens were
obtained from 398 cancer patients who had undergone operations
between 1997 and 2007. The specimens were fixed in formalin for
less than 10 days and excluded stage I and IV patients in
non-radical surgery (Fig. 1).
Pretreatment of FFPE specimens prior to
RNA extraction
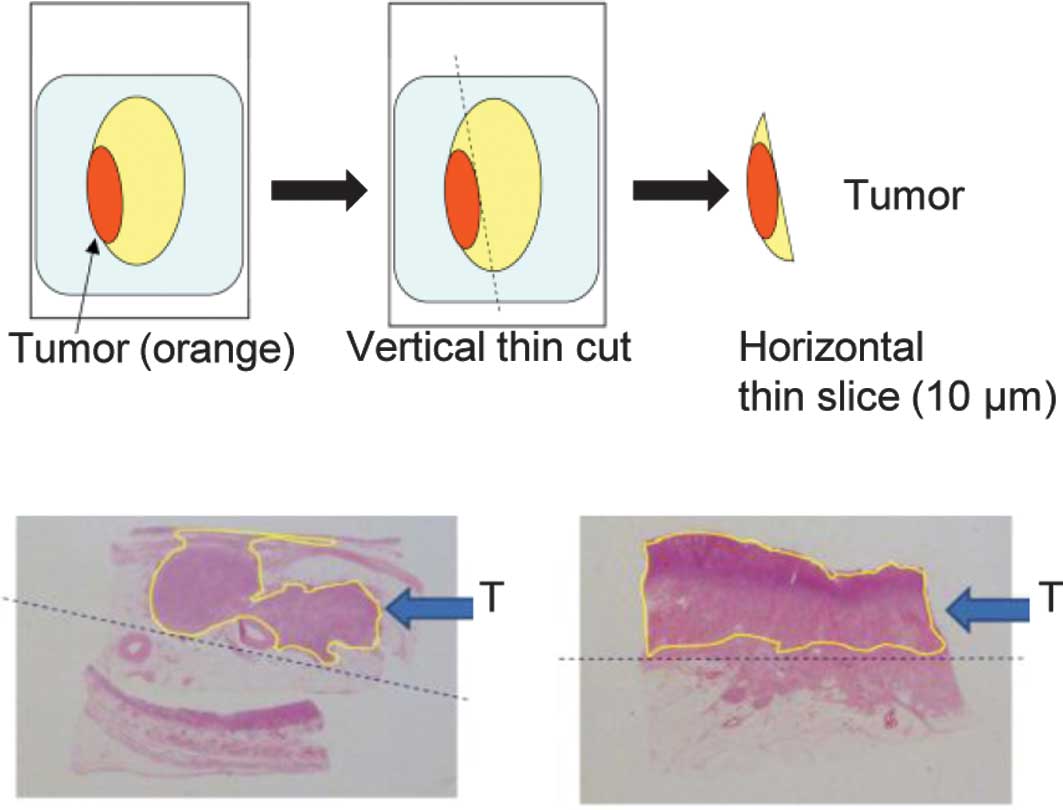
First, we examined the tumor ratio in FFPE blocks
and then compared the miRNA expression in the tumor with that in
normal tissue. Our preliminary study showed that a tumor occupancy
cut-off value of 70% should be used for tumors in this study. FFPE
blocks were cut vertically into thin sections, which were then
sliced into horizontal sections. Samples of 10-μm horizontal slices
were used (Fig. 2).
RNA extraction
Sections (10-μm) were prepared from each FFPE
specimen. Paraffin was removed by xylene treatment and tissues were
washed with ethanol twice to remove xylene. Tissues were then
treated with proteinase K at 37°C overnight. Following
centrifugation, the supernatant was processed with a silica-based
spin column (Toray Industries, Japan) in order to obtain purified
total RNA. The degrees of RNA cross-linking and RNA degradation
were analyzed by electrophoresis using an Agilent 2100 Bioanalyzer
(Agilent Technologies, Santa Clara, CA, USA).
To estimate the possibility of analysis of RNA
extracted from FFPE, selection criteria for RNA quality were
applied. The RNA electrophoresis pattern was found to be crucial
for estimation of the RNA quality for DNA microarray analysis. When
the majority of RNAs were >4000 nucleotides in size due to
cross-linking or when almost all of the RNAs were fragmented (e.g.,
<1000 nucleotides), the RNA quality was considered to be
unsuitable for the miRNA analysis. On the basis of these criteria,
we determined whether RNAs extracted from FFPE could be used for
microarray analysis.
miRNA assays
miRNA profiling was examined using a Toray
3D-Gene® miRNA oligo chip (Toray Industries), which is a
DNA chip. The number of mounted genes on a chip is 885 in
total.
RNA extracted from a sample was processed into an
appropriate form. A solution adjusted for the DNA chip was applied
(including 500 ng of total RNA) and hybridization was performed. A
probe complementary to the target nucleic acids was present on the
DNA chip. The probe forms a double-stranded structure with the
complementary target nucleic acids in the sample solution. The
excess reagents were removed by washing and double strands formed
by hybridization were detected. Of note is that double strands
cannot be verified macroscopically. Therefore, a typical method
involves fluorescent dye incorporation and the use of a detector to
observe fluorescence. In this study, the expression level of each
miRNA was normalized using the median of signal strength of the
entire gene in each chip.
qRT-PCR
cDNA was prepared from miRNA samples using a Taq Man
microRNA reverse transcription kit on the ABI Prism®
7000 real-time PCR system according to the manufacturer’s
instructions (Applied Biosystems, Foster City, CA, USA).
Predesigned Taq Man microRNA assays for hsa-miR-21, hsa-miR-106b,
hsa-miR-133a, hsa-miR-133b and hsa-miR-145 were purchased from
Applied Biosystems. qRT-PCR was performed using a Taq Man universal
PCR master mix, according to the manufacturer’s protocol (Applied
Biosystems). Each miRNA expression in FFPE specimens was estimated
with a standard curve using human gastric reference RNA (Human
stomach tumor total RNA, BD, No. 636629).
Statistical analysis
The expression levels of miRNA in tumor and normal
tissues were analyzed by the t-test. The overall survival time was
calculated from the date of operation until the patient succumbed
to the disease or the last follow-up contact. The Kaplan-Meier
method was used to estimate survival. The differences in survival
for over-expression (T/N ratio >2.0) and reduced expression (T/N
ratio <0.5) were analyzed using the log-rank test. A
multivariate Cox regression analysis was utilized. Characteristics
such as gender, age, TNM classification, histology, adjuvant
chemotherapy and specific microRNA were used to investigate whether
the microRNA signature is an independent predictor of overall
survival in gastric cancer patients. Statistical analyses were
conducted using Dr. SPSS II for Windows and JMP 8. Two-tailed tests
and P<0.05 were used for statistical significance.
Results
Out of 47 paired samples, 37 pairs (78.8%) were
evaluable by quality check. Table I
shows the characteristics of the gastric cancer patients in the
inclusion criteria. The patients of stages III and IV received
post-operative 5FU-based adjuvant chemotherapy and half of the
patients of stage II received the same chemotherapy. Patients did
not receive adjuvant radiotherapy.
 | Table IClinical characteristics of the
evaluable gastric cancer patients. |
Table I
Clinical characteristics of the
evaluable gastric cancer patients.
| Characteristic | No. of patients
(yrs) | % |
|---|
| Age |
| <65 | 10 | 27.0 |
| ≥65 | 27 | 73.0 |
| Gender |
| Female | 11 | 29.7 |
| Male | 26 | 70.3 |
| Tumor site |
| Lower third | 13 | 35.1 |
| Middle third | 11 | 29.8 |
| Upper third | 10 | 27.0 |
| Entire | 3 | 8.1 |
| Tumor diameter
(mm) |
| ≥20 | 5 | 13.5 |
| >20 and ≤50 | 13 | 35.1 |
| >50 and ≤100 | 15 | 40.5 |
| >100 | 4 | 10.8 |
| Histological
type |
|
Differentiateda | 18 | 48.6 |
|
Non-differentiatedb | 19 | 51.4 |
| Depth of
invasion |
| T1 | 3 | 8.1 |
| T2 | 13 | 35.1 |
| T3 | 19 | 51.3 |
| T4 | 2 | 5.4 |
| Lymph node
metastasis |
| Absent | 7 | 18.9 |
| Present | 30 | 81.1 |
| No. of involved lymph
nodes |
| ≤6 | 16 | 53.3 |
| ≥7 | 14 | 46.7 |
| Adjuvant
chemotherapy |
| Absent | 14 | 37.8 |
| Present | 23 | 62.2 |
| Operation method |
| Distal | 21 | 56.8 |
| Total | 16 | 43.2 |
Selection of specific miRNAs in gastric
cancer
A total of 30 miRNAs were significantly
over-expressed (T/N ratio >1.40) in gastric cancer compared with
those in normal gastric tissue (Table
II). On the other hand, the expressions of 11 miRNAs were
significantly reduced (T/N ratio <0.85) in gastric cancer
compared with those in normal gastric tissue (Table III).
 | Table IIOver-expression of miRNAs in gastric
cancers. |
Table II
Over-expression of miRNAs in gastric
cancers.
| ID | Normal | Tumor | T/N ratio | P-value |
|---|
| hsa-miR-185 | 15.6 | 23.9 | 1.53 | 0.0001 |
| hsa-miR-106b | 20.6 | 50.1 | 2.44 | 0.0002 |
| hsa-miR-17 | 48.7 | 88.2 | 1.81 | 0.0002 |
| hsa-miR-425 | 17.9 | 31.6 | 1.76 | 0.0003 |
| hsa-miR-106a | 28.7 | 53.4 | 1.86 | 0.0007 |
| hsa-miR-20a | 13.9 | 28.5 | 2.04 | 0.0010 |
| hsa-miR-181b | 10.1 | 15.3 | 1.51 | 0.0016 |
| hsa-miR-25 | 43.2 | 71.2 | 1.65 | 0.0016 |
| hsa-miR-93 | 52.6 | 91.4 | 1.74 | 0.0019 |
| hsa-miR-21 | 76.3 | 208.6 | 2.73 | 0.0020 |
| hsa-miR-20b | 11.8 | 21.5 | 1.82 | 0.0022 |
| hsa-miR-192 | 104.7 | 205.4 | 1.96 | 0.0032 |
| hsa-miR-130b | 4.7 | 7.2 | 1.55 | 0.0033 |
|
hsa-miR-146b-5p | 25.3 | 46.4 | 1.83 | 0.0035 |
| hsa-miR-103 | 163.4 | 228.7 | 1.40 | 0.0054 |
| hsa-miR-200a | 46.9 | 93.8 | 2.00 | 0.0069 |
| hsa-miR-107 | 129.6 | 182.3 | 1.41 | 0.0075 |
| hsa-miR-96 | 3.9 | 6.2 | 1.59 | 0.0076 |
| hsa-miR-1308 | 2697.8 | 4333.4 | 1.61 | 0.0087 |
| hsa-miR-222 | 58.5 | 88.9 | 1.52 | 0.0104 |
| hsa-miR-194 | 152.4 | 300.8 | 1.97 | 0.0117 |
| hsa-miR-16 | 133.6 | 199.8 | 1.50 | 0.0131 |
| hsa-miR-1290 | 33.1 | 52.4 | 1.58 | 0.0151 |
| hsa-miR-19b | 11.7 | 26.8 | 2.30 | 0.0153 |
| hsa-miR-362-5p | 5.4 | 8.6 | 1.59 | 0.0173 |
| hsa-miR-223 | 34.6 | 67.0 | 1.94 | 0.0204 |
| hsa-miR-337-5p | 3.5 | 6.4 | 1.85 | 0.0205 |
| hsa-miR-15a | 11.9 | 18.7 | 1.57 | 0.0216 |
| hsa-miR-141 | 13.2 | 22.6 | 1.71 | 0.0358 |
| hsa-miR-224 | 4.1 | 12.5 | 3.01 | 0.0497 |
| hsa-miR-34a | 59.1 | 73.7 | 1.25 | 0.0935a |
 | Table IIIReduced expression of miRNAs in
gastric cancers. |
Table III
Reduced expression of miRNAs in
gastric cancers.
| ID | Normal | Tumor | T/N ratio | P-value |
|---|
| hsa-miR-133a | 53.1 | 21.2 | 0.40 | 0.0000 |
| hsa-miR-143a | 34.6 | 18.2 | 0.53 | 0.0000 |
| hsa-miR-133b | 80.9 | 33.9 | 0.42 | 0.0000 |
| hsa-miR-145 | 5925.8 | 2578.6 | 0.44 | 0.0000 |
| hsa-miR-187 | 20.9 | 13.1 | 0.63 | 0.0002 |
| hsa-miR-302b | 35.9 | 18.1 | 0.50 | 0.0024 |
| hsa-miR-29ca | 20.5 | 13.3 | 0.65 | 0.0031 |
| hsa-miR-143 | 2024.1 | 1271.2 | 0.63 | 0.0127 |
| hsa-miR-548m | 8.0 | 3.6 | 0.45 | 0.0139 |
| hsa-miR-29aa | 4.7 | 3.0 | 0.64 | 0.0264 |
|
hsa-let-7f-1a | 7.2 | 6.2 | 0.85 | 0.0345 |
Correlation with clinicopathological
characteristics
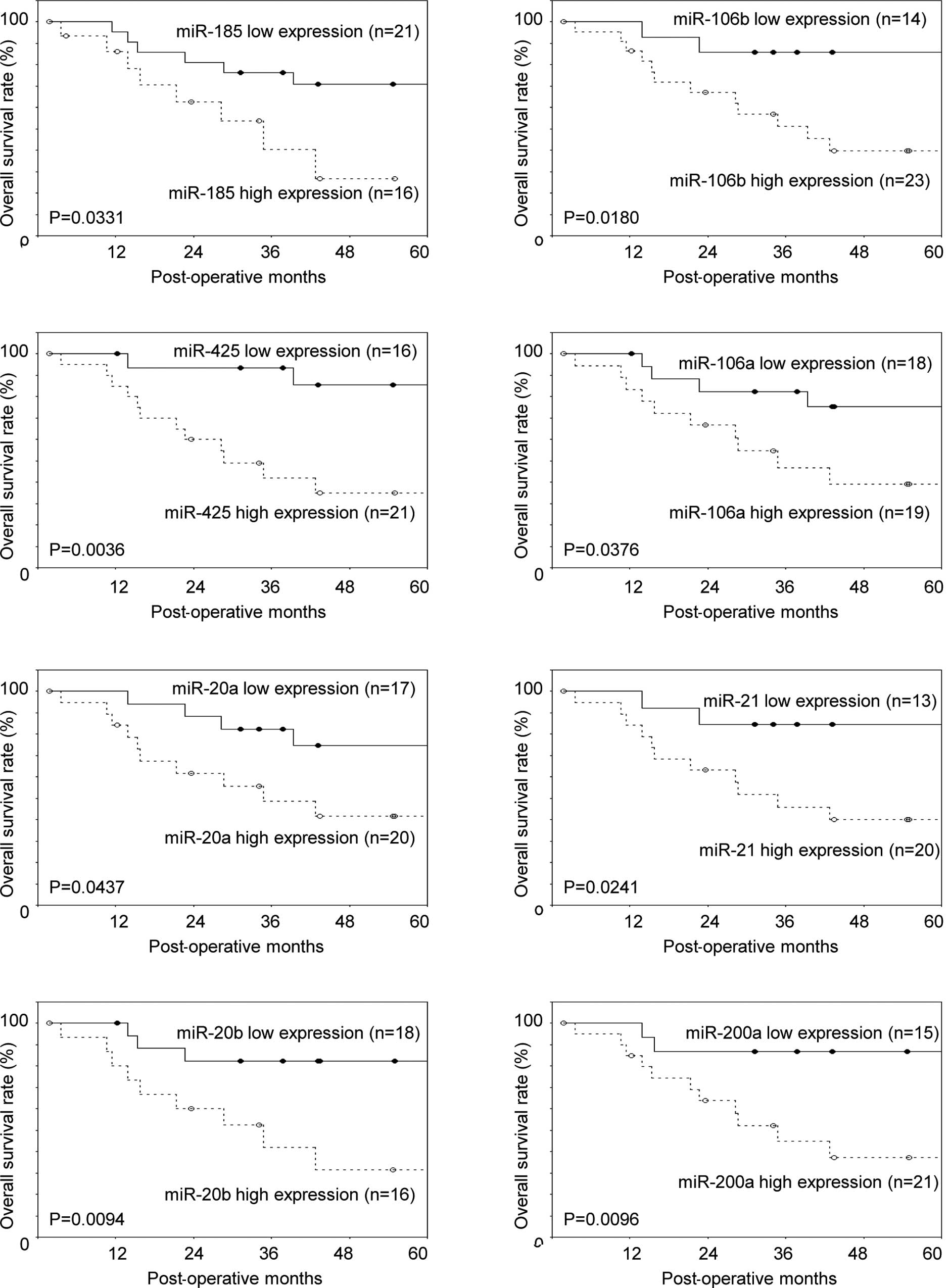
In total, 13 over-expressed miRNAs were found to be
significant prognostic factors by the Kaplan-Meier estimates of
overall survival (median follow-up time, 37.8 months). These miRNAs
were: miR-185, miR-106b, miR-425, miR-106a, miR-20a, miR-21,
miR-20b, miR-200a, miR-15a, miR-103, miR-107, miR-16 and miR-34a.
Fig. 3 shows the Kaplan-Meier
estimates of overall survival and the 13 over-expressed miRNAs.
Although the T/N ratio of miR-34a expression was 1.25, a
significant difference in the Kaplan-Meier estimates of overall
survival associated with its expression was observed (P=0.0076). On
the other hand, miR-143 was the only significant prognostic factor
exhibiting a reduced miRNA expression (Fig. 3). A multivariate Cox proportional
hazard model revealed miR-34a to be an independent prognostic
factor [risk ratio (RR) 7.11]. The prognostic factors are shown in
Table IV.
 | Table IVmiRNAs that were associated with the
prognosis of gastric cancer patients. |
Table IV
miRNAs that were associated with the
prognosis of gastric cancer patients.
| Univariate
analysis | Cox proportional
hazard model |
|---|
|
|
|
|---|
| ID | Log-rank
(P-value) | Risk ratio | 95% | CI P-value |
|---|
| hsa-miR-107 | 0.0007 | 2.05 | 0.40–12.21 | 0.392 |
| hsa-miR-103 | 0.0008 | 2.57 | 0.66–11.47 | 0.174 |
| hsa-miR-143 | 0.0014 | 1.63 | 0.11–22.36 | 0.712 |
| hsa-miR-425 | 0.0036 | 4.80 | 0.80–38.36 | 0.086 |
| hsa-miR-34a | 0.0076 | 7.49 | 1.59–40.19 | 0.011a |
| hsa-miR-20b | 0.0094 | 1.93 | 0.42–11.36 | 0.410 |
| hsa-miR-200a | 0.0096 | 2.19 | 0.33–19.69 | 0.422 |
| hsa-miR-106b | 0.0180 | 1.60 | 0.21–14.39 | 0.643 |
| hsa-miR-16 | 0.0218 | 1.14 | 0.29–4.860 | 0.847 |
| hsa-miR-21 | 0.0241 | 1.44 | 0.18–14.99 | 0.732 |
| hsa-miR-15a | 0.0288 | 1.95 | 0.47–9.130 | 0.357 |
| hsa-miR-185 | 0.0331 | 2.88 | 0.84–10.61 | 0.092 |
| hsa-miR-106a | 0.0376 | 1.73 | 0.38–8.500 | 0.481 |
| hsa-miR-20a | 0.0437 | 1.11 | 0.20–7.140 | 0.907 |
qRT-PCR in FFPE specimens
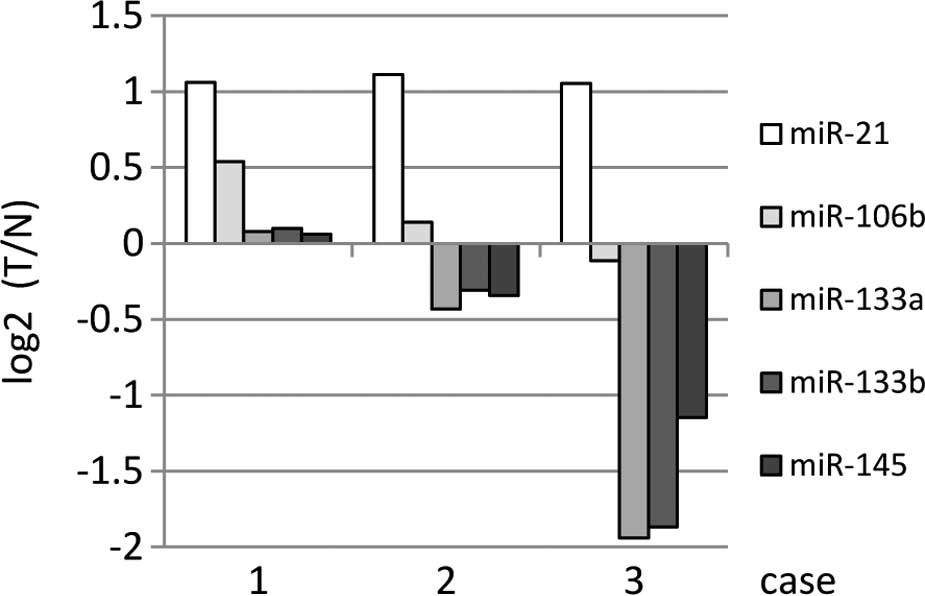
miRNA expression was verified in the three remaining
FFPE specimens using qRT-PCR. In FFPE specimens, the
over-expression of miR-21 and miR106b in DNA chips tended to
increase, while a reduced expression was noted for miR-133a,
miR-133b and miR-145 in DNA chips (Fig.
4).
Discussion
miRNA expression patterns have been described in
various hematological and solid cancers (5,9–11).
Findings of the present study showed that 14 miRNAs were associated
with the prognosis of gastric cancer patients. Among these miRNAs,
miR-21 and miR-20b have already been described in this context in
the literature (9,12–14).
Antiapoptotic miR-21 is up-regulated in gastric
cancer and is related to tumor growth (9,12).
miR-21 targets programmed cell death 4 (PDCD4) and maspin
(SERPINB5), resulting in tumor invasion and metastasis.
miR-20b has been reported to accumulate in tumor
cells and is considered to have an oncogenic role. It plays a
crucial role in fine-tuning the adaptation of tumor cells to oxygen
concentration. The inhibition of miR-20b was found to increase the
protein levels of HIF-1a and VEGF in normoxic tumor cells (13) and it was one of the most highly
expressed miRNAs in gastric cancer tissues (14).
In this study, the results of the multivariate Cox
proportional hazard model showed that only miR-34a was an
independent prognostic factor. In addition, miR-185 and miR-425
tended to have an independent prognostic impact on patient survival
(RR 2.84, P=0.09 and RR 4.88, P=0.07, respectively).
miR-34a is a well-known miRNA. It was identified as
a p53 target by Welch et al (15), who reported that ectopic miR-34a
induces apoptosis when reintroduced into neuroblastoma cell lines.
In cancer, miR-34-mediated apoptosis may be suppressed by the
inactivation of p53 and/or miR-34 genes. miR-34a was found to be
expressed in hepatocellular carcinoma and colon cancer. In gastric
carcinoma, Martin et al (16) reported that these carcinomas
expressed high levels of p53 protein and survival analysis revealed
a strong association between the p53 status of the tumor and
patient survival time after diagnosis. In addition, Qing et
al (17) showed that
restoration of tumor suppressor miR-34 inhibits human p53-mutant
gastric cancer tumorspheres.
A recent report (18) identified miRNAs differentially
expressed in gastric carcinoma tissues. A total of 11 miRNAs were
compatible with our results; however, miR-143, miR-200a and miR-185
were not listed.
In their study, Li et al (19) showed that a seven-microRNA signature
(miR-10b, miR-21, miR-223, miR-338, let-7a, miR- 30a-5p and
miR-126) is closely associated with relapse-free and overall
survival among patients with gastric cancer.
The results of our study showed for the first time
that miR-34a is correlated with the prognosis of gastric cancer
patients.
FFPE tissue samples are an invaluable source for the
study of human disease. FFPE specimens are available in state
hospitals and associated clinical data are usually recorded. These
specimens are regarded as useful in the monitoring of diseases that
have long-term clinical courses of treatment, such as breast and
thyroid cancer. A large number of tissue blocks are archived
worldwide with corresponding well-documented clinical histories and
histopathological reports. In addition, it was reported that there
was a high correlation in miRNA expression between paired FFPE and
fresh frozen material by quantitative RT-PCR (20). Although our data from microarray
analysis do not completely correlate with the results of qRT-PCR
from the remaining FFPE specimens, we believe that miRNA from FFPE
may be a valuable source.
The experimental method established in this study
may be useful for gene expression analysis in translational
research. Although we detected various significant miRNAs in
gastric cancer, the sample size was limited and definite
conclusions could not be drawn. Moreover, we confirmed the
relationships of these miRNAs to biological functions, such as
cellular proliferation, invasion, chemosensitivity and lymph node
metastasis, in gastric cancer.
In conclusion, our results identified miRNAs that
are associated with prognosis in gastric cancer patients. miRNA
profiling using FFPE samples is a useful and promising method of
evaluating samples that are stored in laboratories worldwid and are
accompanied by extremely valuable clinical data.
Acknowledgements
The authors thank Drs Hiroyuki Takahashi and Yasuo
Takano for their technical support.
References
|
1
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Calin GA, Ferracin M, Cimmino A, et al: A
microRNA signature associated with prognosis and progression in
chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Takamizawa J, Konishi H, Yanagisawa K, et
al: Reduced expression of the let-7 microRNAs in human lung cancers
in association with shortened postoperative survival. Cancer Res.
64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bloomston M, Frankel WL, Petrocca F, et
al: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar
|
|
7
|
Roldo C, Missiaglia E, Hagan JP, et al:
MicroRNA expression abnormalities in pancreatic endocrine and
acinar tumors are associated with distinctive pathologic features
and clinical behavior. J Clin Oncol. 24:4677–4684. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lewis F, Maughan NJ, Smith V, et al:
Unlocking the archive–gene expression in paraffin-embedded tissue.
J Pathol. 195:66–71. 2001.
|
|
9
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gramantieri L, Ferracin M, Fornari F, et
al: Cyclin G1 is a target of miR-122a, a microRNA frequently
down-regulated in human hepatocellular carcinoma. Cancer Res.
67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schetter AJ, Leung SY, Sohn JJ, et al:
MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Z, Li Z, Gao C, et al: miR-21 plays
a pivotal role in gastric cancer pathogenesis and progression. Lab
Invest. 88:1358–1366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lei Z, Li B, Yang Z, et al: Regulation of
HIF-1alpha and VEGF by miR-20b tunes tumor cells to adapt to the
alteration of oxygen concentration. PLoS One. 4:e76292009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo J, Miao Y, Xiao B, et al: Differential
expression of microRNA species in human gastric cancer versus
non-tumorous tissues. J Gastroenterol Hepatol. 24:652–657. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Welch C, Chen Y and Stallings RL:
MicroRNA-34a functions as a potential tumor suppressor by inducing
apoptosis in neuroblastoma cells. Oncogene. 26:5017–5022. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Martin HM, Filipe MI, Morris RW, et al:
p53 expression and prognosis in gastric carcinoma. Int J Cancer.
50:859–862. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Qing Ji, Hao Xinbao, Meng Yang, et al:
Restoration of tumor suppressor miR-34 inhibits human p53-mutant
gastric cancer tumorspheres. BMC Cancer. 8:2662008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yoshiyuki T, Chisato N, Tsuyoshi N, et al:
MicroRNA-375 is downregulated in gastric carcinomas and regulates
cell survival by targeting PDK1 and 14–3–3ζ. Cancer Res.
70:23392010.PubMed/NCBI
|
|
19
|
Li X, Zhang Y, Zhang Y, et al: Survival
prediction of gastric cancer by a seven-microRNA signature. Gut.
59:579–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Glud M, Klausen M, Gniadecki R, et al:
MicroRNA expression in melanocytic nevi: the usefulness of
formalin-fixed, paraffin-embedded material for miRNA microarray
profiling. J Invest Dermatol. 129:1219–1224. 2009. View Article : Google Scholar : PubMed/NCBI
|